Abstract
Protein decay rates are regulated by degradation machinery that clears unnecessary housekeeping proteins and maintains appropriate dynamic resolution for transcriptional regulators. Turnover rates are also crucial for fluorescence reporters that must strike a balance between sufficient fluorescence for signal detection and temporal resolution for tracking dynamic responses. Here, we use components of the Escherichia coli degradation machinery to construct a Saccharomyces cerevisiae strain that allows for tunable degradation of a tagged protein. Using a microfluidic platform tailored for single-cell fluorescence measurements, we monitor protein decay rates after repression using an ssrA-tagged fluorescent reporter. We observe a half-life ranging from 91 to 22 min, depending on the level of activation of the degradation genes. Computational modeling of the underlying set of enzymatic reactions leads to GFP decay curves that are in excellent agreement with the observations, implying that degradation is governed by Michaelis–Menten-type interactions. In addition to providing a reporter with tunable dynamic resolution, our findings set the stage for explorations of the effect of protein degradation on gene regulatory and signalling pathways.
Keywords: gene regulation, temporal dynamics
Introduction
A central focus of synthetic biology is constructing isolated gene regulatory networks in living cells in order to determine their dynamical behavior in a given environment. The complexity of naturally occurring gene networks makes this determination difficult, and many investigators have concentrated on smaller artificial ‘circuits' in an attempt to understand fundamental principles (Hasty et al, 2002; Sprinzak and Elowitz, 2005). These smaller networks are more amenable to computational modeling, construction and modification. In addition, the systematic construction of genetic circuits from a set of quantitative design principles will accelerate progress towards therapeutic applications.
Many cellular functions rely on underlying regulatory processes that are highly dynamic. In the context of interconnected gene regulatory and signalling networks, the persistence of mRNA and protein molecules has a large effect on the overall behavior of the cell. This effect often manifests in the underlying network dynamics, where degradation rates can play a central role in mitigating crucial timing events. Naturally occurring components are often adapted for the construction of artificial gene circuits. These components are usually stable, and this stability constrains the ability to monitor highly dynamic circuits and to construct circuits that possess desired response characteristics. For example, GFP is often used as a reporter in native and artificial synthetic genetic networks, due to its ability to be expressed in various hosts without interfering with cellular function. However, GFP is stable in most cells on a time scale of hours to days (Andersen et al, 1998). Once stable GFP is expressed, it is cleared from the system only through growth and subsequent dilution, making measurements at higher temporal resolution difficult to interpret. Temporal resolution can be improved by decreasing the half-life of fluorescent proteins. However, such an increase in temporal responsiveness is not without cost, since it reduces detectable signal. Ideally, one could control the stability of a network component, tuning it to balance the needs of signal detection and dynamical resolution.
There have been several studies that use native protein degradation systems to make network components less persistent. Andersen et al (1998) used the ssrA system in Escherichia coli to target proteins to degradation pathways. In bacteria, polypeptides that stall during translation (e.g., under starvation conditions) have an 11-amino-acid tag added to their C-terminus by a small ssrA molecule. This tag is specifically recognized by the ClpXP proteasome, and tagged proteins are degraded (Gottesman et al, 1998; Karzai et al, 2000). More recently, it was demonstrated that degradation in E. coli could be increased with a modified tag that binds a helper molecule (McGinness et al, 2006). In Saccharomyces cerevisiae, a similar system was described that made translational fusions between a given protein and a domain of CLN2, a yeast protein that is degraded quickly (Mateus and Avery, 2000). It has also been shown that modification of the S. cerevisiae N-degron signal sequence can lead to impressive destabilization of reporters down to a half-life of 2 min (Hackett et al, 2006). While all of these systems produce more dynamic proteins, none focused on developing tunability over a wide range of degradation rates. In addition, the overexpression of native degradation components in the same organism for the purpose of increasing degradation can have undesired pleiotropic effects.
Results and discussion
We have constructed a S. cerevisiae strain (CGD699) that allows tunable degradation of a tagged protein. To accomplish this, we expressed a modified E. coli ClpXP protease in yeast under the control of a repressible promoter. Proteins that are tagged with the ssrA tag are quickly degraded, and this degradation rate is controlled by the induction level of ClpXP. We integrated the two E. coli genes (clpX and clpP) that code for the ClpXP protease into the yeast genome (Figure 1, and Materials and methods). We found that the clpX gene needed to be modified with 10 silent mutations to be expressed in yeast (see Materials and methods). These two genes were placed under the control of two separate copies of the modified ADH1i promoter (Blake, 2003), at two different loci in the yeast genome. Additionally, we integrated mlacI, a mammalian-enhanced version of lacI (Cronin et al, 2001), controlled by a wild-type ADH1 promoter. The wild-type version of the ADH1 promoter exhibits constitutive expression, while the ADH1i promoter is repressed by LacI in the absence of IPTG. Addition of IPTG to the medium results in ClpXP production and degradation of a tagged protein. To demonstrate the utility of this approach, we also integrated a yEGFP gene tagged with an 11-amino-acid ssrA tag (AANDENYALAA), under the control of the GAL1 promoter into CGD699. This promoter is fully induced by 0.5% w/v galactose and repressed by 2% w/v glucose. CGD699 cells grown in the presence of galactose produce GFP and are fluorescent, and GFP production ceases if the carbon source in the media is switched from galactose to glucose. Observations of growth rate and morphology indicate that the exogenous proteases cause no deleterious cellular effects over a wide range of IPTG levels (Figure 2A and B). While coexpression of tagged yEGFP and the degradation machinery resulted in almost complete loss of fluorescence, coexpression of untagged yEGFP with the degradation machinery showed no significant drop in fluorescence (Figure 2C). This confirms that the degradation effect is specific to tagged proteins.
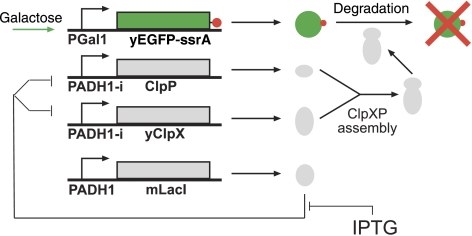
Figure 1.
Network diagram of the degradation module in yeast. The GAL1 production promoter drives repressible expression of yeast-enhanced GFP tagged with the ssrA tag. yEGFP is induced by galactose and repressed by glucose. Two separate copies of the LacI-repressible ADH1i promoter drive expression of E. coli ClpP or E. coli yClpX. ClpXP is induced by the addition of IPTG to bind to mLacI. The proteolytic complex ClpXP degrades ssrA-tagged proteins. A wild-type ADH1 promoter drives the expression of mLacI. All four cassettes are integrated into the yeast genome.
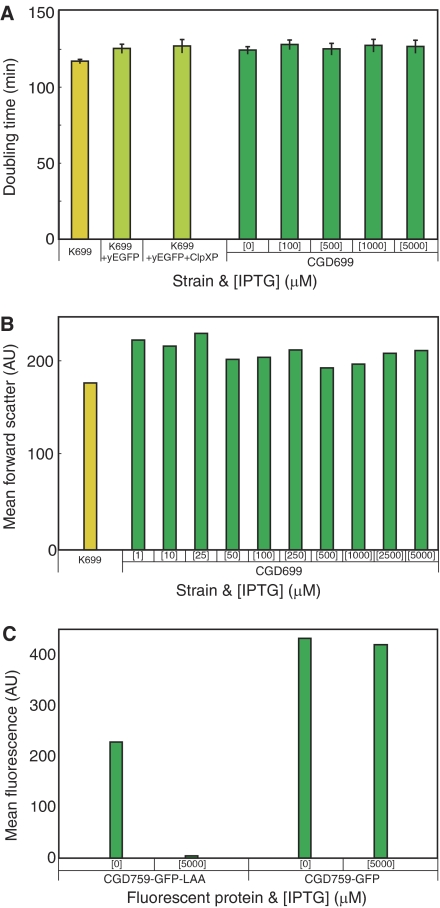
Figure 2.
Effect of the exogenous degradation machinery on CGD699. (A) Doubling times in batch culture for various strains and IPTG concentrations. Shown are the original strain (K699), two intermediate strains (K699 with yEGFP and K699 with yEGFP and ClpXP) and the complete strain (CGD699). (B) Flow cytometry forward scatter means for K699 and CGD699 for various IPTG concentrations. Note that varying the IPTG level (and therefore the resulting concentration of ClpXP) does not significantly affect the forward scatter. (C) Flow cytometry fluorescence means for CGD759 derivatives containing either untagged or ssrA-tagged yEGFP integration cassettes. CGD759 contains integrated clpP, clpX and lacI expression cassettes as in CGD699. Note that the addition of IPTG causes a loss of fluorescence only with ssrA-tagged yEGFP.
We further investigated the ability of CGD699 cells to degrade tagged yEGFP in response to IPTG induction by microscopy in a microfluidic chamber. Prior to imaging, the cells were grown in log phase for 24–36 h in media containing 0.5% galactose and the experimental IPTG concentration. These cells were then loaded into a yeast microfluidic chamber (Cookson et al, 2005) that was designed to allow temporal switching of inducer (Bennett et al, 2007). The growth media was switched to media containing IPTG and 2% glucose to shut off GFP production from the GAL1 promoter. Figure 3A shows a series of bright-field and corresponding fluorescent images for cells exhibiting active degradation in the 1 mM IPTG condition (also see Supplemental Movie). In cells with yeGFP-ssrA but without clpXP incorporation, the half-life of GFP was 104±19 min. This half-life is the result of growth-related dilution (note that the doubling time of the batch culture grown on galactose was approximately 125 min; see Figure 2A). The induction of varied expression of the ClpXP protease allows the reduction of the half-life of GFP to as low as 22 min at 5 mM IPTG.
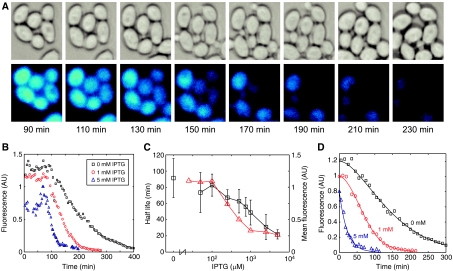
Figure 3.
Degradation of tagged GFP in yeast cells. (A) A series of images in both bright-field (above) and fluorescence (false colored, below) imaging of GFP decaying in yeast cells at 1 mM IPTG induction. The 90 min timestamp of the first image is 30 min after the media switch from galactose to glucose. (B) Representative time fluorescence trajectories of three cells in differing concentrations of IPTG. (C) Experimentally measured half-lives of yEGFP-ssrA as a function of IPTG concentration, compared to mean fluorescence. The black squares (half-lives; left y-axis) are determined from single-cell microscopy time trajectories. The red triangles (mean fluorescence; right y-axis) are derived from flow cytometry experiments. The x-axis is a discontinuous log scale. (D) Comparison of single-cell trajectories (symbols) to time series obtained from numerical integration of equation 1. Each trajectory was numerically fit to the data.
Previous studies have used exponential fits to characterize the half-life of the reporter. We found that modeling decay as arising from a set of enzymatic Michaelis–Menten reactions led to excellent agreement between model and experiment (see below). However, in order to systematically compare with the previous fluorescent reporter degradation studies, we chose to first analyze the fluorescence trajectories with exponential fits that were reasonably accurate (Figure 3B). From these fits, we were able to calculate the mean half-life for each concentration of IPTG, as shown in Figure 3C. The half-life decreases from a value of 91 min for no IPTG to 22 min for media containing 5 mM IPTG.
In addition to the exponential fitting, we used the experimental data to determine how the degradation process should be modeled. For this, we assumed that both the GAL1 and ADH1 promoters are constitutive, and that yEGFP-ssrA, ClpXP and mLacI have reached equilibrium before the introduction of glucose. In this case, the transcription of the GAL1 promoter stops once glucose has been added and its mRNA begins to decay exponentially from its steady-state value. However, the translation of the mRNA continues, leading to an ODE describing the dynamics of the concentration of yEGFP-ssrA:
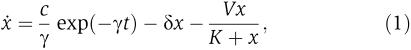 |
where x is the concentration of yEGFP-ssrA, γ is the degradation rate of the GAL1 mRNA, c is a constant related to the transcription and translation rates of GAL1, δ is the dilution rate and V and K are the Michaelis–Menten constants describing the ClpXP-mediated enzymatic decay of yEGFP-ssrA. We assume that glucose is added at t=0, so that
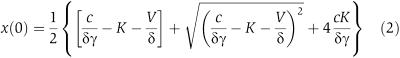 |
is the steady-state initial concentration. Because the constant V is proportional to the concentration of the enzyme (ClpXP), it will increase with increasing levels of IPTG (which activates ClpXP production). Therefore, increasing the concentration of IPTG will increase the degradation rate of the substrate (yEGFP-ssrA). Figure 3D shows a comparison between single-cell data from three trials (symbols) and time series obtained from a numerical best-fit algorithm of equation 1 (solid lines). The excellent agreement implies that ClpXP-mediated degradation is a Michaelis–Menten enzymatic process, as opposed to first-order decay, which is often assumed in models describing signalling or gene-regulatory networks.
We have adapted the prokaryotic ssrA tagging system (Andersen et al, 1998; Gottesman et al, 1998) for use in eukaryotic yeast cells. The CGD699 strain allows for the tunable degradation of any protein by adding a short amino-acid tag. In reporter systems, the competing requirements of signal detection and dynamical resolution can be balanced without the need for additional cloning procedures. This system has several advantages over previously described systems. The degradation is tunable over a range of IPTG concentrations. The degradation tag is small and presumably unlikely to interfere with protein function. The small size of the tag simplifies construction of tagged genes by PCR amplification or use of a tagging vector, and many genes can be tagged in parallel. Several variant ssrA tags have been described (Andersen et al, 1998; McGinness et al, 2006), allowing another level of control. Induction of ClpXP was non-toxic, as we found no significant change in growth rate or morphology at different IPTG concentrations. In addition, although a yeast homologue to ClpX has been identified (van Dyck et al, 1998), it is located in the mitochondria and acts as a chaperone and not a protease. No homologue to ClpP has been found in yeast. The degradation components are stably integrated into the yeast genome, which allows it to be adapted for use with any number of tagged targets, either integrated or expressed from a plasmid. Due to the simple nature of the system, it might be portable into other genetically tractable eukaryotic models. There is evidence that bacterial ClpX and ClpP can interact with mammalian mitochondrial ClpX and ClpP (Kang et al, 2002), but the restricted subcellular localization of these mammalian proteins should limit in vivo interaction.
Increased degradation might be achievable by coexpressing the E. coli sspB gene, which facilitates ssrA tag binding to the ClpXP protease (Hersch et al, 2004). Although we have only used the CGD699 strain with a fluorescent reporter protein, it should be applicable to any protein that can support the short C-terminal ssrA (Huh et al, 2003). We expect that this system will prove useful for a variety of applications where increased temporal resolution is desired, both as a tool in the construction of small-scale synthetic networks (Hasty et al, 2002; Sprinzak and Elowitz, 2005), and in systems biology studies, which seek to unravel network connectivity through perturbation of a given component (Ideker et al, 2001; Faith et al, 2007).
Materials and methods
Growth of microorganisms
S. cerevisiae cells were grown at 30°C with 300 r.p.m. shaking in SD dropout media containing appropriate amino-acid supplements. This was supplemented with 20 mg/l tryptophan, histidine and uracil and 100 mg/l leucine, as appropriate. E. coli DH5α cells were grown at 37°C with shaking at 300 r.p.m. in LB medium supplemented with 100 μg/ml ampicillin to maintain plasmids.
Construction of strains
DNA manipulations were carried out by standard techniques (Sambrook and Russell, 2001). yEGFP (Sheff and Thorn, 2004) was cloned from pRS31-yg (Volfson et al, 2006) into an ssrA tagging vector that attached an ssrA tag (AANDENYALAA) to the C-terminal end, and then cloned back into pRS31-yg and integrated into the HIS3 locus of S. cerevisiae strain K699. clpX and clpP were cloned from E. coli 2.300 by genomic PCR using Phusion polymerase (New England Biolabs). clpP was placed behind the LacI-repressible ADH1i promoter on pRS3GFPi (Blake, 2003), replacing the GFP gene. The promoter–gene–terminator cassette was then cloned into pRS404 and incorporated into the S. cerevisiae TRP1 locus. Yeast-optimized clpX was incorporated into the S. cerevisiae URA3 locus via pRS406 using a similar procedure. Mammalian-enhanced lacI (Cronin et al, 2001) was placed under the wild-type ADH1 promoter on pRS3GFPa, replacing the GFP gene. The promoter–gene–terminator cassette was then cloned into pRS405 and incorporated into the S. cerevisiae LEU2 locus. Integrations were confirmed via selective auxotrophic plates. The resulting strain was designated CGD699, and is available upon request. Other strains were constructed by integrating the expression cassettes in a different order.
Yeast optimization of clpX
Our initial attempts to construct this strain resulted in fluorescent signal but no corresponding drop in fluorescence when clpX and clpP were induced. RT–PCR results showed that the mRNA was transcribed, but Western blotting results showed that the ClpX was not translated (data not shown). We suspected that codon bias between the two organisms was a factor, and optimized clpX for yeast expression.
Sources of possibly deleterious codon bias were identified by comparing codon usages between E. coli and S. cerevisiae using the Codon Usage Database (http://www.kazusa.or.jp/codon/). Arginine codons were commonly problematic. We introduced 10 silent mutations in two rounds of multiple mutagenesis using a previously described method (Sawano and Miyawaki, 2000), and using Phusion polymerase (NEB). We omitted the megaprimer step, instead transforming single-stranded DNA directly into ultracompetent XL-10 Gold cells (Stratagene). The following primers were used (mutations induced are set bold):
Primer 1: GGTCGCGGTATACAACCATTACAAAAGATTGCGCAACGGCGATACCAGCAATGGCGTCGAG
Primer 2: GCTGCTGGCTGAAACGCTGGCTAGATTGCTGGATGTTCCGTTCACCATGGCCGAC
Primer 3: CTATCGCTAAGAAAGCGATGGCTAGAAAAACCGGTGCCCGTGGCCT.
ClpXP-mediated GFP decay analysis
A microfabricated chemostatic growth chamber that contained an on-chip media switch was used to measure ClpXP-mediated GFP decay within single cells. The microdevices were fabricated using well-documented poly-dimethylsiloxane (PDMS, Dow Corning Sylgard 184) replica molding techniques (Xia and Whitesides, 1998). Chambers were fabricated to 4 μm in depth to facilitate quantitative fluorescence imaging of single cells (Cookson et al, 2005).
Before experimentation, cells were grown from plates in appropriate auxotrophic media containing 0.5% galactose, 2% raffinose and the experimental IPTG condition for at least 24 h in batch culture. Each culture was maintained in log-phase growth. Yeast cells were loaded into microfabricated growth chambers at an OD600 of 0.4–0.6 and allowed to grow for 1 h in loading media. They were then transferred to media containing 2% (w/v) glucose and no galactose, the experimental IPTG concentration and a red fluorescent tracer dye (sulforhodamine 101, Sigma).
Cells were imaged over the course of 4–12 h in bright field, green fluorescence and red fluorescence at 5 min intervals. Images were processed by background and camera bias subtraction and flatfield correction. Individual cells were segmented from image backgrounds using bright-field images. Mean pixel intensity within each object was calculated and tracked through each time series using object centroids. Temporal profiles of 15–30 cells that exhibited decay responses for each IPTG concentration were collected. All image processing, segmentation and tracking procedures were performed using software developed in Matlab (The Mathworks Inc.).
Supplementary Material
Supplementary Movie 1
Acknowledgments
We thank Natalie Ostroff and Michael Ferry for extensive assistance with yeast molecular biology. This work was supported by NIH Grant GM69811-01.
References
- Andersen JB, Sternberg C, Poulsen LK, Bjørn SP, Givskov M, Molin S (1998) New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64: 2240–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MR, Pang WL, Ostroff NA, Tsimring LS, Hasty J (2007) Metabolic gene regulation in a dynamically changing environment (submitted) [DOI] [PMC free article] [PubMed]
- Blake WJ (2003) Development of artificial gene regulatory networks in Saccharomyces cerevisiae: genetic switches and noise modulation. PhD Thesis, Boston University [Google Scholar]
- Cookson S, Ostroff N, Pang WL, Volfson D, Hasty J (2005) Monitoring dynamics of single-cell gene expression over multiple cell cycles. Mol Syst Biol 1: 2005.0024 10.1038/msb4100032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin CA, Gluba W, Scrable H (2001) The lac operator-repressor system is functional in the mouse. Genes Dev 15: 1506–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS (2007) Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol 5: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT (1998) The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev 12: 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett EA, Esch RK, Maleri S, Errede B (2006) A family of destabilized cyan fluorescent proteins as transcriptional reporters in S. cerevisiae. Yeast 23: 333–349 [DOI] [PubMed] [Google Scholar]
- Hasty J, McMillen D, Collins JJ (2002) Engineered gene circuits. Nature 420: 224–230 [DOI] [PubMed] [Google Scholar]
- Hersch GL, Baker TA, Sauer RT (2004) SspB delivery of substrates for ClpXP proteolysis probed by the design of improved degradation tags. Proc Natl Acad Sci USA 101: 12136–12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292: 929–934 [DOI] [PubMed] [Google Scholar]
- Kang SG, Ortega J, Singh SK, Wang N, Huang NN, Steven AC, Maurizi MR (2002) Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP. J Biol Chem 277: 21095–21102 [DOI] [PubMed] [Google Scholar]
- Karzai AW, Roche ED, Sauer RT (2000) The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol 7: 449–455 [DOI] [PubMed] [Google Scholar]
- Mateus C, Avery SV (2000) Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast 16: 1313–1323 [DOI] [PubMed] [Google Scholar]
- McGinness KE, Baker TA, Sauer RT (2006) Engineering controllable protein degradation. Mol Cell 9: 701–707 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: a Laboratory Manual. CSHL Press: Cold Spring Harbor, NY, USA [Google Scholar]
- Sawano A, Miyawaki A (2000) Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res 28: e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS (2004) Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21: 661–670 [DOI] [PubMed] [Google Scholar]
- Sprinzak D, Elowitz MB (2005) Reconstruction of genetic circuits. Nature 438: 443–448 [DOI] [PubMed] [Google Scholar]
- van Dyck L, Dembowski M, Neupert W, Langer T (1998) Mcx1p, a ClpX homologue in mitochondria of Saccharomyces cerevisiae. FEBS Lett 438: 250–254 [DOI] [PubMed] [Google Scholar]
- Volfson D, Marciniak J, Blake WJ, Ostroff N, Tsimring LS, Hasty J (2006) Origins of extrinsic variability in eukaryotic gene expression. Nature 439: 861–864 [DOI] [PubMed] [Google Scholar]
- Xia YN, Whitesides GM (1998) Soft lithography. Angew Chem Int Edit 37: 550–575 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1