Abstract
Motility is achieved in most bacterial species by the flagellar apparatus. It consists of dozens of different proteins with thousands of individual subunits. The published literature about bacterial chemotaxis and flagella documented 51 protein–protein interactions (PPIs) so far. We have screened whole genome two-hybrid arrays of Treponema pallidum and Campylobacter jejuni for PPIs involving known flagellar proteins and recovered 176 and 140 high-confidence interactions involving 110 and 133 proteins, respectively. To explore the biological relevance of these interactions, we tested an Escherichia coli gene deletion array for motility defects (using swarming assays) and found 159 gene deletion strains to have reduced or no motility. Comparing our interaction data with motility phenotypes from E. coli, Bacillus subtilis, and Helicobacter pylori, we found 23 hitherto uncharacterized proteins involved in motility. Integration of phylogenetic information with our interaction and phenotyping data reveals a conserved core of motility proteins, which appear to have recruited many additional species-specific components over time. Our interaction data also predict 18 110 interactions for 64 flagellated bacteria.
Keywords: flagellum, motility, protein–protein interactions, swarming assay, yeast two-hybrid
Introduction
Motility in most bacterial species depends on a sophisticated molecular machine called the flagellum. The flagellar apparatus is made of dozens of different proteins and thousands of individual subunits. The bacterial flagellum is actually a mechanical nanomachine with a rotation frequency of 300 Hz, an energy conversion rate of nearly 100%, and the ability to self assemble (Macnab, 1999, 2003; Berg, 2003; Kojima and Blair, 2004).
Various efforts have been made to identify all components required for bacterial motility, resulting in a list of more than 60 proteins in Escherichia coli (Supplementary Table S1, Kanehisa et al, 2004). Functionally, these proteins can be subdivided into several subsets: the chemotaxis system connects environmental stimuli to the direction of flagellar rotation and thus direction of movements. The chemotaxis system is connected to the basal body complex, which anchors the flagellum in the inner membrane and also incorporates a type-III-secretion system necessary for the self-assembly process of the flagellum. Two motor proteins, MotA and MotB, convert an ion gradient (for most bacteria a proton gradient) into rotational energy of basal body components; these components are connected to the rod structure and then via a flexible hook to the filament of the flagellum, which operates like a propeller. However, whereas the overall structure has been known for decades, many of the mechanistic details responsible for the assembly and operation of the motor have yet to be worked out. In fact, it remains unclear whether all the protein components of the flagellar apparatus have been identified. Similarly, whereas at least 51 protein–protein interactions (PPIs) have been described in the literature (Supplementary Table S2), many more interactions are likely to be required for assembly and proper operation.
Despite the vast body of literature about bacterial motility, there have been few systematic attempts to identify the components of the flagellar apparatus and their function besides genome sequencing. Systematic analysis of hundreds of completely sequenced genomes, for example, has predicted many additional motility genes based on their location in flagellar operons or gene clusters, yet their actual roles in motility often remain unknown.
In this study, we systematically identified genes essential for bacterial motility by testing the swarming capability of 3985 gene deletion strains of E. coli (Baba et al, 2006). In addition, we integrated data from similar screens carried out for Bacillus subtilis (Schumann et al, 2001) and mutant screens of Campylobacter jejuni (Golden et al, 2000; Hendrixson et al, 2001) and Helicobacter pylori (Salama et al, 2004).
Second, we screened all motility proteins recovered from the literature for PPIs. We reasoned that unknown motility proteins can be discovered by interactions with known flagellar and chemotaxis components. Protein interactions were identified by screening the proteomes of two small distantly related bacteria, Treponema pallidum and C. jejuni, using comprehensive array-based yeast-two-hybrid screens (Uetz et al, 2000; Parrish et al, in press). In addition, we compared our data to protein interaction data of H. pylori (Rain et al, 2001) and E. coli (Arifuzzaman et al, 2006). Finally, we integrated these experimental data sets with predictions of functional associations from the STRING database (von Mering et al, 2003; Stein et al, 2005). The result is a list of known and new flagellar components, including 23 novel motility proteins (Figure 1 and Table I).
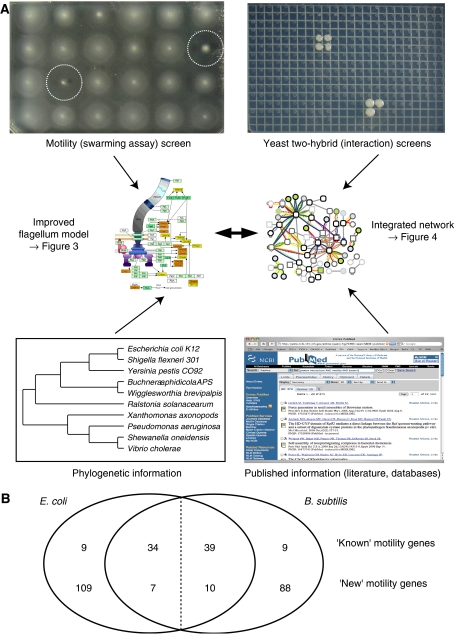
Figure 1.
Strategy used in this study. (A) Motility genes were studied by screening 3985 E. coli mutants for motility defects (24 are shown in the top left panel) and by screening motility proteins for protein interactions (here: bait FlaB1 from T. pallidum returning preys FlaB1 and FliS). These data were integrated with literature interactions and large-scale interaction data sets to refine the current picture of the bacterial flagellum. (B) Comparison of motility genes identified in E. coli (this study) and B. subtilis (Schumann et al, 2001). There are 43 ‘known' motility genes reported in the literature for E. coli, 34 of which have orthologs in B. subtilis. Our mutant screens identified 116 additional ‘new' motility genes of which only 7 have orthologs in B. subtilis. Equivalent numbers are given for B. subtilis (individual genes are listed in Supplementary Tables S1 and S3).
Table 1.
Novel motility proteins
| COG | Gene locus (ORF) | Mutant gene (ortholog) | Interacting proteins | Motility | Notes [Refs] |
|---|---|---|---|---|---|
| COG1463 | CJ1648 HP1464 | — | MotB | Rain et al (2001) | |
| COG0082 | CJ1634C | aroC (ECO) | FlhA, FlgE2, FliE, FlgK, FliD, FlgB | −/+a | See Figure 4 |
| COG0055 | B3732 | atpD (ECO) | CheZ,CheW, Tsr (E. coli) | −/+a | Arifuzzaman et al (2006) |
| COG2194 | CJ0256 | CJ0256, yjgX (ECO) | FlgG-2 | −a | Golden et al (2000) |
| COG0642 | CJ1222c | CJ1222c (CJE) | FliN | Coloniz.b ↓ MacKichan et al (2004) | |
| COG2604 | CJ1318 | CJ1318 (CJE) | FliN, FliQ | − | Golden et al (2000) |
| None | HP0488 | CJ1340c (CJE) | FlgE, FlgB | − | FlhA+FlhFc Rain et al (2001); Niehus et al (2004) |
| COG1923 | B4172 | hfq (ECO) | FliS, FliL | −/+a | Arifuzzaman et al (2006) |
| COG0455 | TP0712 (CJ0063c) | HP1034 (HPY) | TP0464 (flhF [CJE]) | − | van Amsterdam and van der Ende (2004) |
| COG0457 | HP1479 | HP1479 (HPY) | FlgB | − | Rain et al (2001); Salama et al (2004) |
| COG0673 | CJ0504c | idh (BSU) | FlaG | − | Schumann et al (2001) |
| COG0791 | Cj1653c | lytF (BSU) | FliL | − | Schumann et al (2001) |
| None | CJ0055c | rapG (BSU), HP1479 (HPY) | FliM | − | Schumann et al (2001); Salama et al (2004) |
| COG0267 | TP0209 | rpmJ (ECO) | FliG-2, CheR, FliY, FlgE, FliS | −/+a | |
| COG1774 | TP0046 | yaaT (BSU) | CheR,FlgD, MetK, FlaB3, TP0959, CheW-2,Mcp2-3 | −/+ | Schumann et al (2001) |
| COG0084 | TP0979 | ycfH (tatD) (ECO) | FliE | ++a | See Figure 5 |
| COG0346 | CJ1301 | ydfO (BSU) | FliM, FliY | − | Schumann et al (2001) |
| COG1512 | TP0561 | ydjH (BSU) | SigG, FliF, FlhB, FliR, FliQ, FliL | −/+a | |
| COG1664 | TP0048 | yhbE yhbF (BSU) | FliY, FliS | −/+a | See Figure 4B |
| COG2001 | TP0383 | yllB (BSU) | FliS, FlgK | −/+a | Kobayashi et al (2003) |
| COG3334 | CJ1496c | ylxF (BSU) | FliL | − | Operon, domaind Schumann et al (2001) |
| COG3391 | TP0421 | yncE (ECO) | TP0567 | −/+a | |
| COG1699 | TP0658 | yviF (BSU) | FlaB2, FlaB1, FlaB3 | −a | Titz et al (2006) |
Conserved hypothetical proteins in the columns ‘Gene locus (ORF)' and ‘Mutant gene (ortholog)' are predicted to be novel flagellar components, based on their physical interactions with known motility proteins (in T. pallidum, C. jejuni, E. coli, or H. pylori) and their motility phenotype (in E. coli (ECO), H. pylori (HPY), C. jejuni (CJE), or B. subtilis (BSU)). The Motility phenotype refers to the Mutant gene (ortholog): reduced motility ‘−/+', nonmotile ‘−‘, and increased motility ‘++'. Entries are sorted by ‘Mutant gene'.
aThis study (Motility phenotype).
bDiminished capacity to colonize.
cRegulated by FlhA and FlhF.
dFlagellum operon, extracellular sensory domain.
Many features of the bacterial flagellum have changed over the course of evolution. This is reflected in the surprisingly different composition and protein interaction patterns in the flagella of different species, which may reflect adaptations to species-specific motility needs (compare Figure 2 and Supplementary Figure S2). While the overall conservation allows us to predict ∼18 000 interactions for 64 proteomes of flagellated bacteria, it remains to be seen how many of them are functional.
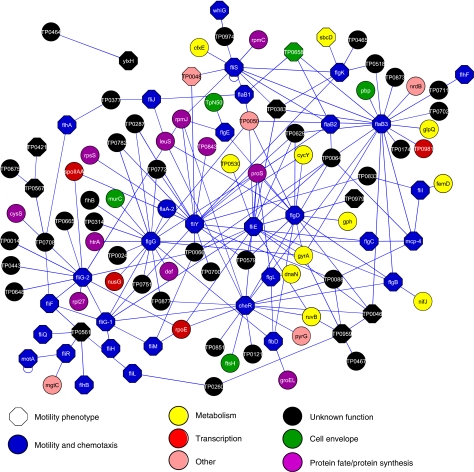
Figure 2.
The motility protein interaction network of T. pallidum. Blue nodes are known motility proteins, whereas black proteins are proteins of yet unknown function. Proteins with a motility phenotype in either E. coli or B. subtilis or H. pylori are indicated as octagons. See legend for other functional assignments. The motility network of C. jejuni is shown in Supplementary Figure S2.
Results
Genes important for bacterial motility
Several systematic mutant screens have been performed to find genes involved in bacterial motility (Hendrixson et al, 2001; Schumann et al, 2001; Golden and Acheson, 2002; Inoue et al, 2007). To generate a comprehensive motility mutant data set for the gram-negative model bacterium E. coli, we have used the gene deletion library constructed by Baba et al (Baba et al, 2006). These mutants were plated out in arrays of 24 colonies on swarming agar and tested for swarming (Figure 1). Of 3985 mutants tested 159 deletions showed a swarming defect (Supplementary Table S3a).
Interestingly, a similar screen in B. subtilis yielded a similar number of 146 motility mutants (Schumann et al, 2001) (Supplementary Tables S3b and 3f). Thus about 4% of the nonessential genes in both species show an effect on motility under the conditions tested. Among them are 43 (30%) and 48 (27%) genes previously annotated as bona fide motility genes in E. coli and B. subtilis, respectively (Figure 1B). The other mutants with motility phenotypes are significantly enriched for proteins involved in ‘motor activity' and ‘macromolecule metabolism' (Supplementary Figure S1). Many of these genes may be required to provide energy to the flagellar motor or may be indirectly involved in the assembly of the flagellar apparatus, for example in restructuring the peptidoglycan to allow penetration of the cell wall.
Unexpectedly, only 7 (10) of the E. coli (B. subtilis) mutant genes that were previously not known to have a motility function, had a homolog with a phenotype in B. subtilis (E. coli) (Figure 1B and Supplementary Table S3g). Thus, there appear to be many proteins with a species-specific role in motility. Examples of such proteins are discussed further below.
Protein interactions of bacterial motility proteins
Decades of research have identified many components of bacterial flagella and their motors (Supplementary Table S1). We have used most of these motility proteins in two-hybrid screens in both T. pallidum and C. jejuni. All known T. pallidum motility proteins were tested as fusions to the Gal4-DNA binding domain (baits) in a systematic array-based yeast-two-hybrid screen against a whole genome prey library (i.e. fusions with the Gal4 activation domain) of T. pallidum. These screens identified 176 PPIs for T. pallidum (TPA, Figures 2 and 3 and Supplementary Table S4a). Similarly, the C. jejuni motility proteins were tested for interactions with most of the C. jejuni proteins in systematic LexA-based Y2H screens (Parrish et al, in press), and a comparable number of 140 high-confidence interactions (CJE HCF) was found among 690 total interactions (CJE All) (Supplementary Figures S2 and S3 and Table S4b-c).
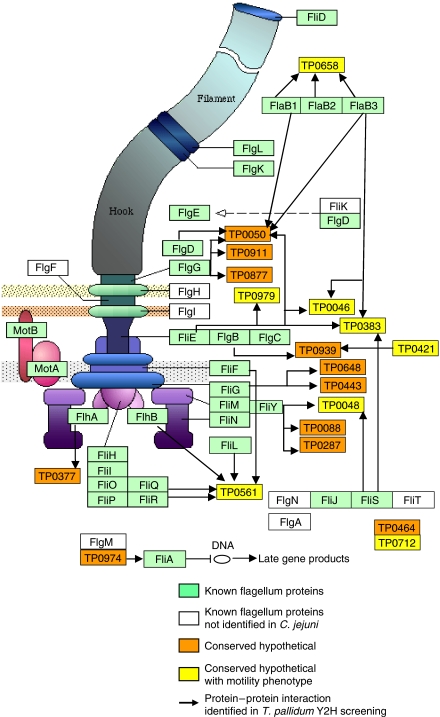
Figure 3.
New motility proteins in the context of the T. pallidum flagellar apparatus. Model of the bacterial flagellar apparatus (based on KEGG; Kanehisa et al, 2006) with putative new motility proteins discovered in this study (yellow and orange). New motility proteins of C. jejuni are shown in Supplementary Figure S3. Compare to Figures 2 and 4.
Additional motility protein interactions were filtered from the Y2H interaction map of H. pylori (HPY, Rain et al, 2001), and from a complex purification study of E. coli (Arifuzzaman et al, 2006) (ECO SPK, ECO SAI, see Materials and methods) (Supplementary Table S2b-d).
Pairwise comparisons of these various interaction data sets revealed only a limited overlap ranging from 2.5% for the E. coli (ECO SPK) versus the Helicobacter data to 25.0% for Helicobacter versus CJE HCF (Supplementary Table S5). Overall, ECO SPK has the weakest pairwise similarities. Thus the overlap between the different data sets appears to reflect both phylogenetic relationships as well as methodological differences between yeast two-hybrid and complex purification data sets.
As might be expected, interactions between motility proteins are common in the motility interaction maps (Figure 2 and Supplementary Figures S1 and S2). An overview of the number of proteins (nodes) and their interactions (edges) and additional properties of these networks can be found in Supplementary Table S6.
Finally, for a comparative analysis of motility interactions, we carried out a comprehensive review of the literature for published flagellar PPIs using PubMed and found 51 unique interactions (Supplementary Table S2a). Of these 51, 39 had interologs in T. pallidum and 38 in CJE ALL, but only 9 and 5 were reproduced in T. pallidum and CJE ALL, respectively (Supplementary Tables S2 and S5a). Only one interaction is common to both T. pallidum and CJE ALL screens, and thus a total of 13 interactions were recovered in either of our screens. That is, sampling of the two species recovered 33% of all published flagellar interactions. One reason for this relatively low coverage may be that most previous studies used different methods that may be better applicable to flagellar proteins. All literature comparisons can be found in Supplementary Table S5.
An integrated view of the flagellum
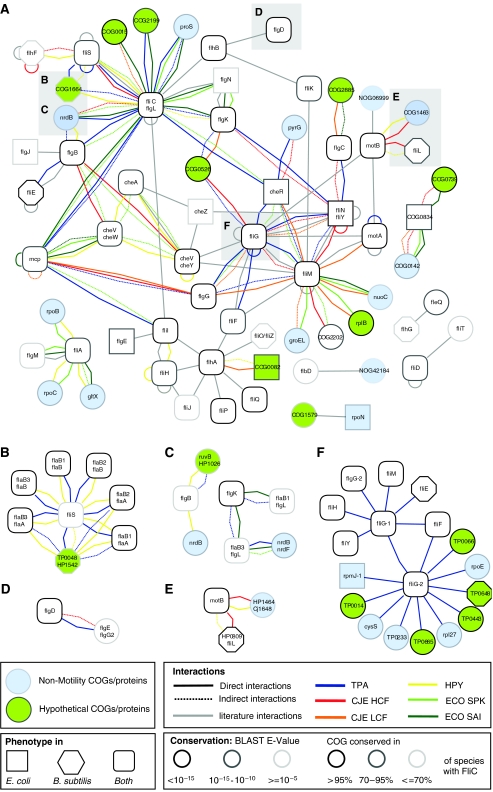
The diversity of information on different genomes, proteins, phenotypes and so on makes it difficult to keep track of all details. Therefore, we generated an integrated motility network, which combines a diverse set of interaction networks as well as phylogenetic and phenotyping data (Figure 4). This network combines protein–protein interactions of T. pallidum, C. jejuni, H. pylori, and E. coli as well as interactions curated from the literature with motility phenotyping data from E. coli and B. subtilis. It also displays clusters of orthologous groups (COGs) rather than individual proteins, which reduces complexity and improves the quality of links. These links represent direct interactions, indirect interactions (if proteins do not interact directly, but via a bridging protein), and literature interactions. Out of all interactions, 73% connect known motility COGs. In addition, 45% were predicted by STRING (highest confidence: S>0.9) to be strongly associated. These numbers indicate that this integrated network is more reliable and biologically relevant than individual networks. In addition, links among orthologous groups can usually be transferred to proteins of other species. However, because of the stringent filtering not all interactions are included in this network.
Figure 4.
Integrated motility network. Multispecies summary of interactions among motility proteins. (A) Homologous protein nodes of pairwise aligned networks were merged into orthologous groups (COGs). Nodes were labelled according to KEGG descriptions (Kanehisa et al, 2006); their shape corresponds to motility phenotypes in E. coli (squares), B. subtilis (octagons), or both (rounded squares). Edges represent either direct, indirect, or literature interactions among orthologous groups and are color-coded (see legend in Figure). The border shading of a COG nodes indicates its conservation ratio among 68 flagellated bacteria. (B–E) Aligned protein interactions showing COG interactions from (A) in more detail: each node represents two homologous proteins from either T. pallidum (TPA), C. jejuni (CJE), H. pylori (HPY), or E. coli (ECO). Edges represent either direct or indirect interactions and are color-coded (see legend in Figure). The border color of a protein node indicates the BLAST E-value of its homologous protein pair. (F) Interactions of paralogous FliG1 and FliG2 proteins taken from T. pallidum network.
Several insights into the internal organization of the bacterial flagellum can be obtained. For example, the aligned network shows that the flagellum filament protein, FliC, and its homolog FlgL, a hook-associated protein, are members of the same COG. FlgL is connected to the second hook-associated protein FlgK and both are stabilized by their export chaperone, FlgN. The interaction of FliC with its chaperone FliS is conserved in all species. The basal body complex with FliN/FliY, FliG, FliM, and FliF, forms another cluster, which is connected to the motor proteins, MotA and MotB, and to rod proteins such as FlgC and FlgG.
The chemotaxis protein cluster in the network is only connected to the flagellum switch complex. The interaction of CheY with FliM depends on CheY's phosphorylation, which is not detected in our yeast two-hybrid assays, because we do not coexpress the pertinent kinase CheA. Nevertheless, the integrated network reflects the fact that external signals are detected by homodimerizing methyl-accepting chemoreceptors (Mcps), which are linked by an adapter protein, CheW, to the kinase CheA, which transfers the phosphate group to CheY. By its interaction with the basal body complex, phosphorylated CheW controls the rotation state of the flagellum. In addition to these previously known interactions, we find conserved links between chemotaxis proteins and rod proteins such as FlgB and FlgG, which are difficult to explain by the standard model of the flagellum, but allow for interesting speculations about the organization of chemotaxis signalling in the cytoplasm.
Another striking connection is the conserved MotB–FliL interaction in C. jejuni and Helicobacter. For Proteus mirabilis, FliL is thought to be involved in sensing of the actual flagellum status (Belas and Suvanasuthi, 2005). Here, we found evidence that this sensing is mediated by a direct interaction with the motor apparatus (Figure 4E).
Discussion
New bona fide motility proteins
A major goal of this study was to find novel flagellar components among the many proteins of still unknown function. In addition, we suspected that there must be previously characterized proteins whose role in motility remained unknown. Indeed, 28% and 33% of the interactions found in T. pallidum and C. jejuni, respectively, connected a known motility protein to a conserved hypothetical protein (Supplementary Table S4), suggesting that there are still unidentified proteins with a motility function. To identify potential novel bona fide motility proteins, we used our integrated data and identified 23 hitherto uncharacterized proteins (Table I).
For example, members of the orthologous group COG1664, such as TP0048 and HP1542 (Figure 4B), show interactions to the FliC–FliS cluster. Additional evidence for a role in motility comes from the double mutant of the B. subtilis orthologs yhbE and yhbF, which also shows reduced motility.
TP0658 (yviF in B. subtilis), a previously uncharacterized protein, was found to interact with all three flagellin proteins (FlaB1-B3) of T. pallidum. The deletion mutants of TP0658 orthologs in both B. subtilis (yviF) and C. jejuni (CJ1075) show a highly reduced motility phenotype (Golden et al, 2000; Titz et al, 2006). We have recently shown that TP0658 and yviF appear to stabilize flagellin in the cytoplasm, thus exhibiting properties of a chaperone (Titz et al, 2006). TP0658 and its orthologs thus appear to be flagellar assembly factors or factors involved in export of the filament protein FliC.
TP0561 is another hitherto uncharacterized protein that appears to be involved in flagellar protein export based on its interaction pattern; it interacts with multiple components of the export machinery such as FliR, FliL, FliQ, and FlhB (Figure 3, bottom left). In addition, a mutation in TP0561 results in significantly reduced motility.
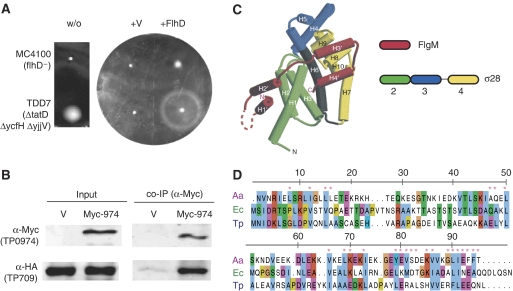
Due to its motility phenotype in E. coli TatD is an exceptional case. Its Treponema homolog TP0979 interacted with FliE. In E. coli, tatD has two paralogs, ycfH and yjjV, which are functionally unassigned, genomically unlinked, and show 29% and 24% amino-acid sequence identity to TatD, respectively. As tatD is localized in an operon with genes of the twin-arginine transport (Tat) system, a transport function of TatD was anticipated. But even a strain with all three TatD paralogs deleted did not show a Tat-related transport deficiency, leaving the question for TatD's function unanswered (Wexler et al, 2000). As we found a very small, but significant, increase in motility for the ycfH single mutant, we tested the previously described triple mutant (all TatD paralogs deleted) for motility (Figure 5A). The triple mutant was constructed in a MC4100 strain background, a strain known to be nonmotile, presumably due to a point mutation in FlhD, a known master regulator for motility (Wexler et al, 2000). Unexpectedly, the triple mutant showed a slight rescue of motility. We investigated the relation between the FlhD point mutation and the TatD paralogs by expressing a functional FlhD construct both in the MC4100 strain and the triple mutant. A strong synergistic effect of FlhD expression and the triple mutation on motility was observed pointing to a regulatory antagonism of FlhD and TatD paralogs (Figure 5A). These findings indicate that TatD and/or its orthologs (COG0084) have a negative role in motility, perhaps mediated by its DNAse activity (Wexler et al, 2000).
Figure 5.
Lessons for individual proteins. (A) E. coli tatD mutants were tested in a swarming assay. The E. coli strain MC4100 (flhD) and a tatD triple mutant (TDD7, all TatD paralogs deleted, i.e., tatD, ycfH and yjjV in a MC4100 background) were left untransformed (w/o) or transformed with an empty expression vector (+V) or the same vector expressing FlhD (+flhD). The tatD triple-mutant was much more motile than the parental strain. (B) TP0974 is the T. pallidum FlgM protein. Myc-tagged TP0974 (Myc-974) or a vector control (V) was co-immunoprecipitated with HA-TP0709 (sigma factor). Note that the results from (A) and (B) are mentioned in the discussion, not results section. (C) Crystal structure of the Aquifex aeolicus σ28/FlgM Complex (Sorenson et al, 2004). The domain architecture and color-coding are illustrated schematically. FlgM is colored red. (D) The homolog of FlgM in T. pallidum (TP0974) aligned to FlgM from A. aeolicus and E. coli using ClustalW (Chenna et al, 2003) (Ae=A. aeolicus, Ec=E. coli, Tp=T. pallidum). The colors of amino acids denote chemical similarity. Numbering is according to the A. aeolicus sequence. A pink asterisk indicates residues mediating the interaction of FlgM with σ28 (showing the conservation of the FlgM interaction epitope). The putative C. jejuni FlgM homolog CJ1464 was left out as its function is still unclear.
A few proteins were previously annotated as having functions unrelated to motility, but appear to be involved in flagellar function based on their motility phenotype and their interactions with other motility proteins. Among these proteins, ribosomal protein RpmJ/L36 (TP0209) is an unexpected case, because it not only interacted with multiple flagellar proteins, but also showed a reduction of motility to about 14% of the wild type when mutated. It is possible that RpmJ links protein synthesis to the flagellar protein export machinery. Another one, Rpe (TP0945) is a ribulose-phosphate 3-epimerase. Surprisingly, this protein also showed a strong reduction in motility when deleted. Rpe could have a role in providing energy for the flagellar motor.
Novel interactions and functions of known motility proteins
Overall, most parts of the flagellum are well conserved in motile bacteria. Nevertheless, evolutionary adaptation of several components can clearly be identified and range from the duplication of proteins, for example of flagellins, to the complete loss or gain of components, for example of export chaperones (Pallen and Matzke, 2006). Here, we will give three examples for such evolutionary processes on the interaction level.
Generation of rotational asymmetry by FliG paralogs
The flagellum of spirochetes possesses several unique features not found in other bacterial species. One unique feature, for example, is the periplasmic localization of two polar flagellum bundles, which perform an asymmetrical rotation (Charon et al, 1992; Li et al, 2000). Interestingly, the molecular basis of this asymmetry is unknown. One hypothesis states that chemotaxis plays an essential role in this asymmetry (Berg and Anderson, 1973; Berg, 1975; Armitage, 1999). Another hypothesis assumes that the motor complexes at both cell poles are somehow differently organized. One candidate that might lead to this asymmetry is FliG, as it is the only duplicated basal body complex protein in spirochetes. The paralogs of FliG in T. pallidum are named FliG-1 (TP0026) and FliG-2 (TP0400). Strikingly, despite significant protein sequence identity (∼30%), both proteins show a very different interaction pattern in our Y2H study (Figure 4F): both proteins interact with each other and with FliF, the flagellum rotor protein. In addition, FliG-1 interacts with FliY, FliM, FliH, and FliE, whereas FliG-2 interacts with TP0014, TP0066, TP0665, TP0443, and TP0648 (all proteins of unknown function), and an anti-sigma factor (TP0233). Although the differential interaction patterns do not clearly explain the asymmetric behavior, they suggest a possible basis for this asymmetry, for example, recruitment of different proteins to the two motor complexes at both ends of the cell. However, it remains to be shown that the two FliG variants are differentially localized and indeed have different effects on motor activity.
Sigma factor and anti-sigma factor interactions
During flagellar assembly in E. coli and Salmonella, the expression of the late flagellar genes is timed by the anti-sigma factor FlgM. At the beginning of assembly, FlgM binds to its sigma factor FliA/σ28 and blocks its function (Figure 5C). Later, the assembled export machinery of the basal body exports FlgM, releases its inhibition of the sigma factor, and the sigma factor becomes free to activate transcription of the late flagellar genes. Initially, no copy of FlgM was identified in the genome of T. pallidum and C. jejuni, which might have implied that the timing of flagellum assembly is differently regulated in these species. Recently, Pallen et al (2005) computationally identified remote FlgM homologs in both species: TP0974 for T. pallidum and CJ1464 in C. jejuni. TP0974 has 23% and 24% sequence identity to its homologs in E. coli and B. subtilis, respectively, and both alignments require multiple gaps, despite its short sequence (around 90 amino acids) (Figure 5D). In our study, we experimentally confirmed the interaction of both putative FlgM homologs with their respective sigma factors: TP0974 interacts with the sigma-factor TP0709, which was confirmed by co-immunoprecipitation (Figure 5B), and CJ1464 interacts with the sigma factor CJ0061c (FliA, σ28). Thus, we provide experimental evidence that FlgM (and its function) is conserved in T. pallidum and C. jejuni. However, it should be noted that flagellin transcription in C. jejuni requires both σ54 and σ28 and that CJ1464 does not appear to have a strong influence on σ28-dependent transcription of flagellin (Hendrixson and DiRita, 2003). Thus, whereas TP0974 may function as an anti-sigma factor, CJ1464 may not. This example shows that large-scale interaction data sets can provide useful initial experimental evidence for functional predictions whose mechanistic details, however, have to be worked out by additional experiments.
Chaperones
Interestingly, two other proteins in the same operon as CJ1464, FlgK (CJ1466) and the remote homolog of FlgN (CJ1465), a chaperone of FlgK, interact with each other, suggesting that CJ1465 likely functions as an FlgK-specific chaperone in C. jejuni.
As judged by the differential phylogenetic distributions of different export chaperones (Pallen et al, 2005), different functions and substrate specificities are likely to be found in different bacterial species. FliS is thought to be a flagellin-specific chaperone (Ozin et al, 2003), whereas FlgN and FliT are substrate-specific flagellar chaperones that prevent oligomerization of the hook-associated proteins, or HAPs, in S. typhimurium. Interestingly, FlgN and FliT orthologs are not present in spirochetes (Pallen et al, 2005). The interactions between FliS and FlgK and between FliS and FlgB are the first experimental evidence that FliS may also function as a chaperone for FlgB and FlgK in spirochetes, partly substituting for FlgN and FliT.
Connections between motility and other functional classes
Whereas the flagellar apparatus is a well-defined nanomachine, it does not act in isolation. Besides the obvious link to the chemotaxis pathway, we noticed several interactions with proteins of other function (Supplementary Figure 1). For example, a link of motility proteins with ‘nucleobase, nucleoside, nucleotide, and nucleic acid metabolism' (GO:0006139) is found in both E. coli and T. pallidum interaction sets, as well as the E. coli mutant phenotyping data. NrdB (ribonucleoside-diphosphate reductase), the key enzyme for the conversion of ribonucleosides into desoxy-ribonucleosides, interacts with two flagellar proteins, FliC and FlgB (Figure 4C). A functional link of NrdB to motility is provided by a study by Nishimura and Hirota (1989), where the authors found a reduction in flagellar protein expression upon nrdB deletion. Although the authors assumed a link on the transcriptional level, an additional post-translational link, as indicated by a direct protein interaction, becomes likely.
The functional link between electron transport (electron transport chain) and motility via a proton gradient (or sodium gradient) is well known and also reflected by an association with ‘transport' (GO:0006810) in a motility gene expression data set (FlhD) (Pruss et al, 2003). We found a direct interaction between NuoC, (NADH dehydrogenase I) and FliM both in the E. coli and C. jejuni data sets (Figure 4A). This enzyme forms complex I of the electron transport chain and converts the oxidation of NADH into an electrochemical proton gradient. At least in these two species (T. pallidum does not have an electron transport chain), motility might be optimized by increasing the local proton concentration.
Motility is known to be regulated by environmental stimuli such as nutrients and this is reflected by the over-representation of ‘response to stimulus' (GO:0050896) proteins among flagellar interactors. For example, motility is controlled by the second messenger cyclic-di-GMP, which is produced by the enzymatic activity of so-called GGDEF domains (Ryjenkov et al, 2005). Here, we find a conserved interaction of the GGDEF COG, COG2199, with FliC in T. pallidum and E. coli (Figure 4A), pointing to an important regulatory role of this interaction.
FliA, the sigma factor for several flagellar operons, interacts with two subunits of the RNA-polymerase (rpoB and rpoC) in the E. coli and the H. pylori interaction sets (Figure 4A). In addition, in the same species, an interaction of FliA with the glutamyl-tRNA synthetase, GltX, was found, suggesting a regulatory role of this interaction.
Thus, a close inspection reveals a number of interesting functional links, which are also supported by the integrated motility network. However, most of these interactions have to be analyzed in more detail to shed more light on their precise biological role and the mechanistic details.
The evolution of the flagellum
Given the amazing complexity of the bacterial motility system, we wondered whether our interaction data and phenotypes can contribute to the understanding of its evolution. As a first step into that direction, we first constructed a phylogenetic supertree of 30 species based on 35 flagellar protein families (Supplementary Figure 4). Our flagellum supertree strongly supports the monophyly of spirochetes, as well as γ and β, ɛ, and α proteobacteria. These relationships are similar to the previously reported phylogenies, for example, an rRNA tree (Olsen et al, 1994) and a tree which was based on 31 highly conserved protein families (Ciccarelli et al, 2006). This shows that the flagellar system evolved together with other cellular systems and not independently.
Evolution of the flagellum is also consistent with the fact that neither any flagellar proteins nor any of their interactions is conserved. In fact, our Treponema data set predicted 173 interactions for C. jejuni, of which we found only 49 (Supplementary Table S4d). This indicates that protein interactions may be evolutionarily less conserved than generally believed.
An evolutionary model also predicts that core proteins, which have been associated with the flagellum, should be tightly integrated, and thus have more interactions than peripheral proteins, which have been only recently recruited to the flagellar machinery. Indeed, we did find a weak, but statistically significant linear relationship between the number of interactions of an orthologous group and its conservation ratio among flagellated bacteria (r=0.43, P<0.005; Supplementary Figure S7). Therefore, our analysis supports the evolution of the flagellum from core components by adding additional ones over time (Pallen and Matzke, 2006).
Conclusions
In this study, we pursue an integrative systems biology approach to assemble a comprehensive picture of bacterial motility. Motility interaction data sets for T. pallidum and Campylobacter pylori and a genome-wide motility data set for E. coli are presented. Our data are combined with functional and interaction data from multiple species to reconstruct an integrated network of bacterial motility. Insights into the internal structure of the flagellum, its connections to other functional classes, and on potentially novel components of the flagellum have been obtained.
Due to the size of our data set, we were able to analyze only a few selected interactions in more detail. We confirmed the presence of anti-sigma factors (FlgM) in T. pallidum and possibly C. jejuni based on their interactions with a flagellum-specific sigma factor. Recently, we have assigned a new function to TP0658 (now called FliW), a conserved protein of previously unknown function. We could show that this protein acts as a molecular chaperone and/or assembly factor of the bacterial flagellum (Titz et al, 2006).
The bacterial flagellum represents an interesting entity to study the evolution of complex biological machines. For an evolutionary view of the flagellum on the protein level, we constructed a phylogenetic supertree solely based on flagellar protein sequences. As anticipated, this tree closely recapitulates phylogenetic relationships identified, employing traditional phylogenetic marker molecules such as rRNAs.
Whereas it is generally believed that the motility machinery evolved from an ancient type III secretion system, the detailed steps leading to current structures have yet to be defined. A prediction from this theory would be that the conserved core proteins should exhibit more interactions than peripheral proteins. Indeed, proteins which are well conserved and part of all flagellar complexes have more conserved interactions (e.g., FliC, FliG, FliY, FliM, FliA, Mcp, CheW, and CheY) than proteins which are found only in a subset of motility complexes (e.g., FlhF or FlgJ; see Figure 4 and Supplementary Figure S7).
Similar to protein sequences and structures, interactions among proteins are often conserved in the course of evolution. In fact, the phylogenetic relationships of different species are partially reflected by the phylogenetic interaction profile of the integrated network (Supplementary Figure S4).
Finally, we could thus use our interaction data sets to predict interactions in other bacterial species. To obtain only high-confidence predictions, we used our integrated motility network, that is, all interactions found in more than one species or supported by other evidence from the literature, and predicted ∼18 000 interactions for 64 flagellated bacteria (Supplementary Table S7). It remains to be seen which of these interactions do indeed occur and what specific role they play in each of these organisms.
Materials and methods
Collection of known motility proteins
We collected motility genes from three major classification systems: KEGG (including motility, chemotaxis, and flagellar assembly; Kanehisa et al, 2006), TIGR (including chemotaxis and motility; Peterson et al, 2001), and GO (including GO:0019861 flagellum; Camon et al, 2004). Data were compiled in March 2007. In total, we identified 293 proteins (in T. pallidum, C. jejuni, H. pylori, E. coli, and B. subtilis) with at least one classification evidence. Among those, 89% were classified by KEGG, 75% by TIGR, and 65% by GOA. As KEGG provides the most comprehensive classification, we have used the KEGG motility collection throughout this study and refer to its proteins as ‘known motility proteins' (Supplementary Table S1).
Orthologous relationships
Throughout this study, we used COGs to infer orthologous relationships between proteins of different species.
COGs were taken from the NCBI COG database (downloaded March 2006) and complemented by COGs from the STRING version 3 database (including nonsupervised COGs) (von Mering et al, 2003).
Swarming (motility) assays and phenotyping data sources
A systematic single-gene knockout collection of E. coli of 3985 individual mutant strains (Baba et al, 2006) was tested for altered motility by a swarming assay. Each gene mutation was tested in two independent strains as provided by the Keio collection. Strains were grown to saturation in LB medium at 37°C (mutants with growth defects were not considered for the motility assay) and transferred to Omnitrays (Nunc) with swarming agar (LB medium with 0.25% Agar) in a 24 colonies per plate format by pin replication with a Biomek 2000 laboratory robot (Beckman-Coulter). The swarming diameters of the mutant strains were compared after ∼8 h incubation at 37°C and mutants with reproducible reduced motility were retested in individual swarming assays. The swarming behavior of each mutant was classified as wild type, reduced (reduction by at least 50%), or nonmotile (reduction by at least 90%), as measured by the diameter of the bacterial colony (Figure 1A).
Additionally, we constructed a few individual gene deletions of E. coli and B. subtilis and tested them for motility: gene disruptions of yjeK, yncE, and ycfH were performed in E. coli, as described by Datsenko and Wanner (2000). The B. subtilis mutants of yviF, ydjH, and yhbE/yhbF (double mutant) were obtained by phleomycin–cassette integration, as described by Fabret et al (2002). The PCR primers used are listed in Supplementary Table S8. Information on gene deletions affecting motility in B. subtilis, H. pylori, and C. jejuni was taken from the literature (Golden et al, 2000; Schumann et al, 2001; Kobayashi et al, 2003; Salama et al, 2004).
The motility Y2H protein–protein interaction network of T. pallidum
Forty-nine T. pallidum proteins, which are part of our KEGG motility collection, were selected (Supplementary Table S1). Bait fusions (Gal4-DNA-binding domain) of these proteins were constructed by Cre-loxP mediated recombination of pUni clones (Liu et al, 2000; McKevitt et al, 2003) with two bait vectors: pAS1 and pLP-GBKT7Amp (created by replacing Kan® by Amp® in pLP-GBKT7 (Clontech). A systematic whole-genome prey library for T. pallidum was created by transferring all ORFs from their original pUni-vector vector (McKevitt et al, 2003) to our prey vector, pLP-GADT7 (Clontech) by Cre-LoxP-mediated recombination. All prey and bait clones were then individually transformed into Y187 (MATa) and AH109 (MATα) (Harper et al, 1993; James et al, 1996) yeast strains, respectively, by a standard LiAc protocol. Prey strains were arrayed onto 384-well formatted Omnitray-agar plates (Nunc) and each bait strain was individually tested against the whole T. pallidum prey array using a previously described array-based Y2H procedure (Cagney et al, 2000).
The motility Y2H protein–protein interaction network of C. jejuni
Interactions involving the 46 C. jejuni proteins assigned to the motility category in the KEGG database (Kanehisa et al, 2002) (Supplementary Table S1) were identified in proteome-wide two-hybrid screens, using a pooled matrix approach as described previously (Zhong et al, 2003) (Parrish et al, in press).
The motility Y2H protein–protein interaction network of H. pylori
The motility PPI set ‘Helicobacter' (HPY) was generated by selecting all interactions of known motility proteins from Rain et al (2001) (also see motility filtering below). Note that Rain et al (2001) tested only 261 bait fusion proteins (out of 1590 ORFs) against a random prey library.
Literature interactions
To mine all the published PPIs of the known flagellum components, we carried out a comprehensive literature review for flagellum PPIs, using the PubMed query ‘(flagellum OR flagella) AND (interaction OR interact OR interacts OR bind OR binds)' on 13 January 2004. This analysis yielded ∼700 abstract/articles from which 51 unique PPIs between flagellar components were manually curated (Supplementary Table S2).
Motility-related interactions derived from E. coli complex purification data
Motility-related protein interactions of E. coli were derived from Arifuzzaman et al (2006), who conducted a comprehensive complex purification study using a His-tagged ORF clone library. A total of 2667 out of 4339 proteins were successfully analyzed and their interacting partners were identified by MALDI-TOF in this study. Complex purification studies do not provide binary interaction data, but only lists of proteins that co-purified with the used bait protein involving both direct and indirect interactions. Arifuzzaman et al (2006) provided their results according to the spoke model. We used two models to predict direct interactions for flagellum/chemotaxis proteins (one among the pair of interacting proteins is a known flagellum/chemotaxis protein) from the complex data. The ‘ECO SPK' interaction set assumes binary interactions between bait proteins and their co-purifying proteins (SPOKE model). The ‘ECO SAI' interaction set is based on a model that has been proposed by Gavin et al (2006) to infer complexes from multiple overlapping purifications. Similar to the matrix model, it predicts PPIs among all proteins. However, the difference is that PPIs are weighted according to the pair's propensity to associate with each other relative to what would be expected from their frequency. Based on the cumulative percentage distribution of socio-affinities, we defined the top 25% of PPIs to be highly associated (socio-affinity score >5). Both ECO sets have been filtered for interactions of known motility proteins (see motility filtering below) (Supplementary Table S2). Data from Butland et al (Nature 433: 531, 2005) have not been considered in this analysis as only one flagellar protein, FliY, appears to have worked as a bait in this study.
Motility filtering
T. pallidum, C. jejuni, H. pylori, and E. coli PPIs were filtered for motility interactions by retaining only PPIs, which contain at least one protein which is part of our KEGG motility collection
Integrated protein interaction network of different species
Construction of aligned protein networks
Pairwise alignments of the PPI sets were performed using the Network Comparison Toolkit (NCT, http://chianti.ucsd.edu/nct/), a Java implementation of the PathBLAST algorithm, as described previously (Kelley et al, 2004). Briefly, the algorithm integrates PPIs from two species with protein sequence homology to generate an ‘aligned network'. Homologous proteins (one from each organism) are merged (aligned) into single nodes. We have defined proteins to be homologs if the geometric mean of their E-values is ⩽10−5 normalized for each genome size based on manual inspections of E-values among orthologous interactions (Supplementary Table S5c ‘Orthology overlap with E-values'). The rule for creating an edge is that one of the pairs of proteins (one species) must represent (have) an interaction (distance 1 edge), whereas the other pair (the other species) can be in one of three states: (i) the other pair is the same protein (distance 0 edge); (ii) the other pair represents an interaction (distance 1 edge); (iii) the two proteins in the other pair do not interact themselves, but interact with a common neighbor (distance 2 edge) also referred to as gap.
Construction of the integrated network
Homologous protein nodes of the pairwise aligned networks were merged into orthologous groups (COGs) if all proteins were members of the same COG. Nodes were labelled according to KEGG (Kanehisa et al, 2006) (Supplementary Table S1 ‘KEGG motility COGs') and by manual inspection of the common names of the merged proteins. Edges were directly transferred from the pairwise aligned networks. Finally, we have incorporated interactions among orthologous groups found in our literature set (Supplementary Table S2). COG conservation is based on a COG's conservation ratio among flagellated species, reported in the STRING database (in total 68 species).
STRING confidence score
The confidence score of the StringDB (S score) is the approximate probability that a predicted link exists between two enzymes in the same metabolic map in the KEGG database. Confidence limits are as follows: low confidence (S score>0.15) 20% (or better); medium confidence (S score>0.4) 50%; high confidence (S score>0.7) 75%; highest confidence (S score>0.9) 95% (from http://string.embl.de).
Co-immunoprecipitation of Myc-TP0974 with HA-TP0709
Myc-tagged TP0974 and HA-tagged TP0709 were cloned into vectors of the pBAD series (Guzman et al, 1995) and were co-transferred into BL21 (DE3) E. coli. Protein expression was induced with 0.2% (w/v) L-Ara for 3 h at 37°C. The co-immunoprecipitation was performed with anti-Myc antibodies (Santa Cruz).
Availability of data
All interaction data from this study can be retrieved from the IntAct database (http://www.ebi.ac.uk/intact/) under the following accession numbers: EBI-1190357 (C. jejuni data set) and EBI-1190361 (T. pallidum data set).
Supplementary Material
Supplementary Figures and Legends
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5
Supplementary Table 6
Supplementary Table 7
Supplementary Table 8
Acknowledgments
Jeffery Errington kindly provided B. subtilis mutant strains. We thank Tanja Kuhn, Sindhu Thomas, and Cathrin Klumpp for technical assistance. This project has been supported by DFG grant Ue 50/4-1. RL Finley has been supported by NIH grant RR18327.
References
- Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, Ara T, Nakahigashi K, Huang HC, Hirai A, Tsuzuki K, Nakamura S, Altaf-Ul-Amin M, Oshima T, Baba T, Yamamoto N, Kawamura T, Ioka-Nakamichi T, Kitagawa M, Tomita M, Kanaya S, Wada C, Mori H (2006) Large-scale identification of protein–protein interaction of Escherichia coli K-12. Genome Res 16: 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage JP (1999) Bacterial tactic responses. Adv Microb Physiol 41: 229–289 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas R, Suvanasuthi R (2005) The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J Bacteriol 187: 6789–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC (1975) Chemotaxis in bacteria. Annu Rev Biophys Bioeng 4: 119–136 [DOI] [PubMed] [Google Scholar]
- Berg HC (2003) The rotary motor of bacterial flagella. Annu Rev Biochem 72: 19–54 [DOI] [PubMed] [Google Scholar]
- Berg HC, Anderson RA (1973) Bacteria swim by rotating their flagellar filaments. Nature 245: 380–382 [DOI] [PubMed] [Google Scholar]
- Cagney G, Uetz P, Fields S (2000) High-throughput screening for protein–protein interactions using two-hybrid assay. Methods Enzymol 328: 3–14 [DOI] [PubMed] [Google Scholar]
- Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R (2004) The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res 32: D262–D266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Greenberg EP, Koopman MB, Limberger RJ (1992) Spirochete chemotaxis, motility, and the structure of the spirochetal periplasmic flagella. Res Microbiol 143: 597–603 [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabret C, Ehrlich SD, Noirot P (2002) A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol Microbiol 46: 25–36 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Golden NJ, Acheson DW (2002) Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect Immun 70: 1761–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden NJ, Camilli A, Acheson DW (2000) Random transposon mutagenesis of Campylobacter jejuni. Infect Immun 68: 5450–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75: 805–816 [DOI] [PubMed] [Google Scholar]
- Hendrixson DR, Akerley BJ, DiRita VJ (2001) Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol Microbiol 40: 214–224 [DOI] [PubMed] [Google Scholar]
- Hendrixson DR, DiRita VJ (2003) Transcription of sigma54-dependent but not sigma28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol Microbiol 50: 687–702 [DOI] [PubMed] [Google Scholar]
- Inoue T, Shingaki R, Hirose S, Waki K, Mori H, Fukui K (2007) Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J Bacteriol 189: 950–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34: D354–D357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Nakaya A (2002) The KEGG databases at GenomeNet. Nucleic Acids Res 30: 42–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32: D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley BP, Yuan B, Lewitter F, Sharan R, Stockwell BR, Ideker T (2004) PathBLAST: a tool for alignment of protein interaction networks. Nucleic Acids Res 32: W83–W88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O'Reilly M, O'Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, Van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100: 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Blair DF (2004) The bacterial flagellar motor: structure and function of a complex molecular machine. Int Rev Cytol 233: 93–134 [DOI] [PubMed] [Google Scholar]
- Li C, Motaleb A, Sal M, Goldstein SF, Charon NW (2000) Spirochete periplasmic flagella and motility. J Mol Microbiol Biotechnol 2: 345–354 [PubMed] [Google Scholar]
- Liu Q, Li MZ, Liu D, Elledge SJ (2000) Rapid construction of recombinant DNA by the univector plasmid-fusion system. Methods Enzymol 328: 530–549 [DOI] [PubMed] [Google Scholar]
- MacKichan JK, Gaynor EC, Chang C, Cawthraw S, Newell DG, Miller JF, Falkow S (2004) The Campylobacter jejuni dccRS two-component system is required for optimal in vivo colonization but is dispensable for in vitro growth. Mol Microbiol 54: 1269–1286 [DOI] [PubMed] [Google Scholar]
- Macnab RM (1999) The bacterial flagellum: reversible rotary propellor and type III export apparatus. J Bacteriol 181: 7149–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM (2003) How bacteria assemble flagella. Annu Rev Microbiol 57: 77–100 [DOI] [PubMed] [Google Scholar]
- McKevitt M, Patel K, Smajs D, Marsh M, McLoughlin M, Norris SJ, Weinstock GM, Palzkill T (2003) Systematic cloning of Treponema pallidum open reading frames for protein expression and antigen discovery. Genome Res 13: 1665–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehus E, Gressmann H, Ye F, Schlapbach R, Dehio M, Dehio C, Stack A, Meyer TF, Suerbaum S, Josenhans C (2004) Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol Microbiol 52: 947–961 [DOI] [PubMed] [Google Scholar]
- Nishimura A, Hirota Y (1989) A cell division regulatory mechanism controls the flagellar regulon in Escherichia coli. Mol Gen Genet 216: 340–346 [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Matsuda H, Hagstrom R, Overbeek R (1994) fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci 10: 41–48 [DOI] [PubMed] [Google Scholar]
- Ozin AJ, Claret L, Auvray F, Hughes C (2003) The FliS chaperone selectively binds the disordered flagellin C-terminal D0 domain central to polymerisation. FEMS Microbiol Lett 219: 219–224 [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Matzke NJ (2006) From The Origin of Species to the origin of bacterial flagella. Nat Rev Microbiol 4: 784–790 [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Penn CW, Chaudhuri RR (2005) Bacterial flagellar diversity in the post-genomic era. Trends Microbiol 13: 143–149 [DOI] [PubMed] [Google Scholar]
- Parrish JR, Liu G, Hines JA, Yu J, Mangolia BA, Zhang H, Chan JE, Ideker T, Pacifico S, Fatouhi F, DiRita VJ, Andrews P, Finley RL (2007) A proteome-wide interaction map for Campylobacter jejuni. Genome Biol, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O (2001) The comprehensive microbial resource. Nucleic Acids Res 29: 123–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss BM, Campbell JW, Van Dyk TK, Zhu C, Kogan Y, Matsumura P (2003) FlhD/FlhC is a regulator of anaerobic respiration and the Entner–Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J Bacteriol 185: 534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schachter V, Chemama Y, Labigne A, Legrain P (2001) The protein–protein interaction map of Helicobacter pylori. Nature 409: 211–215 [DOI] [PubMed] [Google Scholar]
- Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M (2005) Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol 187: 1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama NR, Shepherd B, Falkow S (2004) Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol 186: 7926–7935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann W, Ehrlich SD, Ogasawara N (2001) Functional Analysis of Bacterial Genes (A practical Manual). John Wiley and sons [Google Scholar]
- Sorenson MK, Ray SS, Darst SA (2004) Crystal structure of the flagellar sigma/anti-sigma complex sigma(28)/FlgM reveals an intact sigma factor in an inactive conformation. Mol Cell 14: 127–138 [DOI] [PubMed] [Google Scholar]
- Stein A, Russell RB, Aloy P (2005) 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Res 33: D413–D417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titz B, Rajagopala SV, Ester C, Hauser R, Uetz P (2006) Novel conserved assembly factor of the bacterial flagellum. J Bacteriol 188: 7700–7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- van Amsterdam K, van der Ende A (2004) Helicobacter pylori HP1034 (ylxH) is required for motility. Helicobacter 9: 387–395 [DOI] [PubMed] [Google Scholar]
- von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 31: 258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler M, Sargent F, Jack RL, Stanley NR, Bogsch EG, Robinson C, Berks BC, Palmer T (2000) TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J Biol Chem 275: 16717–16722 [DOI] [PubMed] [Google Scholar]
- Zhong J, Zhang H, Stanyon CA, Tromp G, Finley RL Jr (2003) A strategy for constructing large protein interaction maps using the yeast two-hybrid system: regulated expression arrays and two-phase mating. Genome Res 13: 2691–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Legends
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5
Supplementary Table 6
Supplementary Table 7
Supplementary Table 8