Abstract
Background and aims
Perturbations in intestinal microbiota composition due to lifestyle changes may be involved in the development of atopic diseases. We examined gut microbiota composition in early infancy and the subsequent development of atopic manifestations and sensitisation.
Methods
The faeces of 957 infants aged 1 month and participating in the KOALA Birth Cohort Study were analysed using quantitative real‐time PCR. Information on atopic symptoms (eczema, wheeze) and potential confounders was acquired through repeated questionnaires. Total and specific IgE were measured in venous blood samples collected during home visits when the infant was 2 years old. During these home visits a clinical diagnosis of atopic dermatitis was made according to the UK‐Working Party criteria.
Results
The presence of Escherichia coli was associated with a higher risk of developing eczema (ORadj = 1.87; 95% CI 1.15 to 3.04), this risk being increased with increasing numbers of E coli (pfor trend = 0.016). Infants colonised with Clostridium difficile were at higher risk of developing eczema (ORadj = 1.40; 95% CI 1.02 to 1.91), recurrent wheeze (ORadj = 1.75; 95% CI 1.09 to 2.80) and allergic sensitisation (ORadj = 1.54; 95% CI 1.02 to 2.31). Furthermore, the presence of C difficile was also associated with a higher risk of a diagnosis of atopic dermatitis during the home visit (ORadj = 1.73; 95% CI 1.08 to 2.78).
Conclusion
This study demonstrates that differences in gut microbiota composition precede the development of atopy. Since E coli was only associated with eczema and C difficile was associated with all atopic outcomes, the underlying mechanisms explaining these association may be different.
Keywords: atopy, Clostridium difficile , Escherichia coli , gut microbiota, infant
The prevalence of atopy has been increasing worldwide during the past decades, particularly in the Western world and amongst children.1 An enhanced T helper 2 (Th2) immune response and the elaboration of cytokines such as interleukin (IL)‐4, IL‐13 and IL‐5 contribute to the induction of atopic diseases.2 Although genetic susceptibility plays an important role in atopy, changes in the prevalence of these diseases have been much faster than any possible shift in genetic constitution.3 Therefore, environmental changes associated with a western lifestyles are thought to be involved in the atopic epidemic. In 1989 Strachan introduced the “hygiene hypothesis”, which states that reduced exposure to infections during childhood results in aberrant immune responses to innocuous antigens later in life.4,5 However, an alternative interpretation of this hypothesis is that perturbations in the composition of gastrointestinal microbiota as a result of changed lifestyles (antibiotic use, diet) in westernised countries have disrupted the mechanisms involved in the development of immunological tolerance.6 Regulatory antigen presenting cells (APCreg) and regulatory T cells (Treg) play a crucial role in the development of immunological tolerance. The maturation of these cells might be hampered as a result of reduced exposure to certain microbes (“old friends”) and consequently a person may develop T helper 1 (Th1) or Th2 mediated inflammatory disorders.7
Differences in intestinal microbiota composition have been shown between infants in countries with high (Sweden) and low (Estonia) allergy prevalence and also between allergic and healthy infants.8,9,10,11 Most reports were based on small populations and although the majority of observational studies found an association between the gut microbiota and allergy, no protective or potentially harmful bacteria have yet been identified.12
Further support for the role of the gut microbiota comes from several clinical trials using probiotics in the treatment13,14,15 and prevention16,17 of atopic eczema, although not all studies have shown probiotics to be effective.18,19
In a large prospective birth cohort study in the Netherlands, we examined the composition of the intestinal microbiota of nearly 1000 infants aged 1 month and the subsequent development of atopic manifestations and/or sensitisation within the first 2 years of life.
Methods
Subjects and study design
The KOALA Birth Cohort Study is a prospective birth cohort study in the Netherlands aimed at identifying factors influencing atopic diseases. The design of the KOALA study has been described in detail elsewhere.20 Briefly, from October 2000 until December 2002 we recruited pregnant women with diverse lifestyles at 34 weeks of gestation. Pregnant women with a conventional lifestyle (n = 2343) were recruited from an ongoing prospective cohort study on pregnancy‐related pelvic girdle pain in the Netherlands.21 Additionally, pregnant women with alternative lifestyles (n = 491) as regards child rearing practices, dietary habits (organic, vegetarian), vaccination schemes and/or the restricted use of medication, were recruited through organic food shops, anthroposophic doctors and midwives, Steiner schools and magazines.
During pregnancy and early childhood, data on perinatal determinants of the child's health as well as on hygiene, infections, nutrition, child rearing, other lifestyle characteristics and atopic manifestations were collected for all members of the cohort by repeated questionnaires at 34 weeks of gestation and at 3, 7, 12 and 24 months post partum.
Participants (with both conventional and alternative lifestyles) recruited from January 2002 onwards were asked to sample the infant's faeces at the age of 1 month post partum (n = 1176). Subjects received a faeces tube with a spoon attached to the lid (Sarstedt, Nümbrecht, Germany), together with a sanitary napkin, an instruction form and a brief questionnaire (faeces questionnaire). Parents placed a sanitary napkin in the diaper (to prevent absorption of the faeces by the diaper), collected the faeces out of the napkin into the collection tube and sent it immediately to our laboratory by post. Transport time was minimised by asking the parents to collect the faeces on a Monday, Tuesday or Wednesday, so that the samples did not remain in the mail service over the weekend.
The study cohort comprised 957 infants after exclusion of premature infants, infants who received antimicrobial agents during their first month of life, infants from whom insufficient amounts of faeces (<1 g) were collected, infants whose faeces were not collected between 3 and 6 weeks of age and infants for whom the faeces questionnaire was missing.
Home visits were made by trained nurses when the infant was 2 years of age (n = 607). During these visits a clinical diagnosis of atopic dermatitis was determined using the UK‐Working Party (UK‐WP) criteria.22,23,24 Venous blood was also collected from the infants in order to determine total (n = 590) and specific (n = 583) serum immunoglobulin (Ig)E.
The KOALA study was approved by the Ethics Committee of the University Hospital of Maastricht and all parents signed informed consent for the study.
DNA purification from faeces
At the laboratory faecal samples were diluted tenfold in peptone‐water (Oxoid CM0009) containing 20% v/v glycerol (Merck, Darmstadt, Germany) and stored at −20°C until analysis. For DNA isolation, 0.2 ml of the diluted faeces was added to a 2 ml vial containing approximately 300 mg glass beads (diameter 0.1 mm) and 1.4 ml of ASL‐buffer from the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) and the samples disrupted in a mini‐bead beater (Biospec Products, Bartlesville, OK) at 5000 rpm for 3 min. Subsequently, bacterial DNA was isolated from the samples using the QIAamp DNA Stool Mini Kit according to the instructions of the manufacturer. The DNA was eluted in a final volume of 200 µl.
Microbial analysis of faecal samples
Faecal samples were subjected to real‐time PCR for counting of Bifidobacterium spp, Escherichia coli, Clostridium difficile, Bacteroides fragilis group, Lactobacillus spp, and total bacterial counts as described previously25 (primers and probes are listed in table 1). The log10 colony forming units (CFU) per gram of the bacterial groups and species were calculated for each stool sample from the threshold cycle values using the constructed standard curves. The prevalence of colonisation was expressed as the percentage of infants colonised with a specific bacterial group or species.
Table 1 Primers and probes used in this study.
| Target organisms (amplicon size) | Primer/probe | Sequence (5′‐3′) | Tm (°C) | Reference |
|---|---|---|---|---|
| Bifidobacterium spp (126 bp). | Forward primer | GCGTGCTTAACACATGCAAGTC | 59 | Penders et al26 |
| Reverse primer | CACCCGTTTCCAGGAGCTATT | 59 | Penders et al26 | |
| Probe | TCACGCATTACTCACCCGTTCGCC | 70 | Penders et al26 | |
| Escherichia coli (96 bp) | Forward primer | CATGCCGCGTGTATGAAGAA | 59 | Huijsdens et al27 |
| Reverse primer | CGGGTAACGTCAATGAGCAAA | 59 | Huijsdens et al27 | |
| Probe | TATTAACTTTACTCCCTTCCTCCCCGCTGAA | 68 | Huijsdens et al27 | |
| Clostridium difficile (114 bp) | Forward primer | TTGAGCGATTTACTTCGGTAAAGA | 58 | Penders et al26 |
| Reverse primer | TGTACTGGCTCACCTTTGATATTCA | 59 | Penders et al26 | |
| Probe | CCACGCGTTACTCACCCGTCCG | 69 | Penders et al26 | |
| Bacteroides fragilis group (92 bp) | Forward primer | CGGAGGATCCGAGCGTTA | 58 | Penders et al25 |
| Reverse primer | CCGCAAACTTTCACAACTGACTTA | 59 | Liu et al28 | |
| Probe | CGCTCCCTTTAAACCCAATAAATCCGG | 68 | Penders et al25 | |
| Lactobacillus spp (341 bp) | Forward primer | AGCAGTAGGGAATCTTCCA | 59 | Walter et al,29 Rinttila et al30 |
| Reverse primer | CACCGCTACACATGGAG | 59 | Rinttila et al,30 Heilig et al31 | |
| Total count (467 bp) | Forward primer | TCCTACGGGAGGCAGCAGT | 59 | Nadkarni et al32 |
| Reverse primer | GGACTACCAGGGTATCTAATCCTGTT | 58 | Nadkarni et al32 |
Determination of total and specific IgE
Blood samples collected during home visits when the infant was 2 years of age were analysed for total and specific serum IgE by Sanquin Research (Amsterdam). For total IgE levels, samples were analysed as described earlier.33,34 A sandwich RIA was used for values <150 IU/ml 34 and a competitive RIA for values >150 IU/ml.33 The detection limit for total serum IgE was <0.5 IU/ml. Blood samples were analysed for specific IgE against hen's egg, cow's milk, peanuts, birch pollen, grass pollen, cat, dog and house dust mite using RAST as described previously.33 Calculation was performed by means of a standard curve that was obtained by RAST with a dilution series of a chimeric monoclonal IgE antibody against the major house dust mite allergen Der‐p2 and Sepharose‐coupled recombinant Der‐p2.35 A RAST value of >0.3 IU/ml was regarded as positive.
Definition of atopic manifestations and sensitisation
Our definitions of eczema and wheeze were based on questions adapted from the ISAAC questionnaires.36 In the 7, 12 and 24 months' postpartum questionnaires, parents were asked: “has your child ever had an itchy rash that came and went over the past months?”. If this question was answered affirmatively, infants were defined as having developed eczema in the first 2 years of life. Infants for whom only diaper rash, rash around the eyes and/or scalp scaling was reported were not regarded as having developed eczema.
“Recurrent wheezing” was defined as the reported presence of wheezing with at least four attacks between 0 and 7 months mentioned in the 7 months' postpartum questionnaire and/or between 7 and 12 months of life mentioned in the 12 months' postpartum questionnaire and/or between 13 and 24 months mentioned in the 24 months' postpartum questionnaire.
Besides the eczema reported by the parents, a diagnosis of atopic dermatitis could be made using the UK‐WP criteria for those infants visited at home.22,23,24 Briefly, the presence of atopic dermatitis was determined according to four clinical symptoms: (i) the presence of an itchy rash, (ii) a history of flexural dermatitis, (iii) visible flexural dermatitis, and (iv) onset before the age of 2 years. In this study, infants with a probability of atopic dermatitis of >0.9 were considered to have atopic dermatitis.
Infants with specific serum IgE levels >0.3 IU/ml against one or more of the tested food or inhalant allergens were considered to be sensitised.
Definition of potential confounders
The following variables were included as potential confounders: subcohort (conventional; alternative), parental atopic history defined as self‐reported doctor‐diagnosed eczema, hay fever, asthma, and pet and/or house dust mite allergy (none; at least one parent), siblings with an atopic history defined as parent‐reported doctor‐diagnosed food allergy, eczema, hay fever, asthma, and pet and/or house dust mite allergy (no siblings; ⩾1 sibling, none atopic; or ⩾1 sibling, at least one atopic), age at collection of faecal sample (age in days) and infant's gender (boy; girl). Maternal probiotic use during the last month of pregnancy (never/sporadic; several times a month; several times a week; daily), place and mode of delivery (vaginal delivery at home; vaginal delivery in hospital; artificial delivery in hospital; or caesarean section in hospital), type of infant feeding during the first month (exclusively breast‐fed; exclusively formula‐fed or a combination) were included as a separate set of confounders. These latter variables may be more distal determinants in the association between the gut microbiota and atopic manifestations instead of true confounders, and adjusted analyses were therefore performed with and without adjustment for these variables.
Statistical analyses
Logistic regression analyses were used to test for (unadjusted) associations between colonisation with the gut bacteria (colonised or uncolonised) under study and the development of atopic outcomes (eczema (reported by parent), atopic dermatitis (UK‐WP criteria), recurrent wheeze and/or atopic sensitisation). Adjusted associations were tested by incorporating the potential confounders into the logistic regression models. Two different sets of confounders were used. The first set included the variables subcohort, parental and sibling atopic history, age at collection of the faecal sample and infant's gender, and the second set included these variables as well as maternal probiotic use during pregnancy, place and mode of delivery and type of infant feeding. Since almost all infants were colonised, the association between colonisation with bifidobacteria and atopic outcomes was analysed as “low” (<10.68 log10 CFU/g) versus “high” (⩾10.68 log10 CFU/g) bifidobacterial counts; the few uncolonised infants (n = 12) were added to the low counts group. The cut‐off point of 10.68 log10 CFU/g was chosen to create two equal groups.
Logistic regression analyses were also used to test for associations between the concentration (counts) of gut bacteria and atopic outcomes. We additionally adjusted for total bacterial counts to account for differences in the consistency of faecal samples in the analyses on bacterial numbers, in addition to the confounders described above. To test for trend, bacterial counts were categorised (uncolonised infants were used as the reference category and the remaining colonised infants were accommodated in two (C difficile, lactobacilli) or three equal groups (bifidobacteria, E coli, B fragilis group)).
Linear regression analyses were used to test for associations between the gut bacteria under study and total serum IgE levels, controlling for the same confounders mentioned above. Since separate analyses of the conventional and alternative subcohorts showed that the key findings were similar within these two subcohorts, in the final analyses we combined the two groups adjusting for “subcohort”.
To examine the possibility of selection bias in those infants visited at home, we performed non‐response analyses. Using logistic regression analyses, infants visited at home (n = 607) were compared with infants not visited at home (n = 305) regarding gut microbiota composition and the prevalence of eczema and recurrent wheeze.
Results
Of the 957 infants participating in this study, almost all were colonised with bifidobacteria (98.7%) at the age of 1 month (table 2). Most of the infants were also colonised with E coli (88.6%) and members of the B fragilis group (81.6%), whereas colonisation with lactobacilli (32.2%) and C difficile (25.1%) was less common. Bifidobacteria were detected in the highest numbers, followed by E coli and B fragilis group species.
Table 2 Characteristics of the participants in this study.
| Conventional subcohort, n = 652* | Alternative subcohort, n = 305* | |
|---|---|---|
| Prevalence of colonisation with intestinal bacteria, % | ||
| Bifidobacteria | 98.3 | 99.7 |
| Escherichia coli | 89.5 | 86.6 |
| Clostridium difficile | 24.7 | 25.9 |
| Bacteroides fragilis group | 83.0 | 78.7 |
| Lactobacilli | 33.1 | 30.2 |
| Counts of intestinal bacteria (log10 CFU/g), median (range) | ||
| Bifidobacteria | 10.71 (6.84–11.56) | 10.68 (6.85–11.49) |
| Escherichia coli | 9.45 (5.91–10.79) | 9.12 (5.92–10.62) |
| Clostridium difficile | 5.12 (2.70–8.41) | 5.70 (2.85–8.81) |
| Bacteroides fragilis group | 9.40 (5.74–10.36) | 9.07 (5.79–10.33) |
| Lactobacilli | 8.70 (7.92–10.73) | 8.58 (7.95–10.33) |
| Total counts | 11.15 (9.43–12.14) | 11.08 (9.58–11.98) |
| Age at collection of faecal sample (days), mean (SD) | 31.60 (3.28) | 31.75 (3.31) |
| Parental history of atopic manifestations, % | ||
| At least one parent with atopy | 54.8 | 51.8 |
| Sibling history of atopic manifestations, % | ||
| No siblings | 43.7 | 32.8 |
| ⩾1 siblings, non‐atopic | 40.5 | 46.2 |
| ⩾1 siblings, at least one atopic | 15.6 | 21.0 |
| Sex of infant (boys, percentage) | 49.4 | 53.4 |
| Maternal probiotic use, %† | ||
| Never/sporadic | 78.5 | 79.9 |
| Several times a month | 9.5 | 9.8 |
| Several times a week | 6.7 | 6.2 |
| Daily | 1.4 | 3.0 |
| Place and mode of delivery, % | ||
| Natural delivery at home | 44.0 | 53.8 |
| Natural delivery in hospital | 34.8 | 29.2 |
| Artificial delivery in hospital‡ | 7.5 | 6.9 |
| Caesarean section in hospital | 10.7 | 9.5 |
| Type of infant feeding, % | ||
| Exclusively breast‐fed | 58.3 | 87.9 |
| Exclusively formula‐fed | 29.4 | 7.2 |
| Combination | 12.0 | 4.9 |
| Infants with atopic outcome at age 2 years, % | ||
| Eczema | 32.8 | 31.7 |
| Recurrent wheeze | 12.8 | 6.3 |
| Sensitisation§ | 29.9 | 25.0 |
| Total IgE, median (range)¶ | 10.0 (⩽0.5–5300.0) | 16.5 (⩽0.5–3700.0) |
*Overall numbers are not always 652 for the conventional subcohort and 305 for the alternative subcohort due to missing bacterial count data or outcome data; †consumed during the last month of pregnancy; ‡forceps or vacuum extraction; §only available for those infants visited at home, total numbers are 391 for conventional subcohort and 192 for alternative subcohort; ¶only available for those infants visited at home, total numbers are 396 for conventional subcohort and 194 for alternative subcohort.
More than half of the infants had at least one parent with a positive history of doctor‐diagnosed atopic manifestations (table 2). Almost one third of the infants who had older siblings, had an atopic sibling. Over 30% of the infants had developed eczema at the age of 2 years and recurrent wheeze was reported in approximately 10%. Over a quarter of the infants had circulating IgE antibodies (>0.3 IU/ml) in their blood against one or more of the food and/or inhalant allergens and thus were regarded as sensitised.
Non‐response analyses showed that there were no differences in the composition of gut microbiota or in the prevalence of eczema and recurrent wheeze between infants who were visited at home and those who were not (data not shown).
Table 3 shows the adjusted association between colonisation with the gut bacteria under study at the age of 1 month and the development of atopic manifestations within the first 2 years of life. The risk of eczema was significantly higher in infants colonised with E coli (adjusted odds ratio (ORadj) = 1.87; 95% CI 1.15 to 3.04) compared with infants not colonised with E coli at the age of 1 month. For those infants visited at home, we were also able to define atopic dermatitis according to the UK‐WP criteria. However a higher risk for infants colonised with E coli was not found when atopic dermatitis was defined according to these criteria (ORadj = 1.02; 95% CI 0.49 to 2.10).
Table 3 The adjusted association between colonisation with gut bacteria at 1 month of age and atopic sensitisation and atopic disease manifestation at 2 years of age (n = 957).
| Intestinal bacteria | Eczema† | Recurrent wheeze† | Atopic dermatitis (UK‐WP)‡ | Sensitisation§ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Prevalence (n/N)¶ | ORadj (95% CI)** | Prevalence (n/N)¶ | ORadj (95% CI)** | Prevalence (n/N)¶ | ORadj (95% CI)** | Prevalence (n/N)¶ | ORadj (95% CI)** | ||||
| Bifidobacteria†† | |||||||||||
| Low | 32.4% (148/457) | 1.0 | 9.4% (41/434) | 1.0 | 17.9% (54/302) | 1.0 | 26.5% (76/287) | 1.0 | |||
| High | 32.5% (156/480) | 1.02 (0.77 to 1.35) | 11.8% (55/466) | 1.32 (0.85 to 2.06) | 14.4% (44/305) | 0.79 (0.51 to 1.23) | 30.1% (89/296) | 1.23 (0.85 to 1.77) | |||
| E coli | |||||||||||
| No | 22.2% (24/108) | 1.0 | 5.8% (6/104) | 1.0 | 15.9% (10/63) | 1.0 | 30.6% (19/62) | 1.0 | |||
| Yes | 33.8% (279/826) | 1.87 (1.15 to 3.04)* | 11.1% (88/793) | 1.92 (0.80 to 4.59) | 16.2% (88/542) | 1.02 (0.49 to 2.10) | 28.1% (146/519) | 0.86 (0.48 to 1.54) | |||
| C difficile | |||||||||||
| No | 30.3% (213/702) | 1.0 | 9.4% (63/671) | 1.0 | 14.1% (64/454) | 1.0 | 26.1% (115/440) | 1.0 | |||
| Yes | 38.7% (91/235) | 1.40 (1.02 to 1.91)* | 14.4% (33/229) | 1.75 (1.09 to 2.80)* | 22.2% (34/153) | 1.73 (1.08 to 2.78)* | 35.0% (50/143) | 1.54 (1.02 to 2.31)* | |||
| B fragilis group | |||||||||||
| No | 32.0% (55/172) | 1.0 | 9.0% (15/166) | 1.0 | 12.5% (14/112) | 1.0 | 29.9% (32/107) | 1.0 | |||
| Yes | 32.5% (249/765) | 1.02 (0.71 to 1.47) | 11.0% (81/734) | 1.20 (0.66 to 2.18) | 17.0% (84/495) | 1.41 (0.76 to 2.60) | 27.9% (133/476) | 0.90 (0.57 to 1.43) | |||
| Lactobacilli | |||||||||||
| No | 31.2% (199/637) | 1.0 | 10.3% (63/613) | 1.0 | 15.7% (64/408) | 1.0 | 28.0% (110/393) | 1.0 | |||
| Yes | 35.0% (105/300) | 1.23 (0.91 to 1.65) | 11.5% (33/287) | 1.22 (0.77 to 1.93) | 17.1% (34/199) | 1.14 (0.72 to 1.81) | 28.9% (55/190) | 1.04 (0.71 to 1.53) | |||
†Based on parents' reports in 7, 12 and/or 24 months' questionnaires; ‡as determined by trained nurses during home visits; §specific IgE antibodies to at least one allergen (cow's milk, hen's egg, peanut, birch pollen, grass pollen, cat, dog, house dust mite); ¶numbers in table do not always add up to 957 due to missing bacterial count data or outcome data; **from logistic regression analysis: adjusted for subcohort, parental history of atopy, sibling history of atopy, age at collection of faecal sample and infant's gender; ††colonisation with bifidobacteria in association with atopic sensitisation and disease manifestation presented as low (<10.68 log10 CFU/g) versus high (⩾10.68 log10 CFU/g) counts since almost all infants were colonised; uncolonised infants (n = 12) were added to the low counts group.
*p<0.05 two‐sided.
Infants colonised with C difficile were also at higher risk of eczema (ORadj = 1.40; 95% CI 1.02 to 1.91) compared with uncolonised infants. This association was even stronger for atopic dermatitis according to the UK‐WP criteria (ORadj = 1.73; 95% CI 1.08 to 2.78). Colonisation with C difficile was furthermore associated with a higher risk of developing recurrent wheeze (ORadj = 1.75; 95% CI 1.09 to 2.80) and atopic sensitisation (ORadj = 1.54; 95% CI 1.02 to 2.31).
Colonisation with bifidobacteria, B fragilis group species and lactobacilli was not associated with any of the atopic outcomes. Results of the unadjusted analyses (data not shown) were comparable with the adjusted analyses. Also, the results of the analyses in which we additionally adjusted for maternal probiotic use during pregnancy, place and mode of delivery and type of infant feeding (data not shown) were comparable with the results presented in table 3.
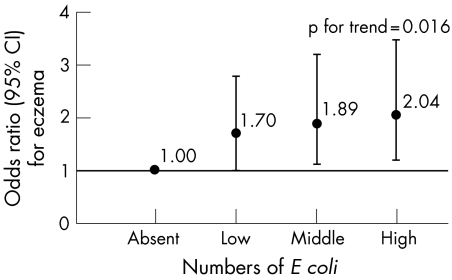
We subsequently analysed the association between the counts of the bacteria under study and the atopic outcomes. Both unadjusted (data not shown) and adjusted analysis revealed that the risk of developing eczema increased with increasing numbers of E coli in the faecal samples (pfor trend = 0.016) (fig 1). Infants with high E coli counts in their stools had a twofold higher risk of developing eczema compared with uncolonised infants. Again this association was not found when atopic dermatitis was defined according to the UK‐WP criteria. The association between C difficile colonisation and eczema, atopic dermatitis, recurrent wheeze and atopic sensitisation was not influenced by the concentration of this bacterium in the faecal samples. Furthermore, counts of bifidobacteria, B fragilis group species and lactobacilli were not associated with atopic disease manifestation or sensitisation.
Figure 1 Adjusted association between numbers of Escherichia coli (log10 CFU/g faeces) in faecal samples of 1 month old infants and the development of eczema in the first 2 years of life (adjusted for subcohort, parental history of atopy, sibling history of atopy, age at collection of faecal sample, infant's gender and total bacterial count). Dots represent the odds ratios, bars represent the 95% confidence intervals. Low: <8.86 log10 CFU; middle: 8.86 to 9.75 log10 CFU; high: >9.75 log10 CFU.
Total serum IgE level at the age of 2 years was not associated with colonisation rates or counts of gut bacteria.
Discussion
This prospective study demonstrates that differences in gut microbiota composition in early infancy are associated with the subsequent development of atopic disease manifestation and sensitisation. The presence of E coli was associated with a higher risk of developing eczema, this risk being increased with increasing numbers of E coli. However, this association was not found when atopic dermatitis was defined according to the UK‐WP criteria. Colonisation with C difficile in early infancy was associated with an increased risk of all atopic outcomes (eczema, atopic dermatitis, recurrent wheeze and atopic sensitisation), independent of the concentration of this bacterium.
This is the first large‐scale prospective study on gut microbiota composition in relation to atopic manifestations. Although previous epidemiological studies were very informative and were the first to suggest a potential role of the gut microbiota in the aetiology of atopic diseases, they were often based on small populations and were not able to adjust for potential confounders. Furthermore, only two previous studies9,11 were prospective and therefore able to determine if differences in the gut microbiota precede the development of atopic symptoms. Another strength of our study is the molecular techniques used, which overcome many of the problems associated with traditional bacteriological culture. Analyses of the gut microbiota using bacteriological culture are biased, since many selective culture media are not absolutely selective. Furthermore, these media do not equally support the growth of the different species comprising a population, and not all bacteria are cultivatable.37,38 The real‐time PCR assays used in the present study are a quantitative culture‐independent approach suitable for high‐throughput analyses of both fresh and frozen samples.
At present, faeces is the only realistic sample available in large non‐invasive epidemiological studies on the gut microbiota. However, a limitation of using faecal samples is that the bacterial composition in the lumen does not reflect the composition of bacteria adhering to the mucosa, and furthermore the composition of bacteria differs throughout the intestinal tract.39 Nevertheless, it can be assumed that although the proportions and activities of the microbiota change with passage through the intestinal tract, most viable and non‐viable intestinal bacteria will still be detectable in the faeces by molecular methods.40 A drawback of this study is the time between collection of the samples by the parents and processing of the samples in the laboratory, which was 1 day for most samples. Ott and colleagues41 demonstrated that the total amount of bacterial DNA as well as the diversity of the microbiota significantly decreased over such a time span. However, they also showed that the similarity (determined by DGGE) of faecal samples processed immediately and processed after 24 h remained high. This means that the dominant microbiota appears to be relatively stable. Furthermore, the aim of the present population‐based study was to examine differences in gut microbiota composition between subjects. It is not likely that the possible change in composition of the samples during transportation was different for infants who developed atopic manifestations later on in life and infants who did not.
In the present study, only one faecal sample per infant was collected at the age of 1 month post partum. There are several reasons why we chose to collect faeces at this age. First, as the gut microbiota is thought to drive the postnatal development of the immune system,12 the first months of life seem to be of major importance. Second, at the age of 1 month colonisation is complete and although the composition may fluctuate, large shifts in the composition do not occur until weaning.42 Finally, as we wanted to exclude the chance of reverse causation, the gut microbiota composition had to be analysed prior to the manifestation of atopic symptoms.
Since manifestation of atopic symptoms and sensitisation to allergens do not always occur together (sensitised infants do not always show symptoms and infants with symptoms are not always sensitised),43 we chose to report the manifestation of atopic symptoms and sensitisation as separate outcome parameters. As a consequence we decided to use the term eczema instead of atopic eczema or atopic dermatitis. However, we did use the term atopic dermatitis for those infants visited at home who fulfilled the UK‐WP criteria, since Williams and colleagues when introducing these criteria originally used the term atopic dermatitis.22
The positive association we found between E coli and eczema is difficult to compare with previous reports, since several of these studies did not determine E coli9 or measured this bacterium as part of total coliforms,8 enterobacteriaceae10 or Gram‐negative rods.11
In contrast to the association between E coli and eczema (based on parents' reports in questionnaires), we did not find an association between E coli and atopic dermatitis based on the UK‐WP criteria. Selection bias could not explain this difference since non‐response analyses showed that the gut microbiota and the prevalence of eczema and recurrent wheeze was similar for those infants who were visited at home and those who were not. The percentage of infants with atopic dermatitis according to the UK‐WP criteria is much lower than the percentage of infants with eczema reported by parents. This can be explained by the fact that eczema reported by parents is based upon the presence of this condition at any time during the infants' first 2 years of life, whereas many of these infants are probably in remission or have already outgrown this condition at time of the home visit. Therefore, it is possible that E coli is only associated with a milder eczematous condition that has already disappeared by the time of the home visit. Another explanation for these discrepant findings is that the UK‐WP criteria included specific predilection sites of atopic dermatitis (flexural involvement),22,23,24 whereas the questionnaire data were based on the presence of an itchy rash anywhere (except diaper rash, rash around the eyes and scalp scaling). The increased risk of eczema in infants colonised with E coli may therefore also be limited to eczema other than at the specific predilection sites for atopic eczema/dermatitis and thus may be non‐atopic. This idea is supported by the fact that we also found no association between E coli and sensitisation.
In contrast, C difficile appears to be associated with a higher risk of atopic eczema since a positive association was also found for atopic dermatitis according to UK‐WP criteria and for sensitisation. Our findings of an association between C difficile and atopy are in agreement with several previous studies. In a study on microflora‐associated characteristics, allergic infants had higher levels of i‐caproic acid in their stools compared with non‐allergic infants. This short chain fatty acid is suggested to indicate the presence of C difficile.44 Two studies used IgG serology against C difficile. Woodcock and colleagues found increased specific IgG against C difficile in sensitised wheezy infants compared with non‐sensitised non‐wheezy infants.45 Linneberg and colleagues found a positive association between IgG seropositivity against C difficile and both allergic rhinitis and sensitisation.46
Furthermore, several studies found an association between high numbers of the genus Clostridium and atopic dermatitis and/or sensitisation,9,11,47 although others did not find such an association.10,48 The genus Clostridium is a very heterogeneous group comprising several different clusters.49 It seems therefore unlikely that the members of such a phyologenetically diverse genus like Clostridium all have the same effect on the human host. It is more likely that certain species (such as C difficile) or a certain cluster of species within this genus is responsible for the increased risk of developing atopic manifestations.
In contrast to several other studies,8,9,10,11,47 we did not find a negative association between allergies and bifidobacteria. A possible explanation is the lack of contrast in our study with respect to bifidobacterial counts, because almost all infants were colonised with high numbers of bifidobacteria. This is probably the consequence of the very young age of our population, an age at which bifidobacteria are known to dominate the gut microbiota.50
Altogether our results support a role of the gut microbiota in the aetiology of atopic diseases. There are several hypotheses by which the associations we found between C difficile and E coli and atopic manifestations could be explained. First of all it should be noted that many of the bacteria in the gut are still unknown; differences in E coli and C difficile colonisation found in our study could therefore also reflect differences in other unknown bacteria.9
The presence of E coli and C difficile could be associated with a decrease in other (unknown) beneficial bacteria. This could result in reduced induction of Treg cells by these beneficial bacteria leading to immune dysregulation. In the absence of optimal levels of immune regulation, an individual may develop a Th1 (such as Crohn's disease or autoimmunity) or Th2 (such as atopic diseases) mediated inflammatory disorder depending on their own Th1/Th2 bias, immunological history and genetic background.7 Secondly, E coli and C difficile could have a direct effect on the production of cytokines by antigen‐presenting cells, thereby affecting the differentiation of T cells.51 Another hypothesis is that E coli and/or C difficile increase the intestinal permeability (for instance by the production of toxins). This increased permeability of the intestinal barrier could facilitate the penetration of innocuous antigens and subsequent sensitisation.46 Indeed it has been shown that C difficile toxins A and B compromise the intestinal cell barrier.52,53 Furthermore, increased intestinal permeability has been described in patients with food allergies, eczema and asthma compared with healthy subjects.54,55,56,57 Finally, it has also been suggested that infants susceptible to the development of allergies are also susceptible to aberrant colonisation of the gut. However, this explanation seems less likely since we have controlled for familial history of atopy. Additionally, the fact that differences in the gut microbiota are already present at such a young age preceding atopic symptoms makes this hypothesis less likely.
The consistent findings of a positive association between C difficile and all atopic symptoms as well as sensitisation strengthen the probability of a causal relationship between the gut microbiota and atopy, and support the potential role of probiotics in the prevention and treatment of these diseases. We have previously examined the external factors influencing the composition of the gut microbiota in early infancy.25 Caesarean section, antibiotic therapy, hospitalisation and formula feeding all caused perturbations in the gut microbiota by increasing colonisation rates and counts of E coli and C difficile and/or by decreasing numbers of bifidobacteria. In particular, when the gut microbiota is disrupted by one or more of these external factors, probiotics may be effective in the treatment or prevention of atopic diseases.
Perturbations in the gut microbiota may also be related to other atopic outcomes which manifest at older ages such as asthma, rhinoconjunctivitis and persistent food allergies; long term follow‐up of cohort studies is necessary to examine whether perturbations are also related to these outcomes.
In conclusion, we demonstrated that differences in the gut microbiota composition precede the manifestation of atopic symptoms and atopic sensitisation. In particular, C difficile was associated with all atopic symptoms and sensitisation, whereas E coli appeared to be only associated with (non‐atopic) eczema. Different immunological mechanisms may underlie the effects of E coli and C difficile. This calls for further research on the mechanisms by which intestinal microbes interfere with our (gastrointestinal) immune system.
Acknowledgements
We thank Chantal Delnoy, Brigitte Winants and Karen Groot for home visits, Cobie Martens and Willeke Hendrikx for assistance with data collection questionnaires, Astrid van Leeuwen (Sanquin) for IgE determination, and last but not least, all mothers and their infants participating in the KOALA study.
Abbreviations
CFU - colony forming units
Ig - immunoglobulin
IL - interleukin
OR - odds ratio
Th1 - Th2, T helper 1, 2
Treg - regulatory T cell
UK‐WP - UK‐Working Party
Footnotes
Funding: This study was supported by grants from the Dutch Asthma Foundation (grant 3.2.03.48) and Royal Friesland Foods (the Netherlands).
Competing interests: None declared.
References
- 1.Beasley R, Keil U, von Mutius E.et al Worldwide variation in prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and atopic eczema: ISAAC. Lancet 19983511225–1232. [PubMed] [Google Scholar]
- 2.Ngoc P L, Gold D R, Tzianabos A O.et al Cytokines, allergy, and asthma. Curr Opin Allergy Clin Immunol 20055161–166. [DOI] [PubMed] [Google Scholar]
- 3.Nowak D, Suppli Ulrik C, von Mutius E. Asthma and atopy: has peak prevalence been reached? Eur Respir J 200423359–360. [DOI] [PubMed] [Google Scholar]
- 4.Strachan D P. Hay fever, hygiene, and household size. BMJ 19892991259–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strachan D P. Family size, infection and atopy: the first decade of the “hygiene hypothesis”. Thorax 200055(Suppl 1)S2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noverr M C, Huffnagle G B. Does the microbiota regulate immune responses outside the gut? Trends Microbiol 200412562–568. [DOI] [PubMed] [Google Scholar]
- 7.Rook G A, Brunet L R. Microbes, immunoregulation, and the gut. Gut 200554317–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjorksten B, Naaber P, Sepp E.et al The intestinal microflora in allergic Estonian and Swedish 2‐year‐old children. Clin Exp Allergy 199929342–346. [DOI] [PubMed] [Google Scholar]
- 9.Bjorksten B, Sepp E, Julge K.et al Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol 2001108516–520. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe S, Narisawa Y, Arase S.et al Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. J Allergy Clin Immunol 2003111587–591. [DOI] [PubMed] [Google Scholar]
- 11.Kalliomaki M, Kirjavainen P, Eerola E.et al Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol 2001107129–134. [DOI] [PubMed] [Google Scholar]
- 12.Bjorksten B. Effects of intestinal microflora and the environment on the development of asthma and allergy. Springer Semin Immunopathol 200425257–270. [DOI] [PubMed] [Google Scholar]
- 13.Majamaa H, Isolauri E. Probiotics: a novel approach in the management of food allergy. J Allergy Clin Immunol 199799179–185. [DOI] [PubMed] [Google Scholar]
- 14.Isolauri E, Arvola T, Sutas Y.et al Probiotics in the management of atopic eczema. Clin Exp Allergy 2000301604–1610. [DOI] [PubMed] [Google Scholar]
- 15.Weston S, Halbert A, Richmond P.et al Effects of probiotics on atopic dermatitis: a randomised controlled trial. Arch Dis Child 200590892–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalliomaki M, Salminen S, Arvilommi H.et al Probiotics in primary prevention of atopic disease: a randomised placebo‐controlled trial. Lancet 20013571076–1079. [DOI] [PubMed] [Google Scholar]
- 17.Kalliomaki M, Salminen S, Poussa T.et al Probiotics and prevention of atopic disease: 4‐year follow‐up of a randomised placebo‐controlled trial. Lancet 20033611869–1871. [DOI] [PubMed] [Google Scholar]
- 18.Boyle R J, Tang M L. The role of probiotics in the management of allergic disease. Clin Exp Allergy 200636568–576. [DOI] [PubMed] [Google Scholar]
- 19.Brouwer M L, Wolt‐Plompen S A, Dubois A E.et al No effects of probiotics on atopic dermatitis in infancy: a randomized placebo‐controlled trial. Clin Exp Allergy 200636899–906. [DOI] [PubMed] [Google Scholar]
- 20.Kummeling I, Thijs C, Penders J.et al Etiology of atopy in infancy: the KOALA Birth Cohort Study. Pediatr Allergy Immunol 200516679–684. [DOI] [PubMed] [Google Scholar]
- 21.Bastiaanssen J M, de Bie R A, Bastiaenen C H.et al Etiology and prognosis of pregnancy‐related pelvic girdle pain; design of a longitudinal study. BMC Public Health 200551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams H C, Burney P G, Hay R J.et al The U.K. Working Party's Diagnostic Criteria for Atopic Dermatitis. I. Derivation of a minimum set of discriminators for atopic dermatitis. Br J Dermatol 1994131383–396. [DOI] [PubMed] [Google Scholar]
- 23.Williams H C, Burney P G, Pembroke A C.et al The U.K. Working Party's Diagnostic Criteria for Atopic Dermatitis. III. Independent hospital validation. Br J Dermatol 1994131406–416. [DOI] [PubMed] [Google Scholar]
- 24.Williams H C, Burney P G, Strachan D.et al The U.K. Working Party's Diagnostic Criteria for Atopic Dermatitis. II. Observer variation of clinical diagnosis and signs of atopic dermatitis. Br J Dermatol 1994131397–405. [DOI] [PubMed] [Google Scholar]
- 25.Penders J, Thijs C, Vink C.et al Factors influencing the intestinal microbiota in early infancy. Pediatrics 2006118511–521. [DOI] [PubMed] [Google Scholar]
- 26.Penders J, Vink C, Driessen C.et al Quantification of Bifidobacterium spp. Escherichia coli and Clostridium difficile in faecal samples of breast‐fed and formula‐fed infants by real‐time PCR. FEMS Microbiol Lett 2005243141–147. [DOI] [PubMed] [Google Scholar]
- 27.Huijsdens X W, Linskens R K, Mak M.et al Quantification of bacteria adherent to gastrointestinal mucosa by real‐time PCR. J Clin Microbiol 2002404423–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C, Song Y, McTeague M.et al Rapid identification of the species of the Bacteroides fragilis group by multiplex PCR assays using group‐ and species‐specific primers. FEMS Microbiol Lett 20032229–16. [DOI] [PubMed] [Google Scholar]
- 29.Walter J, Hertel C, Tannock G W.et al Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group‐specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol 2001672578–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rinttila T, Kassinen A, Malinen E.et al Development of an extensive set of 16S rDNA‐targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real‐time PCR. J Appl Microbiol 2004971166–1177. [DOI] [PubMed] [Google Scholar]
- 31.Heilig H G H J, Zoetendal E G, Vaughan E E.et al Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol 200268114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nadkarni M A, Martin F E, Jacques N A.et al Determination of bacterial load by real‐time PCR using a broad‐range (universal) probe and primers set. Microbiology 2002148257–266. [DOI] [PubMed] [Google Scholar]
- 33.Aalberse R C, Koshte V, Clemens J G. Immunoglobulin E antibodies that crossreact with vegetable foods, pollen, and Hymenoptera venom. J Allergy Clin Immunol 198168356–364. [DOI] [PubMed] [Google Scholar]
- 34.Stallman P J, Aalberse R C. Estimation of basophil‐bound IgE by quantitative immunofluorescence microscopy. Int Arch Allergy Appl Immunol 1977549–18. [DOI] [PubMed] [Google Scholar]
- 35.Schuurman J, Perdok G J, Lourens T E.et al Production of a mouse/human chimeric IgE monoclonal antibody to the house dust mite allergen Der p 2 and its use for the absolute quantification of allergen‐specific IgE. J Allergy Clin Immunol 199799545–550. [DOI] [PubMed] [Google Scholar]
- 36.Asher M I, Keil U, Anderson H R.et al International Study of Asthma and Allergies in Childhood (ISAAC): rationale and methods. Eur Respir J 19958483–491. [DOI] [PubMed] [Google Scholar]
- 37.Furrie E. A molecular revolution in the study of intestinal microflora. Gut 200655141–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tannock G W. Analysis of the intestinal microflora: a renaissance. Antonie Van Leeuwenhoek 199976265–278. [PubMed] [Google Scholar]
- 39.Macpherson A J, Harris N L. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 20044478–485. [DOI] [PubMed] [Google Scholar]
- 40.Mai V, Morris J G., Jr Colonic bacterial flora: changing understandings in the molecular age. J Nutr 2004134459–464. [DOI] [PubMed] [Google Scholar]
- 41.Ott S J, Musfeldt M, Timmis K N.et al In vitro alterations of intestinal bacterial microbiota in fecal samples during storage. Diagn Microbiol Infect Dis 200450237–245. [DOI] [PubMed] [Google Scholar]
- 42.Edwards C A, Parrett A M. Intestinal flora during the first months of life: new perspectives. Br J Nutr 200288(Suppl 1)11–18. [DOI] [PubMed] [Google Scholar]
- 43.Kusel M M, Holt P G, de Klerk N.et al Support for 2 variants of eczema. J Allergy Clin Immunol 20051161067–1072. [DOI] [PubMed] [Google Scholar]
- 44.Bottcher M F, Nordin E K, Sandin A.et al Microflora‐associated characteristics in faeces from allergic and nonallergic infants. Clin Exp Allergy 2000301590–1596. [DOI] [PubMed] [Google Scholar]
- 45.Woodcock A, Moradi M, Smillie F I.et al Clostridium difficile, atopy and wheeze during the first year of life. Pediatr Allergy Immunol 200213357–360. [DOI] [PubMed] [Google Scholar]
- 46.Linneberg A, Ostergaard C, Tvede M.et al IgG antibodies against microorganisms and atopic disease in Danish adults: the Copenhagen Allergy Study. J Allergy Clin Immunol 2003111847–853. [DOI] [PubMed] [Google Scholar]
- 47.Sepp E, Julge K, Mikelsaar M.et al Intestinal microbiota and immunoglobulin E responses in 5‐year‐old Estonian children. Clin Exp Allergy 2005351141–1146. [DOI] [PubMed] [Google Scholar]
- 48.Kirjavainen P V, Apostolou E, Arvola T.et al Characterizing the composition of intestinal microflora as a prospective treatment target in infant allergic disease. FEMS Immunol Med Microbiol 2001321–7. [DOI] [PubMed] [Google Scholar]
- 49.Stackebrandt E, Kramer I, Swiderski J.et al Phylogenetic basis for a taxonomic dissection of the genus Clostridium. FEMS Immunol Med Microbiol 199924253–258. [DOI] [PubMed] [Google Scholar]
- 50.Fanaro S, Chierici R, Guerrini P.et al Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl 20039148–55. [DOI] [PubMed] [Google Scholar]
- 51.Karlsson H, Hessle C, Rudin A. Innate immune responses of human neonatal cells to bacteria from the normal gastrointestinal flora. Infect Immun 2002706688–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pothoulakis C. Effects of Clostridium difficile toxins on epithelial cell barrier. Ann N Y Acad Sci 2000915347–356. [DOI] [PubMed] [Google Scholar]
- 53.Feltis B A, Wiesner S M, Kim A S.et al Clostridium difficile toxins A and B can alter epithelial permeability and promote bacterial paracellular migration through HT‐29 enterocytes. Shock 200014629–634. [DOI] [PubMed] [Google Scholar]
- 54.Majamaa H, Isolauri E. Evaluation of the gut mucosal barrier: evidence for increased antigen transfer in children with atopic eczema. J Allergy Clin Immunol 199697985–990. [DOI] [PubMed] [Google Scholar]
- 55.Benard A, Desreumeaux P, Huglo D.et al Increased intestinal permeability in bronchial asthma. J Allergy Clin Immunol 1996971173–1178. [DOI] [PubMed] [Google Scholar]
- 56.Caffarelli C, Cavagni G, Menzies I S.et al Elimination diet and intestinal permeability in atopic eczema: a preliminary study. Clin Exp Allergy 19932328–31. [DOI] [PubMed] [Google Scholar]
- 57.Pike M G, Heddle R J, Boulton P.et al Increased intestinal permeability in atopic eczema. J Invest Dermatol 198686101–104. [DOI] [PubMed] [Google Scholar]