Abstract
After whole-genome duplication (WGD), deletions return most loci to single copy. However, duplicate loci may survive through selection for increased dosage. Here, we show how the WGD increased copy number of some glycolytic genes could have conferred an almost immediate selective advantage to an ancestor of Saccharomyces cerevisiae, providing a rationale for the success of the WGD. We propose that the loss of other redundant genes throughout the genome resulted in incremental dosage increases for the surviving duplicated glycolytic genes. This increase gave post-WGD yeasts a growth advantage through rapid glucose fermentation; one of this lineage's many adaptations to glucose-rich environments. Our hypothesis is supported by data from enzyme kinetics and comparative genomics. Because changes in gene dosage follow directly from post-WGD deletions, dosage selection can confer an almost instantaneous benefit after WGD, unlike neofunctionalization or subfunctionalization, which require specific mutations. We also show theoretically that increased fermentative capacity is of greatest advantage when glucose resources are both large and dense, an observation potentially related to the appearance of angiosperms around the time of WGD.
Keywords: evolution, genome duplication, metabolism
Introduction
Analyses of several yeast genomes have confirmed the presence of a whole-genome duplication (WGD) in the clade including the bakers' yeast Saccharomyces cerevisiae (Wolfe and Shields, 1997; Kellis et al, 2004). One of the most puzzling aspects of any WGD event is the question of what immediate selective advantage it conferred upon its possessor. Such an advantage would have been necessary to counteract the disadvantages of reproductive isolation (Grieg et al, 2002a, 2002b) and increased metabolic costs (Wagner, 2005; Gerstein et al, 2006) experienced by a post-WGD organism compared to its peers. Here, we try to place the genome duplication into the larger picture of the evolutionary history and ecology (Wagner, 2000; Hittinger et al, 2004) of this species. Several authors have speculated that WGD enhanced S. cerevisiae's ability to metabolize glucose (Wolfe and Shields, 1997; Wolfe, 2004; Liti and Louis, 2005) and/or to grow anaerobically (Kwast et al, 2002; Piškur and Langkjær, 2004; Piškur et al, 2006). We provide evidence that the preservation of some duplicate gene pairs created by the WGD was related to their contribution toward high glycolytic flux. We further consider the possibility that this selection was active soon after the WGD and may be the reason for its survival.
In the presence of oxygen, most eukaryotes fully oxidize glucose to carbon dioxide and water using the TCA cycle, driving mitochondrial ATP synthesis with the accumulated reduced coenzymes. When oxygen is limited, a fermentative pathway is used instead, so that, in yeasts, glucose is converted to ethanol (Pronk et al, 1996). S. cerevisiae is unusual in that it prefers to ferment glucose into ethanol even in the presence of oxygen (the Crabtree effect; Geladé et al, 2003; Johnston and Kim, 2005), despite this pathway's energetic inefficiency. This phenotype is part of a suite of adaptations that allow S. cerevisiae to maintain very high growth rates when glucose is in excess (Piškur et al, 2006).
On the basis of comparative genetics and genomics, some of these Crabtree-related adaptations can be dated to prior to the WGD and some to after it. For instance, the HAP4 gene in S. cerevisiae seems to have acquired a role in the regulation of respiration since the split with the non-WGD species Kluyveromyces lactis (Blom et al, 2000; Buschlen et al, 2003). The alcohol dehydrogenase genes ADH1 and ADH2 in S. cerevisiae are the product of a gene duplication also post-dating the WGD (Thomson et al, 2005). The two resulting gene products allow S. cerevisiae to efficiently use glucose through fermentation (Figure 1). The product of ADH1 is primarily responsible for producing ethanol from acetaldehyde, while ADH2's gene product is optimized to catalyze the reverse reaction (Thomson et al, 2005).
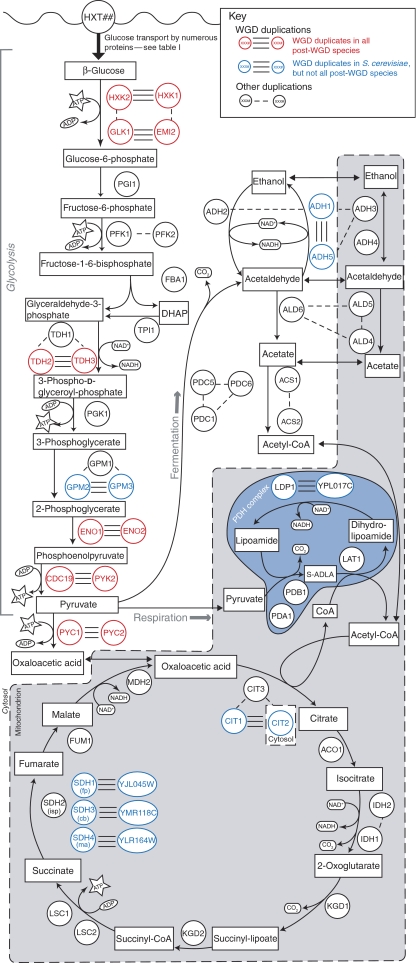
Figure 1.
Overview of three catabolic pathways in S. cerevisiae: glycolysis, alcohol fermentation and the TCA cycle. Enzymes catalyzing each reaction are illustrated by circled gene names. Single lines joining pairs of enzymes indicate paralogous genes. Enzymes joined by three lines indicate paralogous enzymes derived from whole-genome duplication. The WGD pairs shown in red are preserved in double copy in four extant yeast species: S. cerevisiae, S. bayanus, C. glabrata and S. castellii. Protein localization for the CIT, ADH and ALD genes is taken from Huh et al (2003). The multi-enzyme complex which constitutes PDH is illustrated by the darker blue enclosure.
On the other hand, a regulatory circuit that represses pathways that metabolize other sugars when glucose is abundant is conserved in K. lactis. This circuit includes the MIG1 repressor (Dong and Dickson, 1997; Geladé et al, 2003) and the glucose-sensing proteins (RAG4 in K. lactis and SNF3 and RGT2 in S. cerevisiae) that initiate signal cascades that in turn alter gene expression in response to glucose (Özcan et al, 1996, 1998; Betina et al, 2001). SNF3 and RGT2 are WGD paralogs of each other and orthologous to RAG4 in K. lactis. Notably, the two S. cerevisiae paralogs appear to have undergone functional divergence since duplication, with the former signaling low glucose concentrations and the latter higher concentrations (Özcan et al, 1996, 1998). Of course the primary metabolic enzymes of glycolysis, fermentation and respiration are ancient and widely distributed in yeasts (Blank et al, 2005), and the ability to ferment glucose under anaerobic conditions also predates WGD (Visser et al, 1990; Møller et al, 2001).
These observations suggest that the yeast lineage leading to S. cerevisiae has been characterized by a long period of natural selection for rapid growth on substrates such as glucose. Several lines of evidence suggest that WGD may have played a role in this selection. A survey of over 40 yeast species both with and without the WGD indicates that the ability to grow anaerobically on minimal media, the presence of a Crabtree effect and the ability to generate petite mutants are all strongly associated with yeasts possessing the WGD (Merico et al, 2007). Another study also found a general, though weak, trend for higher rates of ethanol production in post-WGD yeasts (e.g., Saccharomyces exiguus and Saccharomyces servazzii) than in non-WGD yeasts (Blank et al, 2005). There is also an excess of energy metabolism genes surviving in duplicate from this event (Kuepfer et al, 2005). In this paper we propose that the WGD had an important impact on gene dosage and that this dosage change had a knock-on effect on how the lineage of post-WGD yeasts (including S. cerevisiae) uses glucose.
We propose three linked hypotheses relating glucose metabolism to the yeast WGD. First, we suggest that the loss of duplicate copies of other genes after WGD increased the concentrations of glycolytic enzymes (which survived in duplicate). Second, we propose that the inherent kinetics of fermentation and respiration meant that this increase in enzyme concentration gave rise to an increased preference for fermentation in the partially polyploid yeast. Finally, we propose that this yeast had a selective advantage because it was able to use glucose more rapidly than its ancestors and hence out-compete other yeasts when glucose was in excess. In the sections below, we briefly introduce each hypothesis in turn.
Hypothesis 1—WGD followed by gene loss raised glycolytic enzyme concentrations
To increase flux through a metabolic pathway is a challenging problem for natural selection. In general, a slow succession of changes in enzyme concentrations (due either to independent gene duplications or to independent changes in gene expression) will be required to increase flux, as (roughly speaking) one reaction after another becomes rate-limiting (Kacser and Burns, 1973). Given that natural selection favors microorganisms with higher growth rates and that gene expression in these organisms can evolve rapidly (Dekel and Alon, 2005), it is not surprising that a laboratory attempt to increase growth rates in S. cerevisiae by overexpression of a single enzyme (pyruvate decarboxylase) was unsuccessful (van Hoek et al, 1998a). It is instead more likely that any increase in the rate of cell division would require more global changes in gene expression such as those seen in experimental evolution studies (Ferea et al, 1999). Duplicate gene pairs produced by WGD have the potential to produce precisely these simultaneous changes in enzyme concentrations, an idea attributable to Ohno (1970). An association between reactions with high flux and an increased frequency of iso-enzymes formed by gene duplication has been found in yeast (Papp et al, 2004), suggesting that duplication can help organisms to adapt their metabolic fluxes. However, that study did not partition the duplicate genes considered into single-gene duplicates and duplicates produced by WGD.
We argue that the process of genome shrinkage following WGD eventually led to a bias in the expression of glycolysis enzymes. In support of this contention, we note that several duplicated genes in S. cerevisiae today have been maintained for reasons of increased gene dosage (Seoighe and Wolfe, 1999; Koszul et al, 2004). We have also previously shown that soon after WGD there was a very rapid loss of duplicate genes (Scannell et al, 2006). It is reasonable to argue this process of gene loss resulted in changes in relative levels of protein expression as part of the cell's protein ‘energy budget' was redirected to the surviving duplicates.
Hypothesis 2—higher enzyme concentrations increased the relative flux through fermentation
If changes in relative gene dosages were able to increase flux through glycolysis, why was the result not simply an overall increase in metabolic rate? We propose that the fermentative and respiratory pathways responded differently to such changes. There are several reasons to think that changes in enzyme concentration should have relatively little impact on respiratory flux. First, respiration depends on the concentration of oxygen in the cell, which, unlike glucose, is difficult or impossible for the cell to alter. Second, because the copy number of the mitochondrial genome is unlikely to have been affected by the WGD, some respiratory proteins would not have seen an increase in concentration. Third, spatial factors such as the number and location of mitochondria may constrain the rate of respiration, as seen in studies of metabolic scaling (West et al, 1999). Although the fact that glycolytic enzymes are observed in association with the mitochondrial surface suggests that spatial constraints also affect glycolysis, not all copies of the requisite enzymes are so localized (Brandina et al, 2006), meaning that this constraint should be weaker for glycolysis than for respiration. Thus, increased dosage from such enzymes may generally increase their dissolved concentration and allow them to route increased flux. Indeed, computational analysis supports a role for increased dosage from duplicates for several reactions in glycolysis (Kuepfer et al, 2005). Glycolytic genes also increase in expression under anaerobic growth relative to aerobic growth in S. cerevisiae (Kwast et al, 2002), presumably because their concentrations are not rate-limiting under respiratory conditions. Moreover, experiments that simultaneously overexpressed several enzymes in the lower half of the glycolytic chain and in the fermentative pathway yielded yeast cells with higher fermentative rates (Smits et al, 2000). Finally, S. cerevisiae strains with mutations in the GCR1 and GCR2 transcription factors show both lowered expression of glycolytic genes and increased reliance on respiratory reactions (Sasaki and Uemura, 2005). Collectively, these points suggest that respiration and fermentation scale differently in yeast.
It might seem surprising that the fermentation enzymes would be in place to route additional flux after an increase in the rate of glycolysis and the consequent saturation of the respiratory apparatus. However, Zeeman et al (1998) found that K. lactis will ferment aerobically after an artificial block of the respiratory apparatus. It thus appears that even before WGD, yeast species may have had some ability to use alcohol fermentation as an overflow pathway, a principle suggested by Käppelli (1986).
At least part of S. cerevisiae's preference for fermentation is due to differences in the enzymes at the branch point between respiration and fermentation. Pyruvate decarboxylase (PDC) is the first step in fermentation, while pyruvate dehydrogenase (PDH, a multicomponent enzyme including PDA1, PDB1, LAT1 and LDP1 in Figure 1) converts pyruvate to acetyl-Coenzyme A as the first step in respiration. These two reactions, which compete for pyruvate as a substrate, differ in their kinetics. The concentration of pyruvate that gives half-maximal activity (Km) of PDH is lower than that for PDC (Kresze and Ronfit, 1981; van Urk et al, 1989). Moreover, PDC exhibits cooperativity (super linear scaling of reaction rate with substrate concentration; Boiteux and Hess, 1970; Hübner et al, 1978) with respect to pyruvate, and the maximal activity of PDC is greater than of PDH (van Urk et al, 1989; Pronk et al, 1996). The net result is to make flux through PDH favored at low pyruvate concentrations and flux through PDC favored at higher ones (Pronk et al, 1996). Thus, for S. cerevisiae, a more rapid rate of glucose consumption should be associated with the routing of an increased proportion of the resulting pyruvate into fermentation as opposed to respiration.
Hypothesis 3—increased fermentation rate conferred a selective advantage
The first two hypotheses do not themselves suggest that any particular changes in gene dosage were advantageous to the ancestors of S. cerevisiae. However, it is plausible that the appearance of fruit-bearing angiosperms opened an ecological niche to which yeasts such as S. cerevisiae were particularly well adapted due to their ability to consume glucose rapidly through fermentation (Ashburner, 1998; Piškur and Langkjær, 2004). Note that this advantage exists in spite of the fact that fermentation yields less ATP per gram of glucose than does respiration. Glucose resources are susceptible to a ‘tragedy of the commons' often seen in competitive situations. In particular, when multiple genotypes compete for glucose, organisms with fast, inefficient metabolism are at a selective advantage relative to their more efficient but slower-growing competitors (Pfeiffer et al, 2001; Pfeiffer and Schuster, 2005; MacLean and Gudelj, 2006).
Results
The above hypotheses led us to examine the genome sequence and metabolic data available for S. cerevisiae and some of its close relatives to see if they showed evidence of selection for such metabolic changes.
Hypothesis 1A—number of glycolytic genes retained in duplicate since WGD
As previously described, it is possible to identify gene duplicates that owe their existence to the WGD by showing that a pair of duplicates lie in paired regions of shared gene order, as inferred by comparing that genome to those of yeast species without the genome duplication (Byrne and Wolfe, 2005). In Figure 1 we find that, of the 10 reactions of glycolysis, five of them maintain WGD duplicates in S. cerevisiae (two duplicate pairs in the case of the first reaction) and moreover that five of these six duplicate gene pairs (excluding the pair GPM2/GPM3) are preserved across four species examined (Saccharomyces cerevisiae, Saccharomyces bayanus, Candida glabrata and Saccharomyces castellii; Byrne and Wolfe, 2005). Given that 551 duplicate gene pairs have survived in S. cerevisiae since genome duplication, one can ask what the chances are that six such pairs would appear in a group of thirteen enzymes (including the ancient duplicate hexokinase, phosphoglycerate mutase and phosphofructokinase enzymes; Figure 1). The hypothesis that the glycolysis genes were preserved in duplicate at the same frequency as the reminder of the genome is rejected by Fisher's exact test (P=0.0014), in agreement with the excess of energy metabolism duplicate genes in S. cerevisiae previously seen (Conant and Wagner, 2002; Kuepfer et al, 2005).
Moreover, given that 239 of the 551 S. cerevisiae duplicate pairs are also duplicated in the other three WGD species (taken from Byrne and Wolfe, 2005), we can ask what would be the chance of seeing as many duplicates preserved across all four species as are seen in glycolysis if that pathway were to follow the pattern of the remainder of the genome. Given than the proportion of WGD duplicate genes in S. cerevisiae that are also preserved in the other three species excluding glycolytic duplicates is (239–5)/(551–6)=0.43, the probability of seeing five or more duplicates preserved in glycolysis is P=0.056 by a binomial test.
One could argue that these results simply reflect an overall preference for retaining duplicate genes of a particular functional class after WGD. To test this possibility, we retrieved all S. cerevisiae genes classified in Gene Ontology (The Gene Ontology Consortium, 2000) as being involved in the biological process ‘catabolism'. We compared the proportion of surviving WGD duplicates in this category excluding glycolysis enzymes to the proportion surviving among the glycolysis enzymes. Significantly more duplicates survive among the glycolytic enzymes (P=0.004, Fisher's exact test, see Supplementary methods for details).
Hypothesis 1B—distribution of hexose transporters
Pritchard and Kell (2002) have shown that hexose transport is the major rate-limiting step in glycolysis. Supporting this observation, Otterstedt et al (2004) found that an S. cerevisiae strain with very limited capacity to transport hexoses does not show a Crabtree effect and under aerobic conditions only respires. Moreover, yeast cells grown in conditions of glucose limitation have been observed to undergo spontaneous duplication of hexose transporters (Brown et al, 1998). This observation implies that the duplication of transporters confers a selective advantage in environments that are otherwise able to support higher growth rates but for which the cells are operating near their maximal glucose uptake rates. If the WGD was fixed in the population in order to allow increased flux through glycolysis, it follows that hexose transport should occur at higher rates in the post-WGD species. Given the lack of experimental data from many of the species studied here, we cannot make a quantitative comparison of hexose transport rates between post-WGD and non-WGD yeasts. However, as a first approximation, we examined the number of hexose transporter genes in these genomes (Table I; see Supplementary methods for details). In agreement with our hypothesis, all of the post-WGD species have at least twice as many hexose transporter genes as the three non-WGD species. Note that this difference is only partially due to WGD—S. cerevisiae in particular has several tandemly duplicated transporters that post-date the WGD (data not shown)—and probably reflects ongoing selection for increased rates of transport.
Table 1.
Number of hexose transport gene paralogs in five species of yeast
| Species | Has WGD? | Number of hexose transport genes |
|---|---|---|
| S. cerevisiae | Yes | 18 |
| S. bayanusa | Yes | 16 |
| C. glabrata | Yes | 11 |
| S. castelliib | Yes | 14 |
| K. lactis | No | 2 |
| S. kluyveri | No | 5 |
| E. gosspyii | No | 5 |
aOne identified Saccharomyces bayanus hexose transporter could not be aligned by GenomeHistory, and is hence omitted from this row.
bThree sequences from Saccharomyces castellii could not be aligned by GenomeHistory, and are hence omitted from this row.
Hypothesis 2A—effects of enzyme concentration changes on the relative fluxes through fermentation and respiration
If our first hypothesis is correct, the yeast ancestor that existed at the time of WGD had lower concentrations of glycolytic enzymes than does the modern S. cerevisiae. To gain insight into how altered enzyme concentrations might change the patterns of carbon flux, we used previously published models of S. cerevisiae metabolism (Teusink et al, 2000; Pritchard and Kell, 2002) with the Jarnac/Jdesigner package (Sauro et al, 2003). We first considered the change in metabolic steady state concentrations that might have resulted from duplication.
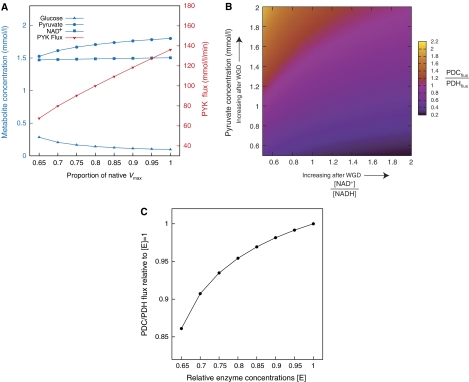
To a first approximation, reaction rates for an enzyme catalyzed reaction depend on two enzyme-specific parameters: Km, the substrate concentration that gives a half maximal reaction rate, which is essentially independent of enzyme concentration, and Vmax, the maximal reaction velocity. Vmax depends on factors such as the activation energy of the reaction and the concentration of the catalyzing enzyme. We modeled the effects of WGD on glycolysis by representing the implied ancestral enzyme concentrations for glycolysis and alcohol fermentation as uniform reductions in their Vmax values. As enzyme concentrations increase from 65 to 100% of their current levels (see Supplementary methods), the concentration of several metabolic intermediates increases, with pyruvate showing a 17% increase in concentration (Figure 2A). Perhaps even more significantly, the flux through the final reaction in glycolysis (i.e., pyruvate kinase, CDC19 and PYK2 in Figure 1) increases by a factor of two across the range of enzyme concentrations considered (red line in Figure 2A).
Figure 2.
(A) Changes in the concentration of key metabolites in response to overall decreases in the Vmax values for the reactions of glycolysis (blue, left axis scale), as well as the change in PYK flux over the same range (red, right axis scale). (B) Ratio of the flux through pyruvate decarboxylase (PDC, fermentative pathways) to the flux through pyruvate dehydrogenase (PDH, respiratory pathway) as a function of pyruvate concentration and the ratio of NAD+ to NADH concentration (because NAD+ and NADH are two oxidation states of the same molecule, their concentrations vary inversely and hence are constrained to sum to 8.01 in B; Theobald et al, 1997). (C) Effect of compartmentalization on the relative fluxes of the first reaction in respiration (PDH) and in fermentation (PDC). On the x-axis is the relative enzyme concentration modeling a change from a rough pre-duplication state of 0.65 to a post-duplicate value of 1.0 (see A). On the y-axis is given the ratio of the fluxes between the two reactions relative to the flux when [E] on the x-axis is equal to 1.0.
We next studied the effect that changes in pyruvate concentration have on the competing reactions PDC and PDH, given the differing kinetics of these two enzymes (see Supplementary methods). Figure 2B shows the ratio of PDC flux over PDH flux. We plot the dependence of this ratio on the concentration of pyruvate and on the ratio of NAD+ to NADH in the mitochondria. Note that we are concerned here with how the inherent kinetics of these two enzymes differ: the relative contours of Figure 2B are independent of the actual concentrations of the two enzymes (so although those concentrations can be changed by regulatory interactions not included in our analysis, the relative behavior of the two enzymes cannot be so easily altered). Increasing the pyruvate concentration increases relative flux through PDC. In principle, this effect could be counteracted by the increased ratio of cytosolic NAD+ to NADH that is also seen when glycolytic enzyme concentrations increase (the slight increase in NAD+ concentration shown in Figure 2A as Vmax increases is matched by an equal decrease in the concentration of NADH). However, this counter effect is probably quite weak, both because the increase in NAD+ concentration seen is small and because the mitochondrial and cytosolic concentrations of these cofactors may not be in equilibrium (Bunoust et al, 2005).
Hypothesis 2B—compartmentalization of respiration
Alcoholic fermentation takes place in the cytosol, whereas respiration is carried out exclusively in the mitochondria of yeast cells. As a result, the ratio of the surface area of mitochondria to the volume of the cytosol imposes a spatial constraint on the rate at which pyruvate enters the mitochondria. Indeed, it appears that during aerobic respiration yeast mitochondria are larger and nearer the cell membrane than during anaerobic growth, possibly because this location is more efficient for oxygen uptake (Hoffmann and Avers, 1973; Jensen et al, 2000). If mitochondrial position is indeed spatially optimized in this manner it is unlikely that, after WGD, the cell would be able to rapidly adapt its mitochondria to accept any increased flux from glycolysis. Thus, the first reaction of respiration (PDH) cannot scale after dosage change in the same way that the first reaction of fermentation (PDC) does. This effect is illustrated in Figure 2C. We first added the PDH reaction to the previous model of glycolysis (see Supplementary methods for details) and then simultaneously lowered the Vmax of all of the reactions, including PDH, as described above. However, the rate of diffusion of pyruvate into the mitochondria was left unchanged. We find that, at these lower Vmax values, the relative contribution of PDH to total flux is increased (Figure 2C). This effect is independent of the kinetics of the enzymes at the branch point (data not shown). A natural result of increased glycolytic activity is thus a decreased flux through the TCA cycle relative to that through fermentation.
Hypothesis 2C—effects of single enzyme concentration changes on glycolytic flux
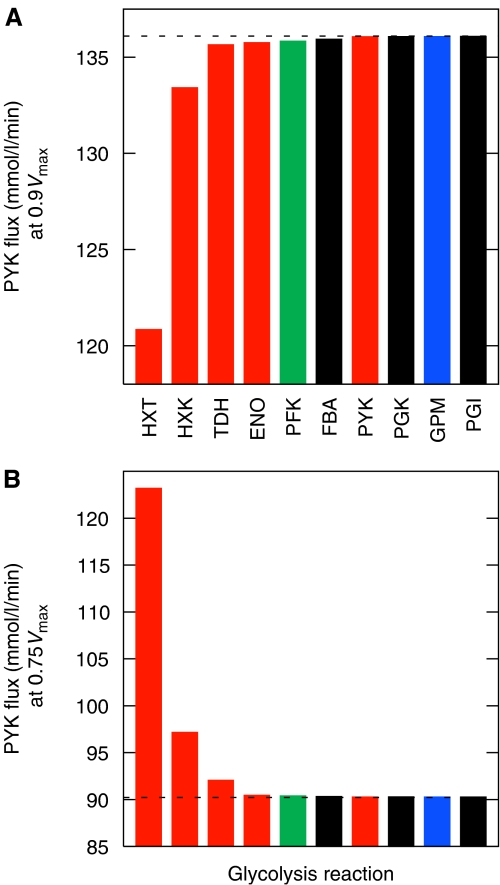
Under our first two hypotheses, the glycolytic enzymes whose genes were retained in duplicate after WGD were preserved to increase the flux through glycolysis (i.e., for dosage reasons). We would thus expect a strong relationship between whether an enzyme is present in duplicate and that enzyme's impact on flux. To test this hypothesis, we studied the effect on PYK flux of individually reducing the Vmax of all the glycolytic enzymes (and the hexose transporters) to 90% of their current values. As Figure 3A shows, the genes that remain in duplicate in all four post-WGD species (HXT, HXK/GLK, TDH, ENO and PYK) are, with the exception of PYK, also those enzymes that cause the greatest reduction in flux if their Vmax values (a proxy for concentration) are reduced. CDC19/PYK2 is an exception to this rule, probably because these enzymes are strongly induced in a feed-forward mechanism by fructose-1-6-bisphosphate (Hess and Haeckel, 1967), a metabolic intermediate whose steady-state concentration increases when the Vmax of the PYK reaction is reduced (data not shown).
Figure 3.
(A) Effect on flux through glycolysis (PYK flux) when the maximal enzymatic rates (Vmax) for the 10 relevant enzymes are individually reduced. Note that the TPI reaction is assumed to be at equilibrium and is not included in this analysis. On the x-axis are plotted the 10 reactions in question sorted in order of their effect on flux. On the y-axis is plotted the reduction in flux when Vmax is reduced by 10% for the reaction in question. Red bars indicate genes preserved in duplicate since WGD as well as the HXT genes (see text). PFK values are shown in green, as this reaction is catalyzed by a pair of more ancient duplicates. The blue bar indicates a WGD pair of enzymes in S. cerevisiae that is not maintained across all four post-WGD species (GPM). Bars in black are single-copy genes in S. cerevisiae. Flux through PYK for the unaltered model was 136.1 mmol/l/min (dashed line). (B) Effect on flux through glycolysis (PYK flux) when Vmax is first reduced to 75% of the current value for all reactions, and then individually increased to 100% of the current value for a single reaction. Thus the y-axis gives the flux through the pathway when all reactions except the one indicated have had their Vmax reduced to 75% of the current value. Reactions are shown in the same order as in panel A for comparison. The dashed line indicates the flux through PYK of 90.2 mmol/l/min seen when all enzymes have their Vmax values reduced to 75% of the current value.
It is also relevant to ask if the same result of increased glycolytic flux could be achieved by single gene duplications as opposed to by WGD. To address this question, we started with a hypothetical pre-duplication state and modeled the effect of single enzyme concentration changes. For computational reasons (see Supplementary methods) we chose to consider the effect of reducing the global Vmax values to 75% of their current value and then individually increasing each enzyme's Vmax to its present value and computing flux. As Figure 3B suggests, the hexose transporters are again the reaction that gives the largest change in flux when Vmax is changed. Nonetheless, no single enzyme change is able to produce the current flux (136.1 mmol/l/min); it is also important to note that the hexose transporters are already a large gene family: single duplications are unlikely to have the large effect on flux depicted in Figure 3B.
Hypothesis 3—yeast ecology and natural selection
Our results suggest that the post-WGD yeasts have been under selection for the rapid consumption of glucose. In the section below, we illustrate one set of circumstances under which this usage pattern is advantageous. Our analysis is inspired by elegant theoretical treatments of this topic (Pfeiffer et al, 2001; Pfeiffer and Schuster, 2005; Gilchrist et al, 2006): we consider a much-simplified model of pseudo-yeast populations growing in a defined volume of liquid (a ‘patch'), avoiding issues such as colony area, density and oxygen availability, that arise for solid cultures. This simplification also neglects the possibility of cooperation among cells for more efficient glucose usage, a phenomenon that has recently been demonstrated in the laboratory (MacLean and Gudelj, 2006). In our model, two pseudo-cells, one from each population, inoculate the patch at the same time. Growth is assumed to follow the Monod equation (Monod, 1942):
 |
where μ is the growth rate at a specific concentration of glucose ([S]) (Walker, 1998) and μmax gives the maximal growth rate given unlimited resources. Rs gives the resource concentration at which half maximal growth is seen.
We first compare two populations growing by fermentation and differing in their maximal fermentation rate. Maximal growth rates (μmaxs) were taken from van Hoek et al (1998b) as was the metabolic efficiency (the mass in grams of dry yeast cells obtained from a fixed mass of input glucose). The resource affinity Rs for respiring populations was taken from Walker (1998). Since we do not have an equivalent value for fermentatively growing yeasts, for illustrative purposes we have assumed that Rs in this case is an order of magnitude higher, similar to the difference in Km observed for the PDC and PDH enzymes (Pronk et al, 1996). Note that the exact magnitude of this difference is not critical: respiring populations will always have the selective advantage seen at lower concentrations of glucose in Figure 4B if their Rs is less than that of fermenting populations. That this is the case in real yeasts is clear from the fact that S. cerevisiae switches to respiratory growth when the concentration of glucose is sufficiently low, indicating that fermentation is not an effective growth strategy at these resource concentrations.
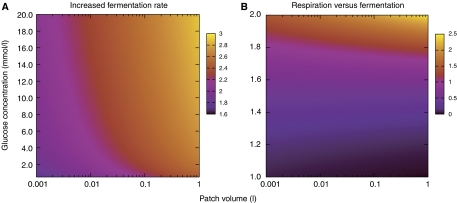
Figure 4.
Relative growth advantage of one population over another for a range of potential resource distributions. On the x-axis is the volume of a resource patch (l); on the y-axis is the concentration of glucose (mmol/l) in that patch. The total mass of glucose is the product of the axes, but note that because metabolic rate does not scale linearly with concentration, equivalent masses of glucose at differing concentrations will give rise to differing competitive advantages. The scale indicates the ratio of the cell mass for each population when resources are exhausted. Thus, values greater than 1.0 indicate regions where a rapidly fermenting population has a competitive advantage. (A) Ratio of final cell masses between two populations, one of which has a 5% advantage in maximal fermentation rate (at a cost of ∼10% loss of efficiency in terms of grams of cell mass produced per gram of glucose consumed). (B) Comparison of a respiring and a fermenting yeast population. Blue regions (ratio<1.0) correspond to conditions under which respiration is favored; orange, where fermentation is favored.
We compared two simulated yeast populations, one of which was given a 5% advantage in maximal fermentative rate (μmax) at the cost of roughly a 10% loss in efficiency per gram of glucose used. We allowed growth until the glucose concentration reached 10−6 M and then compared the ratio of the masses of the two populations as a proxy for selective advantage (Figure 4A).
We find the advantages of faster fermentation are greatest in the presence of large patches with very dense resources (Figure 4A). Although the faster fermenting yeast always has a growth advantage in our model, this is probably because no cost was imposed for increased protein synthesis in the faster fermenting strain. Thus, Figure 4A should be interpreted not to give actual selective advantages but a sense of where those advantages are likely to be the largest. Figure 4A also shows that selection will allow reduced efficiency if the result is a correspondingly greater increase in growth rate.
We also compared a fermenting population to a respiring one (Figure 4B). Once again, the greater efficiency of the respiring population imparts no direct benefit. However, because the affinity (1/Rs) for glucose of the respiring population is greater than that of the fermenting population, there is a range of resource densities for which respiration is selectively favored (ratio values <1.0; Figure 4B).
For the two competing fermenting populations (Figure 4A) the variable that determines selective advantage is the overall quantity of glucose present, while, for competition between respiring and fermenting populations, selective advantage is dependant primarily on the glucose concentration (Figure 4B). Since the quantity of glucose is function of both volume and concentration, we see an interaction of these two terms to produce the selective advantage in Figure 4A. On the other hand, for Figure 4B, the concentration will largely determine which population has the advantage, meaning that the volume of the patch mostly determines the relative magnitude of that advantage.
Taken together, Figure 4A and B can be seen to indicate that the evolutionary appearance of large, resource-dense fruits could have made the fixation of a WGD for increased glucose metabolism more favorable. The timing of the WGD in yeasts corresponds approximately with the first appearance of angiosperms in the fossil record (Crane, 1987; Wolfe and Shields, 1997). We speculate that the selective environment that existed around the time of the appearance of angiosperms may have been especially favorable to the survival of a partly polyploid yeast because a new ecological niche of glucose-rich fruits had appeared. Existing yeast species were likely not adapted to thrive in this novel environment, meaning that even with the ‘baggage' of many duplicated genes, the ancestral yeast could have had a selective advantage for the reasons shown above. Of course, further regulatory changes, which yielded S. cerevisiae's current glucose response would also have evolved, in some cases using the WGD as raw material, such as is seen with SNF3 and RTG2.
Discussion
In this work, we propose a link between whole-genome duplication, increases in enzyme concentrations and the preference of modern S. cerevisiae to ferment glucose in the presence of oxygen. We find evidence for the preferential retention of duplicate genes from WGD in the glycolytic pathway, as well as evidence that, at least for the kinetic constants measured in modern S. cerevisiae, increases in enzyme concentrations both tend to increase glycolytic flux and to favor ethanol fermentation over oxidative respiration. We also confirm previous work showing that organisms with fast but inefficient metabolisms can have a selective advantage over their more efficient kin under certain conditions. Collectively, these results tend to support the hypotheses proposed, although of course much remains to be done in order to fully understand the effect of WGD on metabolism and on glycolysis in particular.
If the above hypotheses are borne out by further analysis, they will help integrate a number of facts regarding the biology of yeasts including S. cerevisiae, such as the origins and phylogenetic distribution of the Crabtree effect (Merico et al, 2007), the evolutionary rationale for the patterns of duplicate gene retention in yeast (Kuepfer et al, 2005) and the nature of ecological competition among microbes (Pfeiffer et al, 2001; Pfeiffer and Schuster, 2005). It will also be interesting to study the effect of WGD events in other taxa on glycolysis. Duplicate copies of the glycolytic genes in vertebrates have been studied, but uncertainties in phylogenetic reconstructions involved made it difficult to determine if observed duplications among these genes owed their origins to WGDs (Steinke et al, 2006), leaving open the possibility of future analyses with gene order data to clarify the nature of the genes surviving from these WGDs.
The chief question raised by the above data is whether dosage selection for increased glycolytic flux was itself the reason for the survival of this WGD in the first place. This intrinsically attractive hypothesis is very difficult to test, and data that both support and undermine its plausibility can be found in the literature. Speaking against this possibility is the view that the uniform doubling in gene content through WGD should not change relative enzyme concentrations. Thus, tetraploids of wild-type S. cerevisiae cells have relative gene expression profiles that are essentially identical to diploid cells (Storchová et al, 2006). Other data, such as that regarding cell size, is ambiguous with respect to the idea of dosage selection. If cell volume does not also double after WGD, even identical relative gene dosages can still yield changes in absolute enzyme concentrations. Artificial tetraploid strains of S. cerevisiae actually showed more than doubling of cell volume relative to diploids, in theory implying a general decrease in enzyme concentrations (Galitski et al, 1999), which would tend to speak against our hypotheses. As a further complication, it is currently an open question whether this WGD was a true duplication of chromosomal content (autopolyploid) or a hybridization between two related species (allopolyploid). In the case of allopolyploidy, others have argued that the scaling of gene expression after hybridization will in general not be uniform and linear (Veitia, 2005).
On the other hand, there are known cases where duplications are associated with apparent selection for gene dosage. These include aneuploidies observed in vineyard and deletion mutant strains of S. cerevisiae (Bakalinsky and Snow, 1990; Guijo et al, 1997; Hughes et al, 2000) as well as in clinical and laboratory isolates of Candida albicans (Perepnikhatka et al, 1999; Wellington and Rustchenko, 2005; Selmecki et al, 2006). In the case of deletion mutants in S. cerevisiae, several aneuploid chromosomes were observed to carry a close homolog of the deleted gene and in some cases showed clear growth advantages as a result (Hughes et al, 2000). Drug-resistant strains of C. albicans carry multiple copies of chromosomes or chromosome arms where genes conferring drug-resistance reside (e.g., Perepnikhatka et al, 1999; Wellington and Rustchenko, 2005; Selmecki et al, 2006). It is also suggestive that genes that survive in duplicate from a WGD in the ciliate Paramecium tetraurelia are enriched for highly expressed genes (Aury et al, 2006). Collectively, these points argue that large-scale duplications can indeed be associated with selection for increased dosages of certain genes.
Although existing data give only mixed support to the idea of dosage selection preserving a WGD, the hypothesis has some very attractive features. One of the most important is the relatively simple nature of the changes required to produce it. We argue that even if relative gene dosages were unchanged immediately after genome duplication, the rapid gene loss that followed the WGD (Scannell et al, 2006) would have quickly altered this situation. It is reasonable to argue that those duplicate pairs that survived this loss would experience an increase in relative expression as a result. Other mechanisms of fixing gene duplications, such as neofunctionalization or subfunctionalization, require specific mutations in one or both of the duplicate genes to create a selective pressure for duplicate maintenance. However, when some fraction of genes are under selection for dosage, mutations leading to loss of ANY other genes elsewhere in the genome will be sufficient to yield a selective advantage. Since this possibility massively increases the mutational ‘target' that can yield the beneficial phenotype, it follows that such a beneficial mutation will arise much more rapidly than is the case for neo- or subfunctionalization.
Of course, it seems rather unnecessary to duplicate an entire genome in order to change the flux in a single pathway. However, we suspect that the overall picture is more complicated than we have described here, with other duplicate genes (such as the previously mentioned pair of glucose-sensing genes) preserved as part of an adaptation to growth on glucose (Özcan et al, 1996, 1998; Conant and Wolfe, 2006). For example, four enzymes of the pentose phosphate pathway also maintain WGD duplicates in S. cerevisiae. This pathway has an important role in biosynthesis, and it would be reasonable to expect that increasing growth rates would also require increasing its flux.
It is clear that this WGD was only one event in a long process of adaptation leading to modern bakers' yeast. It did, however, have lasting consequences, since a number of the duplicated genes have since evolved new or more specialized roles. For instance, the HXK and PYK gene pairs appear to have partitioned their ancestral functions to roles in high and low glucose levels (Boles et al, 1997; Rodríguez et al, 2001; Conant and Wolfe, 2006). Equally intriguingly, two glycolytic duplicates may have acquired roles in cell proliferation: i.e. in meiosis for EMI2 (Enyenihi and Saunders, 2003) or mitosis for CDC19 (Hartwell et al, 1973; Aon et al, 1995). Under our model, duplicate copies of glycolysis genes were initially maintained for dosage reasons, but subsequent tuning of enzyme expression levels may have later freed one paralog to innovate (Blanchard and Lynch, 2000).
More generally, our argument suggests that the niche inhabited by S. cerevisiae is only one of many such niches about which we know very little. To properly understand the genetics and metabolisms of yeasts, it will therefore be vital to appreciate the role played by natural selection in adapting each species to a particular niche. Behavior seen in the laboratory, under conditions very different from the wild, can then be understood in the light of this (currently unknown) ecology.
Supplementary Material
Supplementary methods
Acknowledgments
We thank K Byrne for assistance with the Yeast Gene Order Browser and M Gilchrist for insights into modeling yeast populations. We also thank B Cusack, J Gordon, N Khaldi, J Mower, D Scannell, M Sémon, M Webster, and M Woolfit for helpful discussions during preparation of this manuscript. This work was supported by Science Foundation Ireland.
References
- Aon MA, Mónaco ME, Cortassa S (1995) Carbon and energetic uncoupling are associated with block of division at different stages of the cell cycle in several cdc mutants of Saccharomyces cerevisiae. Exp Cell Res 217: 42–51 [DOI] [PubMed] [Google Scholar]
- Ashburner A (1998) Speculations on the subject of alcohol dehydrogenase and its properties in Drosophila and other flies. BioEssays 20: 949–954 [DOI] [PubMed] [Google Scholar]
- Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, Camara F, Duharcourt S, Guigo R, Gogendeau D, Katinka M, Keller AM, Kissmehl R, Klotz C, Koll F, Le Mouel A, Lepere G, Malinsky S, Nowacki M, Nowak JK, Plattner H, Poulain J, Ruiz F, Serrano V, Zagulski M, Dessen P, Betermier M, Weissenbach J, Scarpelli C, Schachter V, Sperling L, Meyer E, Cohen J, Wincker P (2006) Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444: 171–178 [DOI] [PubMed] [Google Scholar]
- Bakalinsky AT, Snow R (1990) The chromosomal constitution of wine strains of Saccharomyces cerevisiae. Yeast 6: 367–382 [DOI] [PubMed] [Google Scholar]
- Betina S, Goffrini P, Ferrero I, Wésolowski-Louvel M (2001) RAG4 gene encodes a glucose sensor in Kluyveromyces lactis. Genetics 158: 541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard JL, Lynch M (2000) Organellar genes: why do they end up in the nucleus? Trends Genet 16: 315–320 [DOI] [PubMed] [Google Scholar]
- Blank LM, Lehmbeck F, Sauer U (2005) Metabolic-flux and network analysis of fourteen hemiascomycetous yeasts. FEMS Yeast Res 5: 545–558 [DOI] [PubMed] [Google Scholar]
- Blom JM, Teixeira de Mattos MJ, Grivell LA (2000) Redirection of the respiro-fermentative flux distribution in Saccharomyces cerevisiae by overexpression of the transcription factor Hap4p. Appl Environ Microbiol 66: 1970–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux A, Hess B (1970) Allosteric properties of yeast pyruvate decarboxylase. FEBS Lett 9: 293–296 [DOI] [PubMed] [Google Scholar]
- Boles E, Schulte F, Miosga T, Freidel K, Schlüter E, Zimmermann FK, Hollenberg CP, Heinisch JJ (1997) Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1-6-biphosphate. J Bacteriol 179: 2987–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandina I, Graham J, Lemaitre-Guillier C, Entelis N, Krasheninnikov I, Sweetlove L, Tarassov I, Martin RP (2006) Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochim Biophys Acta 1757: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Todd KM, Rosenzweig RF (1998) Multiple duplications of yeast hexose-transport genes in response to selection in a glucose-limited environment. Mol Biol Evol 15: 931–942 [DOI] [PubMed] [Google Scholar]
- Bunoust O, Devin A, Avéret N, Camougrand N, Rigoulet M (2005) Competition of electrons to enter the respiratory chain: a new regulatory mechanism of oxidative metabolism in Saccharomyces cerevisiae. J Biol Chem 280: 3407–3413 [DOI] [PubMed] [Google Scholar]
- Buschlen S, Amillet J-M, Guiard B, Fournier A, Marcireau C, Bolotin-Fukuhara M (2003) The S. cerevisiae HAP complex, a key regulator of mitochondrial function, coordinates nuclear and mitochondrial gene expression. Comp Funct Genomics 4: 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH (2005) The yeast gene order browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 15: 1456–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wagner A (2002) GenomeHistory: a software tool and its application to fully sequenced genomes. Nucleic Acids Res 30: 3378–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH (2006) Functional partitioning of yeast co-expression networks after genome duplication. PLoS Biol 4: e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane PR (1987) Vegetational consequences of the angiosperm diversification. In The Origins of Angiosperms and Their Biological Consequences, Friis EM, Chaloner WG, Crane PR (eds), pp 107–144. Cambridge: Cambridge University Press [Google Scholar]
- Dekel E, Alon U (2005) Optimality and evolutionary tuning of the expression level of a protein. Nat Genet 436: 588–592 [DOI] [PubMed] [Google Scholar]
- Dong J, Dickson RC (1997) Glucose represses the lactose-galactose regulon in Kluyveromyces lactis though a SNF1 and MIG1-dependent pathway that modulates galactokinase (GAL1) gene expression. Nucleic Acids Res 25: 3657–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyenihi AH, Saunders WS (2003) Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163: 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferea TL, Botstein D, Brown PO, Rosenzweig RF (1999) Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci USA 96: 9721–9726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR (1999) Ploidy regulation of gene expression. Science 285: 251–254 [DOI] [PubMed] [Google Scholar]
- Geladé R, Van de Velde S, Van Dijck P, Thevelein JM (2003) Multi-level response of the yeast genome to glucose. Genome Biol 4: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Chun H-J, Grant A, Otto SP (2006) Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet 2: e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist MA, Sulsky DL, Pringle A (2006) Identifying fitness and optimal life-history strategies for an asexual filamentous fungus. Evolution 60: 970–979 [PubMed] [Google Scholar]
- Grieg D, Borts RH, Louis EJ, Travisano M (2002a) Epistasis and hybrid sterility in Saccharomyces. Proc R Soc Lond B Biol Sci 269: 1167–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieg D, Louis EJ, Borts RH, Travisano M (2002b) Hybrid speciation in experimental populations of yeast. Science 298: 1773–1775 [DOI] [PubMed] [Google Scholar]
- Guijo S, Mauricio JC, Salmon JM, Ortega JM (1997) Determination of the relative ploidy in different Saccharomyces cerevisiae strains used for fermentation and ‘Flor' film ageing of dry sherry-type wines. Yeast 13: 101–117 [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Mortimer RK, Culotti J, Culotti M (1973) Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics 74: 267–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Haeckel R (1967) Interaction between potassium-, ammonium- and frutose-1, 6-diphosphate activation of yeast pyruvate kinase. Nature 214: 848–849 [DOI] [PubMed] [Google Scholar]
- Hittinger CT, Rokas A, Carroll AS (2004) Parallel inactivation of multiple GAL pathway genes and ecological diversification in yeasts. Proc Natl Acad Sci USA 101: 14144–14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HP, Avers CJ (1973) Mitochondrion of yeast: ultrastructural evidence for one giant, branched organelle per cell. Science 181: 749–751 [DOI] [PubMed] [Google Scholar]
- Hübner G, Weidhase R, Schellenberger A (1978) The mechanism of substrate activation of pyruvate decarboxylase: a first approach. Eur J Biochem 92: 175–181 [DOI] [PubMed] [Google Scholar]
- Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, Friend SH, Marton MJ (2000) Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet 25: 333–337 [DOI] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–690 [DOI] [PubMed] [Google Scholar]
- Jensen RE, Hobbs AE, Cerveny KL, Sesaki H (2000) Yeast mitochondrial dynamics: fusion, division, segregation, and shape. Microsc Res Tech 51: 573–583 [DOI] [PubMed] [Google Scholar]
- Johnston M, Kim J-H (2005) Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem Soc Trans 33: 247–252 [DOI] [PubMed] [Google Scholar]
- Kacser H, Burns JA (1973) The control of flux. Symp Soc Exp Biol 27: 65–104 [PubMed] [Google Scholar]
- Käppelli O (1986) Regulation of carbon metabolism in Saccharomyces cerevisiae and related yeasts. Adv Microb Physiol 28: 181–209 [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624 [DOI] [PubMed] [Google Scholar]
- Koszul R, Caburet S, Dujon B, Fischer G (2004) Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J 23: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresze GB, Ronfit H (1981) Pyruvate dehydrogenase complex from baker's yeast. 1. Purification and some kinetic and regulatory properties. Eur J Biochem 119: 573–579 [DOI] [PubMed] [Google Scholar]
- Kuepfer L, Sauer U, Blank LM (2005) Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res 15: 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwast KE, Lai L-C, Menda N, James DT III, Aref S, Burke PV (2002) Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J Bacteriol 184: 250–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Louis EJ (2005) Yeast evolution and comparative genomics. Annu Rev Microbiol 59: 135–153 [DOI] [PubMed] [Google Scholar]
- MacLean RC, Gudelj I (2006) Resource competition and social conflict in experimental populations of yeast. Nature 441: 498–501 [DOI] [PubMed] [Google Scholar]
- Merico A, Sulo P, Piškur J, Compagno C (2007) Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. FEBS J 274: 976–989 [DOI] [PubMed] [Google Scholar]
- Møller K, Olsson L, Piškur J (2001) Ability for anaerobic growth is not sufficient for development of the petite phenotype in Saccharomyces kluyveri. J Bacteriol 183: 2485–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J (1942) Recherches sur la croissance des cultures bactériennes. Paris: Hermann & cie [Google Scholar]
- Ohno S (1970) Evolution by Gene Duplication. New York: Springer [Google Scholar]
- Otterstedt K, Larsson C, Bill RM, Ståhlberg A, Boles E, Hohmann S, Gustafsson L (2004) Switching the mode of metabolism in the yeast Saccharomyces cerevisiae. EMBO Rep 5: 532–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Dover J, Johnston M (1998) Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J 17: 2566–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Dover J, Rosenwald AG, Wölfl S, Johnston M (1996) Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA 93: 12428–12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD (2004) Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature 429: 661–664 [DOI] [PubMed] [Google Scholar]
- Perepnikhatka V, Fischer FJ, Niima M, Baker RA, Cannon RD, Wang Y-K, Sherman F, Rustchenko E (1999) Specific chromosome alterations in fluconazole-resistant mutants of Candida albicans. J Bacteriol 181: 4041–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T, Schuster S (2005) Game-theoretical approaches to studying the evolution of biochemical systems. Trends Biochem Sci 30: 20–25 [DOI] [PubMed] [Google Scholar]
- Pfeiffer T, Schuster S, Bonhoeffer S (2001) Cooperation and competition in the evolution of ATP-producing pathways. Science 292: 504–507 [DOI] [PubMed] [Google Scholar]
- Piškur J, Langkjær RB (2004) Yeast genome sequencing: the power of comparative genomics. Mol Microbiol 53: 381–389 [DOI] [PubMed] [Google Scholar]
- Piškur J, Rozpedowska E, Polakova S, Merico A, Compagno C (2006) How did Saccharomyces evolve to become a good brewer? Trends Genet 22: 183–186 [DOI] [PubMed] [Google Scholar]
- Pritchard L, Kell DB (2002) Schemes of flux control in a model of Saccharomyces cerevisiae glycolysis. Eur J Biochem 269: 3894–3904 [DOI] [PubMed] [Google Scholar]
- Pronk JT, Steensma HY, van Dijken JP (1996) Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12: 1607–1633 [DOI] [PubMed] [Google Scholar]
- Rodríguez A, de la Cera T, Herrero P, Moreno F (2001) The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HKX2 genes of Saccharomyces cerevisiae. Biochem J 355: 625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Uemura H (2005) Influence of low glycolytic activities in gcr1 and gcr2 mutants on the expression of other metabolic pathway genes in Saccharomyces cerevisiae. Yeast 22: 111–127 [DOI] [PubMed] [Google Scholar]
- Sauro HM, Hucka M, Finney A, Wellock C, Bolouri H, Doyle J, Kitano H (2003) Next generation simulation tools: the systems biology workbench and BioSPICE integration. OMICS 7: 355–372 [DOI] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH (2006) Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440: 341–345 [DOI] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J (2006) Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313: 367–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C, Wolfe KH (1999) Yeast genome evolution in the post-genome era. Curr Opin Microbiol 2: 548–554 [DOI] [PubMed] [Google Scholar]
- Smits HP, Hauf J, Müller S, Hobley TJ, Zimmermann FK, Hahn-Hägerdal B, Nielsen J, Olsson L (2000) Simultaneous overexpression of enzymes of the lower part of glycolysis can enhance the fermentative capacity of Saccharomyces cerevisiae. Yeast 16: 1325–1334 [DOI] [PubMed] [Google Scholar]
- Steinke D, Hoegg S, Brinkmann H, Meyer A (2006) Three rounds (1R/2R/3R) of genome duplications and the evolution of the glycolytic pathway in vertebrates. BMC Biol 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová Z, Breneman A, Cande J, Dunn J, Burbank K, O'Toole E, Pellman D (2006) Genome-wide genetic analysis of polyploidy in yeast. Nature 443: 541–547 [DOI] [PubMed] [Google Scholar]
- Teusink B, Passarge J, Reijenga CA, Esgalhado E, van der Weijden CC, Schepper M, Walsh MC, Bakker BM, van Dam K, Westerhoff HV, Snoep JL (2000) Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur J Biochem 267: 5313–5329 [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2000) Gene ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald U, Mailinger W, Baltes M, Rizzi M, Reuss M (1997) In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae: I. Experimental observations. Biotechnol Bioeng 55: 305–316 [DOI] [PubMed] [Google Scholar]
- Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, Benner SA (2005) Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet 37: 630–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoek P, Flikweert MT, van der Aart QJM, Steensma HY, van Dijken JP, Pronk JT (1998a) Effects of pyruvate decarboxylase overproduction on flux distribution at the pyruvate branch point in Saccharomyces cerevisiae. Appl Environ Microbiol 64: 2133–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoek P, van Dijken JP, Pronk JT (1998b) Effect of specific growth rate on fermentative capacity of baker's yeast. Appl Environ Microbiol 64: 4226–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Urk H, Schipper D, Breedveld GJ, Mak PR, Scheffers WA, van Dijken JP (1989) Localization and kinetics of pyruvate-metabolizing enzymes in relation to aerobic alcoholic fermentation in Saccharomyces cerevisiae CBS 8066 and Candida utilis CBS 621. Biochim Biophys Acta 992: 78–86 [DOI] [PubMed] [Google Scholar]
- Veitia RA (2005) Paralogs in polyploids: one for all and all for one? Plant Cell 17: 4–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser W, Scheffers WA, Batenburg-van der Vegte WH, van Dijken JP (1990) Oxygen requirements of yeasts. Appl Environ Microbiol 56: 3785–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A (2000) Inferring lifestyle from gene expression patterns. Mol Biol Evol 17: 1985–1987 [DOI] [PubMed] [Google Scholar]
- Wagner A (2005) Energy constraints on the evolution of gene expression. Mol Biol Evol 22: 1365–1374 [DOI] [PubMed] [Google Scholar]
- Walker GM (1998) Yeast Physiology and Biotechnology. Chichester: John Wiley and Sons [Google Scholar]
- Wellington M, Rustchenko E (2005) 5-Fluoro-orotic acid induces chromosome alterations in Candida albicans. Yeast 22: 57–70 [DOI] [PubMed] [Google Scholar]
- West GB, Brown JH, Enquist BJ (1999) The fourth dimension of life: fractal geometry and allometric scaling of organisms. Science 284: 1677–1679 [DOI] [PubMed] [Google Scholar]
- Wolfe KH (2004) Evolutionary genomics: yeasts accelerate beyond BLAST. Curr Biol 14: R392–R394 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713 [DOI] [PubMed] [Google Scholar]
- Zeeman AM, Luttik MAH, Thiele C, van Dijken JP, Pronk JT, Steensma HY (1998) Inactivation of the Kluyveromyces lactis KIPDA1 gene leads to loss of pyrvuate dehydrogenase activity, impairs growth on glucose and triggers aerobic alcoholic fermentation. Microbiology 144: 3437–3446 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods