Abstract
The Shk1 protein kinase, a homolog of Saccharomyces cerevisiae Ste20 and mammalian p21Cdc42/Rac-activated kinases, is an essential component of a Ras- and Cdc42-dependent signaling cascade required for cell viability, normal morphology, and mitogen-activated protein kinase-mediated sexual responses in the fission yeast, Schizosaccharomyces pombe. To identify S. pombe proteins that modulate or mediate Shk1 functions, we conducted a two-hybrid screen for Shk1-interacting proteins. One of the genes identified as a result of this screen was skb1. We show that Skb1 interacts with a region of the N-terminal regulatory domain of Shk1 distinct from that to which Cdc42 binds, and that Shk1, Cdc42, and Skb1 are able to form a ternary complex in vivo. S. pombe cells carrying an skb1 null mutation are less elongate in morphology than wild-type cells and exhibit a moderate growth defect. The morphology defect of the skb1 deletion mutant is suppressed by overexpression of Shk1. Overexpression of Skb1 causes wild-type S. pombe cells to become hyperelongated. Additional genetic analyses described herein suggest that Skb1 is a component of the morphology control branch of the Ras signaling cascade in S. pombe and that it positively modulates Shk1 function. Homologs of Skb1 are encoded by open reading frames in the genomes of S. cerevisiae and Caenorhabditis elegans and by an uncharacterized human cDNA sequence. Thus, skb1 may be the first well-characterized member of a highly conserved family of genes encoding potential p21Cdc42/Rac-activated kinase regulators.
Keywords: Ras, Cdc42, p21-activated kinases, signal transduction, cell morphology
The small GTP-binding proteins encoded by members of the highly conserved ras gene family are essential components of signal transduction pathways regulating cell growth and differentiation in eukaryotes from yeasts to mammals (1). It has been firmly established that a key function of Ras proteins in mammalian cells is in the regulation of mitogen-activated protein kinase/extracellular-regulated kinase (MAPK/ERK) cascades (2). The best-characterized Ras effector is the serine/threonine kinase, Raf, which phosphorylates and activates the MAPK/ERK kinase MEK. However, multiple lines of evidence indicate that there are additional Ras effectors besides Raf in mammalian cells. For example, Ras proteins have been shown to bind phosphatidylinositol-3-OH kinase (3), Ral-GDS (4), and MEKK, a MAPK kinase kinase (MAPKKK) that is structurally unrelated to Raf (5). In addition, Ras appears to act through MAPKKKs distinct from Raf in some types of mammalian cells (6, 7, 8) and a mutant Ras protein, H-RasT35S, that does not bind to Raf strongly induces MAPK activation in Xenopus oocyte extracts (8).
Perhaps the most significant unanswered questions pertaining to Ras function revolve around its role in regulation of the actin cytoskeleton. Treatment of mammalian cells with growth factors or with oncogenic Ras proteins rapidly induces actin reorganization, membrane ruffling, and alterations in cell morphology (9). Joneson et al. (10) recently used mutants of H-Ras to show that membrane ruffling and MAPK signaling occur through distinct Ras effector pathways in mammalian cells. Although the molecular details have yet to be fully elucidated, various lines of evidence indicate that members of the Rho family of Ras-related GTP-binding proteins, which includes Rho, Cdc42, and Rac proteins, are key intermediates in the Ras-induced cytoskeletal/morphological control pathways in higher eukaryotes (11). Protein kinases that are activated specifically by Rho family members have been identified and include the Rho-activated kinases ROKα and PKN (12, 13), and several structurally related p21Cdc42/Rac-activated kinases (PAKs) (14, 15, 16). However, it has yet to be demonstrated whether any of these kinases link the Rho-related proteins to cytoskeletal regulatory pathways.
The product of the single known ras homolog, ras1, in the fission yeast Schizosaccharomyces pombe is required for two distinct cellular functions that closely parallel Ras functions in higher organisms. First, Ras1 regulates a pheromone-induced MAPK cascade composed of the protein kinases Byr2 (MAPKKK), Byr1 (MAPKK), and Spk1 (MAPK) (17). Ras1 binds to the N-terminal regulatory domain of Byr2 in a GTP-dependent fashion (18, 19). This interaction is believed to be analogous to the interaction between Ras and Raf in mammalian cells. The second, genetically separable function of Ras1 is in the control of cell morphology. Wild-type S. pombe cells are rod-shaped, whereas ras1null mutants are ellipsoidal in morphology (20, 21). Neither the mating pheromone receptors, Gpa1, nor the components of the MAPK module are required for normal morphology in S. pombe (22, 23, 24, 25, 26, 27). A fission yeast homolog of mammalian Cdc42 (28) acts downstream from Ras1 in controlling cell morphology (29). Ras1 and Cdc42 are part of a multiprotein complex that includes Scd1, the putative Cdc42 guanine nucleotide exchange factor, and Scd2, an SH3 domain-containing protein of unknown function (29). We recently showed that the Shk1 protein kinase is an essential component of the Ras1/Cdc42 signaling module in S. pombe (30). Genetic analyses demonstrated that Shk1 is essential for cell viability and participates both in the regulation of cell morphology and in the Ras1-dependent mating response pathway (30). Shk1 is a homolog of mammalian PAKs (14, 15, 16) and the Ste20 protein kinase, which is required for pheromone response, localization of cell growth, and induction of filamentous growth in the budding yeast, Saccharomyces cerevisiae (31, 32, 33, 34).
As a step toward further elucidating the function and regulation of Shk1, we conducted a two-hybrid screen for S. pombe cDNAs encoding Shk1-interacting proteins. In this report, we describe the cloning and characterization of the skb1 gene, which was identified as a result of this screen. We provide genetic evidence that the Skb1 protein functions in the morphology control branch of the Ras1 signaling cascade in S. pombe and that it positively modulates Shk1 function. Skb1 was found to be homologous to proteins encoded by open reading frames (ORFs) in the genomes of S. cerevisiae and Caenorhabditis elegans and by an uncharacterized human cDNA sequence, suggesting that skb1-related genes have been highly conserved through evolution.
MATERIALS AND METHODS
Yeast Strains, Manipulation, and Genetic Analysis.
S. pombe strains used in this study were SP870 (h90 ade6-210 leu1-32 ura4-D18) (from D. Beach, Cold Spring Harbor Laboratory), SP870D (h90 ade6-210 leu1-32 ura4-D18/h90 ade6-210 leu1-32 ura4-D18) (from V. Jung, Cold Spring Harbor Laboratory), SP870M3 (h90 ade6-210 leu1-32 ura4-D18 scd1-1) (29), SP42N17 (h90 ade6-216 leu1-32 ura4::adh1-cdc42N17) (30), SPGLD (h90 ade6-210 leu1-32 ura4-D18 gpa1::LEU2/h90 ade6-210 leu1-32 ura4-D18 gpa1::LEU2) (17), SPRN1 (h90 ade6-210 leu1-32 ura4-D18 ras1Δ) (35), SPRN1D (h90 ade6-210 leu1-32 ura4-D18 ras1Δ/h90 ade6-210 leu1-32 ura4-D18 ras1Δ) (see below), SPSCD2L (h90 ade6-210 leu1-32 ura4-D18 scd2::leu2) (29), SPSKB1U (h90 ade6-210 leu1-32 ura4-D18 skb1::ura4) (see below), and SPSUD (h90 ade6-210 leu1-32 ura4-D18 byr2::ura4/h90 ade6-210 leu1-32 ura4-D18 byr2::ura4) (26). S. cerevisiae strains used were L40 (MATa ade2 his3 leu2 trp1 LYS2::lexA-HIS3 URA3::lexA-lacZ) (37) and HF7c (MATa ade2-101 his3-200 leu2-3, 112 lys2-801 trp1-901 ura3-52 gal4-542 gal80-538 LYS2::GAL1UAS-GAL1TATA-HIS3 URA3:: GAL417 mers(x3)-CYC1TATA-lacZ) (36). Standard yeast culture media and genetic methods were used (37, 38). S. pombe cultures were grown on either rich medium (YEA) or synthetic minimal medium (EMM) with appropriate auxotrophic supplements (38). S. cerevisiae cultures were grown on either rich medium (YPD) or drop-out medium with appropriate auxotrophic supplements (37). Yeast were transformed by the lithium acetate procedure (38). The skb1::ura4 strain, SPSKB1U, was constructed by transformation of SP870D with a 2.5-kb SpeI–EarI skb1::ura4 fragment from the plasmid pBSskb1::ura4. Diploid transformants carrying a single disrupted and a single wild-type copy of skb1 were identified by Southern blot analysis. The ras1Δ/ras1Δ diploid strain, SPRN1D, was isolated by subjecting the ras1Δ haploid strain, SPRN1, to the lithium acetate transformation process, followed by selection of diploids on YEA plates containing phloxin B.
Plasmids.
The two-hybrid plasmids pGADGH (for expression of GAD fusions), pHP5 and pGBT9 (for expression of GBD fusions), and pBTM116 and pVJL11 (for expression of LBD fusions) have been described previously (18, 29, 39, 40). The plasmids pGADCdc42, pGADRas1, pGADShk1, pGADScd1, pGADScd2, pGBDByr1, pLBDByr2, pLBDCdc42, pLBDlamin, pLBDMek1, pLBDRas1, pLBDSte7, pLBDSte11, pLBDSte20, pAAUCMShk1, and pALR have also been described (29, 30, 39, 40). pLBDRac1 was a gift from J. Camonis (Faculte de Medecine Lariboisiere, Paris). pLBDRaf1 was a gift from L. Van Aelst (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). The S. pombe cDNA library constructed in the plasmid pGADGH was a gift from G. Hannon (Cold Spring Harbor Laboratory). The S. pombe-Escherichia coli shuttle vectors pAAUCM (30) and pART1 (41) were used for high level expression of coding sequences from the S. pombe adh1 promoter. pREP1 (42) was used for expressing coding sequences from the S. pombe nmt1 promoter. The S. cerevisiae-E. coli shuttle vector pAUD6 (40) was used for expressing coding sequences from the S. cerevisiae ADH1 promoter. pLBDShk1 was obtained by subcloning a BamHI–SalI fragment of shk1 from pGADShk1 into the corresponding sites of pVJL11. An HpaI–BamHI fragment of shk1 (encoding amino acid residues 189–540) was isolated from pBSIIShk1 (30) and cloned into the SmaI–BamHI sites of pBTM116, generating plasmid pLBDShk1ΔN. Polymerase chain reactions (PCRs) were used to generate DNA fragments encoding Shk1ΔC (residues 1 to 262), Shk1 R2 (residues 29-85), and Shk1 R3 (residues 86-262) subdomains for subcloning into pGADGH, pHP5, and pVJL11 to produce the plasmids pGADShk1ΔC, pGBDShk1R2, and pLBDShk1R3, respectively. A BamHI fragment of Shk1 from pAAUCMShk1 was cloned into the BamHI sites of pAUD6 to produce pAUD6Shk1. pAUD6Shk1-ADE2 was constructed by cloning a SacI fragment of the ADE2 gene into the corresponding site in pAUD6Shk1. pREP1Shk1 was constructed by cloning a SalI–SacI fragment of shk1 from pAAUCMShk1 into the corresponding sites of pREP1. A BamHI–KpnI fragment of skb1-1 from pGADSkb1-1 was cloned into the corresponding sites of pBluescript II SK− (Stratagene) to produce pBSskb1-1. A PCR-derived BamHI–NdeI fragment of the skb1 gene was used to replace the BamHI–NdeI fragment of skb1-1 in pBSskb1-1 to produce pBSskb1. pGADSkb1 was constructed by cloning a BamHI–XhoI fragment of skb1 from pBSskb1 into the corresponding sites of pGADGH. A BamHI–KpnI fragment of skb1 from pBSskb1 was cloned into pAAUCM producing pAAUCMSkb1. A BamHI–SacI fragment of skb1 was isolated from pAAUCMSkb1 and cloned into pREP1 and pART1 to produce pREP1Skb1 and pART1Skb1, respectively. pGBDSkb1-1 was constructed by cloning a BamHI–XhoI fragment of skb1 from pGADSkb1-1 into the BamHI–SalI sites of pHP5. pGBDSkb1ΔN72 was constructed by cloning an EcoRI–XhoI fragment of skb1 into the EcoRI–SalI sites of pGBT9. pGBDScd1 was constructed by cloning a BamHI fragment of Scd1 from pGADScd1 into pHP5. pGBDScd2 was constructed by cloning a BamHI fragment of Scd2 from pGADScd2 into pHP5. A BamHI fragment of shk1 was isolated from pBSIIShk1 and cloned into pHP5 to produce pGBDShk1. pLBDScd1 was constructed by cloning a BamHI fragment of scd1 from pGBDScd1 into pVJL11. The plasmid pBSskb1::ura4 was constructed in three steps. First, a 3′ skb1 fragment was amplified by PCR using pGADSkb1-1 as template and the primer pair 5′-AAACCCAAGCTTCCAATGCCTATCTGGTGT and 5′-GTAATACGACTCACTATAGGGC. The resulting product was digested with HindIII and XhoI and cloned into the corresponding sites of pBluescript II SK− to create pBSskb1-3′. Next, a 5′ fragment of skb1 was amplified by PCR using pGADSkb1-1 as template and the oligonucleotide primer pair 5′-GATACCCCACCAAACCCAAAA and 5′-AAACCCAAGCTTGGGTGAAGTTGGACCCAT. The resulting product was digested with BamHI and HindIII and cloned into the corresponding sites of pBSskb1-3′. The resulting plasmid, pBSskb1Δ, was digested with HindIII, then ligated to a HindIII fragment of the ura4 gene to produce pBSskb1::ura4. An skb1::ura4 knockout fragment, which carries a 1.4-kb deletion of the skb1 ORF, is released by digesting pBSskb1::ura4 with EarI and SpeI.
β-Galactosidase (β-gal) Assays.
The 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) filter assay for determining two-hybrid interactions was performed as described (18). The liquid assay for β-gal activity was performed as described (37). β-Gal activity was calculated using the following formula: (OD420 × 1.7)/(0.0045 × protein conc. × extract volume × time). Protein conc. is expressed as mg/ml, extract volume in ml, and time as minutes.
RESULTS
Cloning and Sequence Analysis of the skb1 Gene.
A TRP1-based plasmid was constructed for expressing Shk1 as a fusion to the C terminus of the LexA DNA-binding domain (LBD) (see Materials and Methods). LBD-Shk1 formed complexes with Cdc42 and Rac Gal4 activation domain (GAD) fusion proteins, but did not form detectable complexes with any of several other proteins implicated in MAPK signaling and/or morphological control in S. pombe, S. cerevisiae, and mammalian cells (some shown in Table 1). An S. cerevisiae two-hybrid host strain, L40, which carries lexA-HIS3 and lexA-lacZ reporter genes (ref. 39; see Materials and Methods), was cotransformed with pLBDShk1 and a S. pombe cDNA library constructed in a LEU2-based plasmid, pGADGH (18), for expression of GAD fusion proteins. From a screen of approximately 360,000 Leu+/Trp+ transformants, 216 His+ colonies were isolated, 33 of which produced strong β-gal signals. Nine of the His+/β-gal+ transformants carried pGADGH clones expressing proteins that interacted with LBD-Shk1, but not with LBD-lamin. Eight of the nine Shk1-interacting clones carried various fragments of the same cDNA sequence, which we have designated skb1 (for Shk1 kinase-binding protein 1). A ninth clone carried a distinct sequence, designated skb2, that will be described elsewhere.
Table 1.
Detection of Skb1–Shk1 and Skb1–Skb1 interactions in the two-hybrid system
| DNA-binding domain fusions | Activating domain
fusions
|
||||||
|---|---|---|---|---|---|---|---|
| Skb1 | Shk1 | Pak1 | Cdc42 | Ras1 | Scd1 | Scd2 | |
| LBD fusions | |||||||
| Shk1 | + | + | ND | + | − | − | − |
| Ste20 | − | ND | ND | + | − | − | − |
| Cdc42 | − | + | + | ND | − | − | + |
| Rac1 | − | + | + | ND | ND | ND | ND |
| Ras1 | − | − | − | − | ND | ND | ND |
| Gpa1 | − | − | ND | ND | − | ND | ND |
| Byr2 | − | − | ND | ND | + | ND | ND |
| Ste11 | − | − | ND | ND | − | ND | ND |
| Raf1 | − | − | ND | ND | + | ND | ND |
| Ste7 | − | − | ND | ND | − | ND | ND |
| Mek1 | − | ND | ND | ND | − | ND | ND |
| Scd1 | − | − | ND | ND | − | ND | + |
| GBD fusions | |||||||
| Skb1 | + | + | − | − | − | − | − |
| Shk1 | + | ND | ND | + | − | ND | ND |
| Gpa1 | − | − | ND | ND | − | ND | ND |
| Scd2 | − | − | ND | + | − | ND | ND |
| Byr1 | − | ND | ND | − | ND | ND | ND |
Values represent the presence of transformed colonies that expressed detectable β-gal activity (+) or not (−). ND, not determined. Activating domain fusions were to the activating domain of S. cerevisiae Gal4 (GAD). GBD–GAD combinations were expressed in the Gal4 two-hybrid tester strain, HF7c. LBD–GAD combinations were tested in the LexA tester strain, L40. At least eight independent transformants were tested for each determination.
The full-length skb1 gene was cloned by screening a S. pombe genomic library (43) for sequences that hybridized to one of the isolated cDNA clones of skb1, skb1-1. The full-length skb1 gene (GenBank accession no. U59684U59684) contains two introns and encodes a predicted protein 646 amino acids in length (Fig. 1A). Searches of computer data bases revealed that Skb1 lacks significant structural similarity to any characterized protein, but is highly homologous to proteins encoded by ORFs in the genomes of S. cerevisiae and C. elegans and by an uncharacterized partial human cDNA sequence (Fig. 1B). We conclude from these analyses that skb1-related genes have been highly conserved through evolution.
Figure 1.
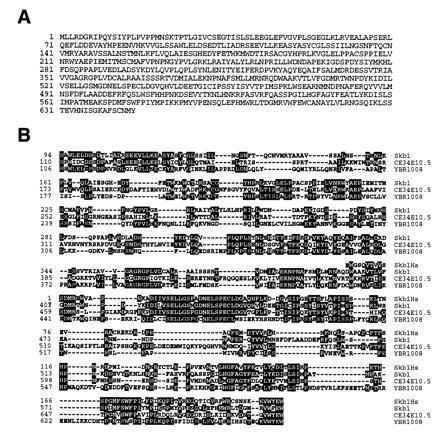
Deduced amino acid sequence of Skb1 and alignment of Skb1-related proteins encoded by open reading frames in the genomes of C. elegans and S. cerevisiae and by an uncharacterized human cDNA sequence. (A) Deduced amino acid sequence of the skb1 gene product. (B) blast searches of nucleic acid data bases revealed the existence of a human cDNA sequence, which we have named HsSkb1, and open reading frames in C. elegans (CE34E10.5) and S. cerevisiae (YBR1008) encoding predicted polypeptides with significant structural homology to Skb1. Alignments were generated using the LaserGene megalign program (DNAStar). Identical amino acid residues are indicated by black boxes. The HsSkb1 sequence was derived from four overlapping human expressed-sequence tag cDNA clones (GenBank accession nos. F12149F12149, H25612H25612, R65681R65681, and T69495T69495). The polypeptide encoded by the entire partial length HsSkb1 cDNA is shown. Residues 110–690 of the 1281 amino acid protein predicted by the CE34E10.5 ORF are shown, while residues 106-677 of the 827 amino acid protein predicted by YBR1008 are shown. Skb1 and HsSkb1 exhibit 47% identity over a span of 213 amino acids. Over its entire length of 646 amino acids, Skb1 is 35% identical to C34E10.5 and 33% identical to YBR1008.
Skb1 Interacts with the R3 Subdomain of Shk1 and Forms a Homomeric Two-Hybrid Complex.
We used the two-hybrid system to examine both the specificity of Skb1 protein–protein interactions and whether Skb1 interacts with other components of the Ras1/Shk1-dependent signaling pathways in S. pombe (Table 1). Skb1 did not form detectable complexes with S. cerevisiae Ste20 or mammalian p65Pak (Pak1). Skb1 also did not interact detectably with the S. pombe proteins Ras1, Gpa1, Cdc42, Scd1, Scd2, Byr2, or Byr1, nor with any of several other proteins involved in Ras and/or MAPK signaling in mammalian cells and S. cerevisiae. However, interaction between GBD-Skb1 and GAD-Skb1 fusion proteins was detected, suggesting that Skb1 is capable of forming homomeric complexes in vivo.
Shk1 is composed of a C-terminal catalytic domain and an N-terminal regulatory domain (Fig. 2A). The regulatory domain can be further divided into at least three subdomains, which we have designated R1, R2, and R3 (Fig. 2A). The R2 subdomain is the Cdc42/Rac-binding site of Shk1 (Fig. 2A) and is highly homologous to Cdc42/Rac-binding sites found in mammalian and S. cerevisiae PAKs. In contrast, the R1 and R3 subdomains are not highly conserved among PAKs across species. Skb1 formed two-hybrid complexes with the complete Shk1 regulatory domain and with the R3 subdomain, but did not form detectable complexes with either the catalytic domain or the R2 subdomain of Shk1 (Fig. 2A). These results demonstrate that Skb1 interacts with a regulatory subdomain of Shk1 distinct from that to which Cdc42 binds, and that Shk1 kinase activity is not required for interaction between Skb1 and Shk1.
Figure 2.
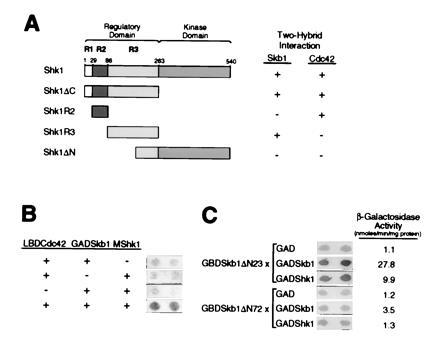
Analysis of two-hybrid interactions between Skb1, Shk1, and Cdc42. (A) Skb1 forms a complex with the R3 subdomain of Shk1. Pairs of two-hybrid fusion proteins were tested for the ability to form complexes as indicated. The domains of Shk1 tested were as follows: Shk1ΔC, residues 1 to 262; Shk1R2, residues 29 to 85; Shk1R3, residues 86 to 262; and Shk1ΔN, residues 189-540. Shk1, Shk1R3, and Shk1ΔN were expressed as LBD fusions; Shk1R2 was expressed as a GBD fusion; and Shk1ΔC was expressed as a GAD fusion. Skb1 and Cdc42 were expressed as GAD fusions for testing against LBD-Shk1, LBD-Shk1R3, and LBD-Shk1ΔN. For testing against GAD-Shk1ΔC, Cdc42 and Skb1 were expressed as LBD and GBD fusions, respectively. The X-Gal filter assay was used for measuring β-gal activity. The detection (+) or not (−) of β-gal is indicated. Two-hybrid interactions were detected between Skb1 and Shk1, Shk1R3, and Shk1ΔC, and between Cdc42 and Shk1, Shk1R2, and Shk1ΔC. (B) Evidence that Skb1, Cdc42, and Shk1 form a ternary complex in vivo. S. cerevisiae strain L40 was transformed (+) or not (−) with plasmids expressing Skb1 fused to GAD (GAD-Skb1), Cdc42 fused to LBD (LBD-Cdc42), and/or c-myc epitope-tagged Shk1 (MShk1) and assayed for β-gal expression by filter assay (shown at right). Complex formation between Skb1 and Cdc42 is bridged by MShk1, suggesting that the three proteins form a ternary complex in vivo. (C) Evidence that Skb1 homomerization is required for interaction with Shk1. Skb1ΔN23 (Skb1-1) and Skb1ΔN72 GBD fusion proteins were tested for their ability to form complexes with GAD, GAD-Skb1, or GAD-Shk1 as indicated. β-gal activity was measured using both X-Gal filter assays and liquid assays of cell extracts. Values at right of panel indicate β-gal activity (nmoles/min per mg protein) as measured by liquid assay.
Having determined that Skb1 and Cdc42 bind to separate subdomains of Shk1, we asked whether the three proteins could form a ternary complex. To do this, we expressed Cdc42 as a fusion to LBD (LBD-Cdc42) and expressed Skb1 as a fusion to GAD (GAD-Skb1). We then determined whether Shk1, overexpressed from a third plasmid, could promote complex formation between LBD-Cdc42 and GAD-Skb1. Interaction between LBD-Cdc42 and GAD-Skb1 was detected only when Shk1 was coexpressed, suggesting that Cdc42, Skb1, and Shk1 are able to form a ternary complex in vivo (Fig. 2B).
We next sought to determine whether Skb1 homomerization is required for interaction with Shk1. Seven of the eight skb1 cDNAs isolated from our Shk1 two-hybrid screen were truncated such that they encoded Skb1 proteins lacking only the first 24 amino acids of full-length Skb1, while the eighth isolate encoded a protein lacking only the first 33 amino acids of Skb1. These results suggested that significant loss of N-terminal Skb1 sequence might render the protein defective in its ability to form a complex with Shk1. Indeed, a truncated protein, Skb1ΔN72, lacking the N-terminal 72 amino acids of full-length Skb1 failed to form a detectable two-hybrid complex with Shk1 (Fig. 2C). In addition, Skb1ΔN72 was significantly impaired in its ability to form a homomeric two-hybrid complex with full-length Skb1 protein (Fig. 2C). This was not due to decreased protein stability, as GBD-Skb1ΔN72 was judged to be as stable as GBD-Skb1 by Western blot analysis (data not shown). These results suggest that the N-terminal domain of Skb1 is required for formation of Skb1–Shk1 and Skb1–Skb1 complexes and that Skb1 homomeric complex formation may be important for interaction with Shk1.
Genetic Evidence for Functional Interaction Between Skb1 and Shk1 in S. pombe.
An skb1 deletion plasmid was generated in which the majority of the skb1 coding sequence was replaced with the ura4 gene (Fig. 3A; see Materials and Methods). The skb1::ura4 fragment was used to transform the S. pombe diploid strain, SP870D. Two independent skb1+/skb1::ura4 diploids were sporulated and asci containing four spores were dissected for tetrad analysis. Most asci produced two viable Ura+ spores and two viable Ura− spores. Southern blotting was used to confirm that cells derived from Ura+ spores contained a disrupted copy of skb1 and lacked the wild-type skb1 gene (data not shown). These results demonstrated that skb1 is not an essential gene.
Figure 3.
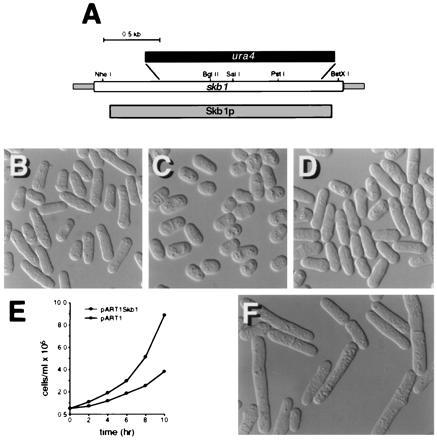
S. pombe cells carrying a deletion of the skb1 gene are aberrant in morphology and slightly defective for growth. (A) Map of the skb1 gene showing the fragment deleted by ura4 in construction of the skb1::ura4 mutant strain, SPSKB1U. The shaded bar corresponds to the sequence encoding the Skb1 protein; the open bar represents the skb1 sequence shown in Fig. 1. (B) Photomicrograph of wild-type (SP870) S. pombe cells. (C) Photomicrograph of skb1::ura4 (SPSKB1U) cells. Note that skb1::ura4 cells are less elongate than wild-type cells. (D) skb1::ura4 cells overexpressing shk1 from the plasmid pREP1Shk1. Cells were grown at 30°C in EMM to about 107 cells per ml prior to photomicrography. (E) Growth curves for SPSKB1U cells transformed with pART1Skb1, for expression of Skb1 (○), and pART1 (•). S. pombe cells carrying the skb1 deletion exhibit a slower growth rate than skb1+ cells. (F) Wild-type (SP870) S. pombe cells overexpressing Skb1 from the plasmid pREP1. Note that cells are abnormally elongate in morphology, compared with control cells in B.
Microscopic analysis of skb1− cells (Fig. 3C) revealed that they are less elongate in morphology than wild-type S. pombe cells (Fig. 3B). This phenotype is similar to, though substantially less severe than, the morphology defects of shk1−, ras1−, and cdc42− mutants, which are spheroidal in shape. Like ras1 null mutants, skb1 mutants exhibited the most marked degree of morphological aberrancy as cultures approached the later phase of exponential growth. As shown in Fig. 3D, overexpression of shk1 restored elongate morphology to the skb1 null mutant. Overexpression of skb1, on the other hand, did not bypass loss of shk1 (data not shown). We observed further that overexpression of skb1 causes wild-type S. pombe cells to become hyperelongated (Fig. 3F). We conclude from these results that Skb1 participates in the regulation of cell morphology in S. pombe and that it acts upstream of Shk1.
Shk1 and Cdc42, but not Ras1, are essential for cell viability in S. pombe (20, 21, 28, 30). As shown in Fig. 3E, skb1− cells have a slower growth rate than skb1+ cells. On the other hand, skb1 null mutants exhibited no obvious mating or sporulation defects (data not shown), suggesting that Skb1 is not required for sexual differentiation in S. pombe.
Genetic Evidence Linking Skb1 to a Ras1-Dependent Morphology Control Pathway in S. pombe.
S. pombe cells carrying a deletion of the ras1 gene are ellipsoidal in morphology (Fig. 4A). This defect is not markedly suppressed by overexpression of Shk1 (Fig. 4C). However, as shown in Fig. 4D, we observed that overexpression of skb1 does partially restore elongate morphology to a S. pombe ras1− mutant. More significantly, we observed that ras1 mutants that cooverexpressed both skb1 and shk1 (Fig. 4E) are substantially more elongate in morphology than ras1− cells overexpressing skb1 or shk1 alone. Overexpression of skb1 also partially restored elongate morphology to an scd2 deletion mutant (data not shown). However, neither overexpression of skb1 alone or in combination with shk1 suppressed the morphology defects of an scd1 deletion mutant or of an S. pombe strain expressing a dominant inhibitory allele of cdc42, cdc42T17N (data not shown). These results suggest that Skb1 participates in the morphological control branch of the Ras1 signaling cascade at a level downstream from Ras1 and Scd2 and at the same level as Cdc42. In addition, these results are consistent with a role for Skb1 functioning as a positive modulator of Shk1 function.
Figure 4.
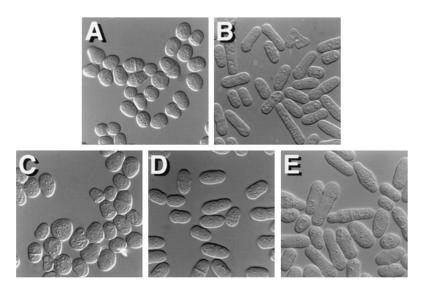
Evidence that Skb1 participates in the Ras1-dependent morphology control pathway in S. pombe. (A–E) Photomicrographs of a S. pombe ras1− mutant, SPRN1, transformed with the control plasmids pAAUCM and pREP1 (A); pALR, which carries the wild-type ras1 gene (B); pAAUCMShk1, for overexpression of Shk1 (C); pREP1Skb1, for overexpression of Skb1 (D); and both pAAUCMShk1 and pREP1Skb1 (E). Overexpression of skb1 partially rescues the morphology defect of ras1-deleted cells. However, ras1− cells overexpressing both Skb1 and Shk1 are substantially more elongate than cells overexpressing Skb1 or Shk1 alone. S. pombe transformants were patched onto EMM and grown overnight at 30°C prior to photomicroscopy.
We performed additional genetic analyses to determine if Skb1 participates in the pheromone-induced MAPK branch of the Ras1 signaling cascade. Overexpression of Skb1, either alone or in combination with Shk1, did not provide detectable suppression of either the conjugation or sporulation defects of S. pombe mutants carrying deletions of ras1, gpa1, or byr2 (data not shown). These results, combined with the fact that skb1− cells lack discernible sexual response defects, suggest that Skb1 function is restricted to the morphological control branch of the Ras1 signaling cascade in S. pombe.
DISCUSSION
We have described the cloning and characterization of the fission yeast skb1 gene, which we identified from a two-hybrid screen for proteins that interact with the Ste20/PAK homolog Shk1. Our genetic analyses indicate that Skb1 is a component of the morphological control branch of the Ras1 signaling cascade in S. pombe and that it positively modulates Shk1 function. Skb1 is not required for either mating or sporulation in S. pombe. Our results are consistent with a role for Skb1 functioning as a Shk1 activator, although we have thus far been unable to prove this hypothesis biochemically using epitope-tagged or glutathione S-transferase fusion proteins. Further insights into the nature of the Skb1–Shk1 interaction may be provided by the identification of additional factors that interact with each protein.
Based on our genetic analyses and two-hybrid experiments, we propose the model for Skb1 function depicted in Fig. 5 in which Skb1 functions downstream from Ras1 and Scd2, at the same level as Cdc42, and upstream of Shk1. Our results are also consistent with an alternative model for Skb1 function in which Skb1 participates in a morphological control pathway that parallels the Ras1 → Scd1 → Cdc42 pathway to Shk1. By identifying additional factors that regulate or physically associate with Skb1, it may become possible to distinguish between these two models for Skb1 function.
Figure 5.
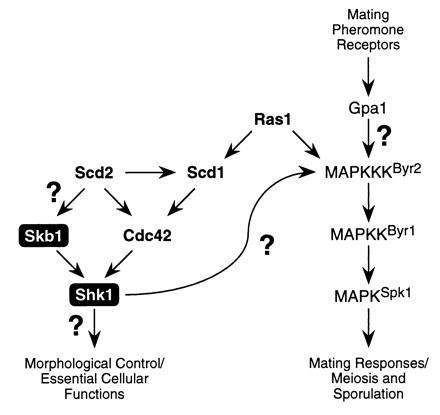
Model for Skb1 function in S. pombe. Our results suggest that Skb1 is a component of the morphological control branch of the Ras1 signaling cascade in S. pombe and that it positively modulates Shk1 function. Genetic analyses suggest that Skb1 acts downstream from Ras1 and Scd2, at the same level as Cdc42, and upstream of Shk1. Question marks denote uncertainties in the Ras1-dependent signaling cascade. Shk1 and Cdc42 are essential for cell viability in S. pombe. See text for details.
Our results indicate that Skb1 is only partially required for function of the Ras1 signaling cascade in S. pombe. Cells carrying a deletion of the skb1 gene exhibit only moderate defects in growth and morphology and no discernible defects in sexual responses. One explanation for this is that Cdc42 provides the dominant input to Shk1, while Skb1 fine-tunes Shk1 function. However, it should be noted that the relative contributions of other components of the S. pombe Ras1 cascade vary significantly (refs. 20, 21, 22, 24, 25, 26, 29, and 30; see Fig. 5). For example, with respect to MAPK functions, while Gpa1, Byr2, Byr1, and Spk1 are each essential for sporulation, Ras1 function is partially dispensable. On the other hand, Scd1 and Scd2 are not required for sporulation at all, even though each is essential for mating. With respect to morphological functions, ras1− cells are ellipsoid in morphology, whereas scd1− and shk1− mutants are much more spherical. scd2− mutants more closely resemble skb1− cells in morphology (stumpy, rather than ellipsoidal). With respect to cell growth, ras1−, scd1−, and scd2− cells do not exhibit obvious growth defects, whereas skb1− cells grow more slowly than wild-type cells and cdc42 and shk1 are each essential for cell viability. These and numerous other observations may provide an indication that the interactions among components of the Ras1 signaling cascade in S. pombe are substantially more complex than those depicted in Fig. 5. It is also possible, if not likely, that additional components of the cascade remain to be identified.
Searches of computer data bases revealed that Skb1 is homologous in structure to proteins encoded by ORFs in the genomes of S. cerevisiae and C. elegans and by an uncharacterized human cDNA sequence. It will be of great interest to determine whether these proteins are also functionally related. Preliminary characterization of the S. cerevisiae Skb1 homolog, YBR1008, suggests that this may turn out to be the case. In collaborative studies, we have determined that null mutations in the YBR1008 gene cause readily discernible changes in cell morphology (C. Inouye, S.M., M. Shulewitz, P.Y., M.G., and J. Thorner, unpublished results). This phenotype resembles that of S. cerevisiae mutants carrying a null mutation of the PAK-encoding gene CLA4 (34). Thus, skb1 may be the first well-characterized member of a highly conserved family of genes encoding potential PAK regulators. Proteins like Skb1 could potentially function to impart functional specificity to PAKs—a specificity that might not be attained by interaction with Cdc42 and/or Rac proteins alone. The identification of novel PAK regulators such as Skb1 should provide additional insights into the function and regulation of PAKs in both yeasts and in higher organisms.
Acknowledgments
We thank Maureen Barr, Heithem El-Hodiri, Karen Marcus, William Mattox, Erin Mooney, Aaron Neiman, and Ruth Pimental for critical comments and suggestions on the manuscript. DNA sequencing was performed by the University of Texas M.D. Anderson Cancer Center Core DNA Sequencing Facility, which is supported by National Institutes of Health (NIH) Grant P30CA16672. This work was supported in part by NIH grants to M.H.C. (R01GM53032) and S.M. (R01GM53239). M.G. is supported by an NIH predoctoral training grant.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
Abbreviations: GAD, Gal4 transcriptional activation domain; GBD, Gal4 DNA binding domain; LBD, LexA DNA-binding domain; MAPK, mitogen-activated protein kinase; MAPKKK, MAPK kinase kinase; β-gal, β-galactosidase; PAK, p21Cdc42/Rac-activated kinase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank data base (accession no. U59684U59684).
References
- 1.Lowy D R, Willumsen B M. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 2.Avruch J, Zhang X F, Kyriakis J M. Trends Biochem Sci. 1994;19:279–283. doi: 10.1016/0968-0004(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 3.Kikuchi A, Demo S D, Ye Z H, Chen Y W, Williams L T. Mol Cell Biol. 1994;14:7483–7491. doi: 10.1128/mcb.14.11.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez-Viciana P, Warne P H, Dhand R, Vanhaesebroeck B, Gout I, Fry M J, Waterfield M D, Downward J. Nature (London) 1995;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 5.Russell M, Lange-Carter C-A, Johnson G L. J Biol Chem. 1995;270:11757–11760. doi: 10.1074/jbc.270.20.11757. [DOI] [PubMed] [Google Scholar]
- 6.Wood K W, Sarnecki C, Roberts T M, Blenis J. Cell. 1992;68:1041–1050. doi: 10.1016/0092-8674(92)90076-o. [DOI] [PubMed] [Google Scholar]
- 7.Porras A, Muszynski K, Rapp U R, Santos E. J Biol Chem. 1994;17:12741–12748. [PubMed] [Google Scholar]
- 8.White M A, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler M H. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 9.Prendergast G C, Gibbs J B. Adv Cancer Res. 1993;62:19–65. doi: 10.1016/s0065-230x(08)60314-0. [DOI] [PubMed] [Google Scholar]
- 10.Joneson T, White M A, Wigler M H, Bar-Sagi D. Science. 1996;271:810–812. doi: 10.1126/science.271.5250.810. [DOI] [PubMed] [Google Scholar]
- 11.Chant J, Stowers L. Cell. 1995;81:1–4. doi: 10.1016/0092-8674(95)90363-1. [DOI] [PubMed] [Google Scholar]
- 12.Leung T, Manser E, Tan L, Lim L. J Biol Chem. 1995;270:29051–29054. doi: 10.1074/jbc.270.49.29051. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe G, Saito Y, Madaule P, Ishizaki T, Fujisawa K, Morii N, Mukai H, Ono Y, Kakizuka A, Narumiya S. Science. 1996;271:645–648. doi: 10.1126/science.271.5249.645. [DOI] [PubMed] [Google Scholar]
- 14.Manser E, Leung T, Salihuddin H, Zhao Z-S, Lim L. Nature (London) 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 15.Martin G A, Bollag G, McCormick F, Abo A. EMBO J. 1995;14:1970–1978. doi: 10.1002/j.1460-2075.1995.tb07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagrodia S, Taylor S J, Creasy C L, Chernoff J, Cerione R A. J Biol Chem. 1995;270:22731–22737. doi: 10.1074/jbc.270.39.22731. [DOI] [PubMed] [Google Scholar]
- 17.Neiman A M, Stevenson B J, Xu H-P, Sprague G F, Jr, Herskowitz I, Wigler M, Marcus S. Mol Biol Cell. 1993;4:107–120. doi: 10.1091/mbc.4.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. Proc Natl Acad Sci USA. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuda T, Kariya K-I, Shinkai M, Okada T, Kataoka T. J Biol Chem. 1995;270:1979–1982. doi: 10.1074/jbc.270.5.1979. [DOI] [PubMed] [Google Scholar]
- 20.Fukui Y, Kozasa T, Kaziro Y, Takeda T, Yamamoto M. Cell. 1986;44:329–336. doi: 10.1016/0092-8674(86)90767-1. [DOI] [PubMed] [Google Scholar]
- 21.Nadin-Davis S A, Nasim A, Beach D. EMBO J. 1986;5:2963–2971. doi: 10.1002/j.1460-2075.1986.tb04593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadin-Davis S A, Nasim A. EMBO J. 1988;7:985–993. doi: 10.1002/j.1460-2075.1988.tb02905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitamura K, Shimoda C. EMBO J. 1991;10:3743–3751. doi: 10.1002/j.1460-2075.1991.tb04943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obara T, Nakafuku M, Yamamoto M, Kaziro Y. Proc Natl Acad Sci USA. 1991;88:5877–5881. doi: 10.1073/pnas.88.13.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toda T, Shimanuki M, Yanagida M. Genes Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Xu H-P, Riggs M, Rodgers L, Wigler M. Mol Cell Biol. 1991;11:3554–3563. doi: 10.1128/mcb.11.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka K, Davey J, Imai Y, Yamamoto M. Mol Cell Biol. 1993;13:80–88. doi: 10.1128/mcb.13.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller P J, Johnson D I. Mol Cell Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang E, Barr M, Wang Y, Jung V, Xu H-P, Wigler M. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 30.Marcus S, Polverino A, Chang E, Cobb M H, Wigler M. Proc Natl Acad Sci USA. 1995;92:6180–6184. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leberer E, Dignard D, Harcus D, Thomas D Y, Whiteway M. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramer S W, Davis R. Proc Natl Acad Sci USA. 1993;90:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, Styles C A, Fink G R. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 34.Cvrcková F, Virgilio C D, Manser E, Pringle J R, Nasmyth K. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- 35.Xu H-P, Rajavashisth T, Grewal N, Jung V, Riggs M, Rodgers L, Wigler M. Mol Biol Cell. 1992;3:721–734. doi: 10.1091/mbc.3.7.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feilotter H E, Hannon G J, Ruddell C J, Beach D. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 38.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. [Google Scholar]
- 39.Vojtek A B, Hollenberg S M, Cooper J A. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 40.Marcus S, Barr M, Polverino A, Wigler M. Proc Natl Acad Sci USA. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCleod M, Stein M, Beach D. EMBO J. 1987;6:729–736. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maundrell K. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 43.Wright A, Maundrell K, Heyer W-D, Beach D, Nurse P. Plasmid. 1986;15:156–158. doi: 10.1016/0147-619x(86)90051-x. [DOI] [PubMed] [Google Scholar]


