Abstract
Four full-length and one partial cDNA clones encoding four different A-type cyclins were isolated from a tobacco S-phase-specific library. The corresponding mRNAs displayed sequential appearance and disappearance during the cell cycle of highly synchronized suspension-cultured tobacco cells. Sequence analysis showed that the plant A-type cyclins can be subdivided into three distinct structural groups that are likely to be represented in every plant species. Two of the isolated tobacco cyclins belonging to the same group were highly expressed throughout S and G2 phases but showed different kinetics of induction at the G1/S transition. Another one belonging to a second group was induced at mid-S phase and expressed until mid-M phase. A similar expression pattern was previously reported for a tobacco cyclin belonging to the third group. This sequential expression of multiple A-type cyclins in one type of plant cells makes a clear distinction from the situation in animal cells in which only one A-type cyclin exists in a given species. Furthermore, the expression of the different A-type cyclin genes responded differently upon a block at mid-S phase by DNA synthesis inhibitors. These results suggest that the multiple A-type cyclins act at different steps of the plant cell cycle and, therefore, exert distinct functions. In contrast, the expression of B-type cyclins was restricted to a narrow window corresponding to the M phase.
Keywords: tobacco suspension, synchronized cells
Our knowledge of the eukaryotic cell cycle has progressed considerably during the last few years, by the finding that protein kinases play a central role in the cell cycle. The activity of the protein kinases depends on the association between a cyclin-dependent serine/threonine kinase (cdk) as the catalytic moiety and a cyclin as a regulatory subunit. The activity of the complexes is further regulated through phosphorylation/dephosphorylation mechanisms and binding to inhibitors (1). In animal cells, distinct cdks associating sequentially with different cyclins determine the substrate specificity of the complexes and monitor the cell cycle transitions at the G1/S and G2/M major checkpoints (2, 3). These sequential associations result from stage-specific synthesis and degradation of the cyclins while the catalytic subunit is present throughout the cell cycle. The animal cyclins have been subdivided into mitotic (A- and B-type) and G1 (C-, D-, and E-type) cyclins according to sequence homologies and expression patterns. While the G1 cyclins are involved in progression through G1 phase and at the G1/S transition, the mitotic A- and B-type cyclins are essential in progression through G2 phase and at the G2/M transition. In addition, the A-type cyclin has also been shown to play essential functions in DNA replication during S phase (4, 5, 6).
It has been recently shown that the basic components of the cell cycle machinery are similar in plants and animals (7). cdk-like cDNAs and genes have been isolated from several mono- and dicotyledonous plants and shown to share about 60% sequence homology with animal cdc2 and cdk2. Using polymerase chain reaction (PCR), reduced-stringency hybridization, or complementation of yeast mutants, cDNAs corresponding to plant mitotic cyclins have also been isolated from several plants (7, 8). More recently isolation of cDNAs corresponding to G1 cyclins (D-type) has been reported from two plant species (9, 10). However, due to the lack of a highly synchronizable plant system, assignment of plant cyclin expression to a particular stage of the cell cycle has only been roughly determined, essentially by using inhibitors arresting the cell cycle at different points or by differential in situ hybridization patterns.
Herein we report the isolation of four full-length and one partial cDNAs corresponding to four different A-type cyclins from the highly synchronizable tobacco BY2 cells. We show that the different A-type cyclins expressed sequentially during S and G2 phases of an unperturbed cell cycle and displayed different responses toward inhibitors of DNA replication, suggesting their different functional implications during cell cycle progression. Moreover, expression of two B-type cyclins appeared to be strictly limited to the M phase.
MATERIALS AND METHODS
Cloning of Tobacco Cyclins.
PCR fragments corresponding to mitotic cyclins were obtained from cDNA from exponentially growing suspension-cultured tobacco cells by using degenerate primers in the cyclin box corresponding to amino acid sequences M(R/S)AI(L/F)(V/I/M)DW and KYEE(I/M)Y(A/P/S/T)P. A BY2 S-phase-specific cDNA library was constructed from 1 μg of poly(A)+ RNA in the vector λZAPII (Stratagene) and screened with the PCR fragments corresponding to A-type cyclins. DNA inserts were excised according to the protocol from the manufacturer and sequenced by the dideoxynucleotide method (11).
Synchronization and Analysis of Cyclin Gene Expression.
Synchronization of tobacco BY2 (Nicotiana tabacum L. cv. Bright Yellow 2) suspension-cultured cells was achieved by a 24-hr subculture of stationary phase cells (7 days old) in a medium containing aphidicolin (Sigma; 3–6 mg/liter) followed by extensive washes, as described (12). Measurement of DNA synthesis, mitotic index, and isolation of RNA were as in ref. 13. Total RNA (25 μg) was electrophoresed on formaldehyde/agarose gels and transferred to Hybond-N membranes (Amersham). Hybridization was under standard high-stringency conditions (11). Transcript levels were quantified from the blots using a PhosphorImager (Molecular Dynamics).
RESULTS
Isolation of Tobacco A-Type Cyclin cDNAs.
Five distinct DNA fragments were produced by PCR amplification of tobacco cDNAs from exponentially growing N. tabacum suspension-cultured cells using degenerate oligonucleotides corresponding to conserved motifs within the cyclin box of mitotic cyclins. The deduced amino acid sequences of three of them were found homologous to the cyclin box of A-type cyclins, and the other two to those of B-type cyclins (data not shown). The fragments corresponding to A-type cyclins were used to screen a tobacco BY2 S-phase-specific cDNA library. Each of the three probes identified 7–15 positive clones from about 4 × 105 plaques screened, among which several had the same sequence but exhibited variation in length up to 100 nt in their 3′ regions. According to sequence conservation, the cDNA clones could be classified into four families. Each family contained two different cDNA sequences that shared more than 90% sequence identity at the 5′ ends and that might represent allelic variants of the same protein, since N. tabacum is an amphidiploid genome resulting from the combination of the genomes of Nicotiana sylvestris and Nicotiana tomentosiformis. The two cDNAs were later confirmed by Southern blot analysis to originate each from one of the ancestral genomes (data not shown).
cDNAs NtcycA59 and NtcycA105, representing two different families, and NtcycA19 and NtcycA30, two alleles both belonging to the third family were completely sequenced. The cDNAs were, respectively, 1665, 1666, 1907, and 1843 bp long and contained open reading frames (ORFs) encoding proteins of 384, 371, 483, and 482 aa long, respectively. In-frame termination codons were present upstream of the translation initiation codons of all these ORFs. In NtcycA59 cDNA, however, a second methionine codon present 9 aa downstream of the first one was embedded in sequences showing high similarity to the plant start codon consensus sequence (14). Thus, NtcycA59 cDNA may only encode a protein of 374 aa rather than 384 aa. The cDNA insert of NtcycA13, which represents the fourth family, was found to be chimeric and thus the sequence of the cyclin cDNA part was only 1072 bp long and encoded 314 aa of the N terminus of the protein.
The amino acid sequences of the five cyclins are presented in Fig. 1. As previously noticed for cyclins of other plants, the cyclin boxes showed segments homologous to both the consensus of animal A and B cyclins. However, analysis of the overall homology level clearly indicated that the cyclins described herein belong to the class II of plant cyclins, which, according to phylogeny, has been proposed as the plant counterpart of the animal A cyclins (15, 16). Sequence homology was high (around 95%) between cyclins NtcycA19 and NtcycA30, the two cyclins originating from the two ancestral genomes. Two A-type cyclin cDNAs have recently been reported from another tobacco line (17), of which one encodes a cyclin (Ntcyc27) different from those presented here (Fig. 1). The other cyclin (Ntcyc25) shows extensive sequence homology (99%) to NtcycA19, suggesting that they are allelic variants of the same cyclin in the two different tobacco lines and that their differences correspond to the evolution of the N. tabacum genome.
Figure 1.

Amino acid comparison of tobacco A-type cyclins. The tobacco cyclin sequences presented in this paper are aligned with a previously published tobacco A-type cyclin (Ntcyc27; ref. 17) and with consensus sequences of the cyclin boxes of A- and B-type cyclins (above and below the sequences, respectively). Destruction boxes are shaded.
Phylogenetic analysis of the sequences of 20 cyclins belonging to class II so far isolated from plants indicates that they fall into three groups that were named CycA1, CycA2, and CycA3 (Fig. 2; ref. 16). It is interesting to note that representative members of each group were found in tobacco as well as in soybean (18), the two plants from which the highest number of A-type cyclins has been identified. This suggests that each group might be represented in every plant species and, therefore, possibly fulfill distinct functions. The consensus sequence of the destruction box differs from one group to another: relatively loose in A1 (RxxLxNL/IxN), moderate in A2 (RAV/ILxDxxN), and highly conserved in A3 (RVVLGEL/IxN). The position of the destruction box also varies, CycA2 having the longest region upstream of the destruction box (about 20 aa longer than in the other groups) and cycA3 having the shortest region between the destruction box and the cyclin box (60–80 aa shorter). These characteristics suggest specific degradation mechanisms to exist for each group.
Figure 2.

Phylogenetic tree of plant A-type (class II) cyclins. The tree was constructed from an alignment of 250 residues of the cyclin core, using the must package. The three groups CycA1, -A2, and -A3 are shown. The GenBank accession numbers of the cyclin sequences are indicated. For more detailed sequence information, see ref. 16.
The two DNA fragments homologous to B-type cyclins did not cross-hybridize and were found to belong to classes I and III of plant cyclins, the plant counterparts of animal and yeast cyclin B (15).
Genomic Organization of the A-Type Cyclin Genes.
Both the entire cDNAs and the PCR fragments corresponding to the cyclin box of the tobacco A-type cyclins led to similar results when used as probes in DNA gel blot analyses. Those obtained using the entire cDNAs as probes are shown in Fig. 3. The discrete patterns of hybridization indicate that the cyclins belonging to the three different families are not tandemly arranged in the genome and do not cross-hybridize to each other under standard high-stringency conditions. The hybridization patterns revealed one or two major bands with each cyclin probe. This is in agreement with the fact that two highly homologous cDNAs, each originating from one of the two ancestors of N. tabacum, have been cloned for each family of cyclins. In addition to the major bands, some weak hybridization bands were also observed, indicating that additional related genes may exist in the genome.
Figure 3.
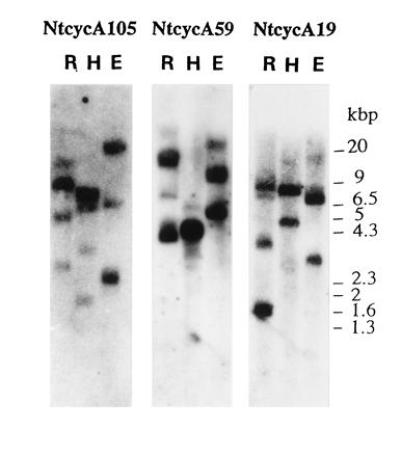
DNA gel blots of A-type tobacco cyclin genes. Restriction digests (E, EcoRI; H, HindIII; R, EcoRV) of tobacco genomic DNA (10 μg) were analyzed by Southern blot hybridization with the indicated cDNAs as probes.
Expression Pattern of the Cyclin Genes During the Cell Cycle.
Suspension-cultured tobacco BY2 cells were synchronized by a 24-hr treatment with aphidicolin, an inhibitor of DNA polymerases, as described (12). After release from the aphidicolin block, DNA synthesis, mitotic index, and cyclin mRNA levels were monitored over almost two complete cell cycles (Fig. 4). As shown in Fig. 4A, a high proportion of cells proceeded through the different phases of the cell cycle in a synchronous manner. As previously shown, the expression of the histone H4 gene used as a marker of S phase was parallel to the DNA synthesis (Fig. 4B; ref. 13). In agreement with the size of the isolated cDNA, a unique hybridization band at about 1.7 kb was detected for cyclin NtcycA59. Surprisingly, two hybridization bands of which only the largest one corresponded to the size of the isolated cDNA were observed when NtcycA19 or NtcycA105 were used as probes. Hybridization bands corresponding to unidentified RNA of smaller size were also previously noticed for some types of cyclins (19). The accumulation patterns of NtcycA59 and NtcycA105 mRNAs were quite similar to that of the histone H4, except that their high steady-state level extended from S phase until the end of G2 phase whereas that of the histone mRNA was restricted to S phase. The NtcycA19 mRNA amount was low after the aphidicolin treatment, began to rise in mid-S phase, peaked in G2 phase, and declined at the end of mitosis. When either of the two PCR fragments corresponding to the cyclin box of B-type cyclins were used as probes, mRNAs of about 2 kb long referred to as NtcycB were detected within a rather narrow window of expression coinciding with the mitotic phase. In agreement with previous studies (20), the level of the mRNA encoding the translation elongation factor (EF-1α) remained relatively constant throughout the cell cycle, thus providing an internal loading control.
Figure 4.
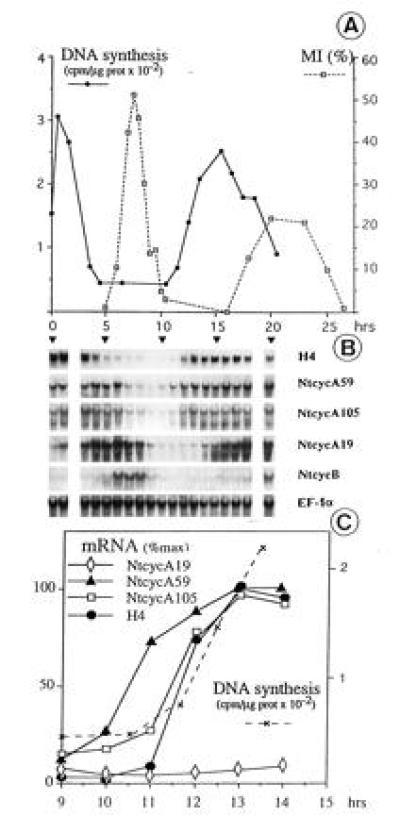
Accumulation patterns of tobacco cyclin mRNAs during the cell cycle. Cells were synchronized as described (12). (A) DNA synthesis and mitotic index (MI) were measured. (B) RNA (25 μg) gel blots were hybridized with the indicated cDNA probes. For cyclins B, hybridization was with the two cloned PCR fragments. As the same pattern was observed with these two probes, only one is presented. RNA lanes are aligned below the corresponding sampling times in A; RNA were not analyzed at points for 2 and 19 hr. (C) mRNA levels were quantitated throughout the G1/S transition of the second cycle shown in B and plotted relative to their maximal level in the second S phase. Large and small mRNAs for NtcycA105 display identical kinetics and thus only one is presented.
To locate more precisely the time points of transcriptional activation at the onset of S phase, the mRNA levels of the A-type cyclins as well as that of the histone gene were quantitatively analyzed during the transition from G1 to early S phase of the second cell cycle (Fig. 4C). The induction of NtcycA105 (both the large and short mRNAs) was parallel to both the induction of the histone gene and the onset of DNA synthesis. Interestingly, the induction of NtcycA59 was earlier and somewhat preceded the onset of DNA synthesis. This quantitative analysis also confirmed that NtcycA19 mRNA amount was low in G1 and early S phases.
Response Pattern of Cyclin Genes to Inhibitors of DNA Replication.
To further investigate relationships between cyclin gene expression and DNA replication, mRNA levels of cyclin genes were studied from cells treated with the two DNA synthesis inhibitors, aphidicolin, which was previously used at a moderate dose in the synchronization procedure, and hydroxyurea, which inhibits the synthesis of dNTPs by interfering with the ribonucleotide reductase (21). After a 24-hr treatment of stationary-phase cells subcultured in fresh medium containing whatever inhibitor, NtcycB transcripts were hardly detectable (data not shown), thus confirming that their synthesis occurs later in the cell cycle. The aphidicolin treatment resulted in accumulation of NtcycA59 and NtcycA105 transcripts (Fig. 5) and the amount of these transcripts remained constant regardless of the concentration of aphidicolin used, even that which completely inhibits the DNA synthesis. In contrast, the mRNA amount was reduced in proportion to the DNA synthesis rate upon increasing concentrations of hydroxyurea (Fig. 5). This behavior toward the two inhibitors was identical to that previously reported for histone genes and assumed to result from the arrest of the cell cycle by the two inhibitors at two different points, one (hydroxyurea) blocking before and the other (aphidicolin) blocking after the point of induction of these genes (13). The basal level of NtcycA19 mRNAs was unaffected by the aphidicolin treatment but decreased upon hydroxyurea treatment (Fig. 5). This might indicate a two-step mechanism for the regulation of the NtcycA19 mRNA abundance, one operating at the entry of cells into the cell cycle and providing a basal level of transcripts in cycling cells and the other one operating at mid-S phase and leading to a considerable increase in the amount of transcripts (see Fig. 4B). These two levels of regulation may result from either transcriptional or posttranscriptional mechanisms.
Figure 5.
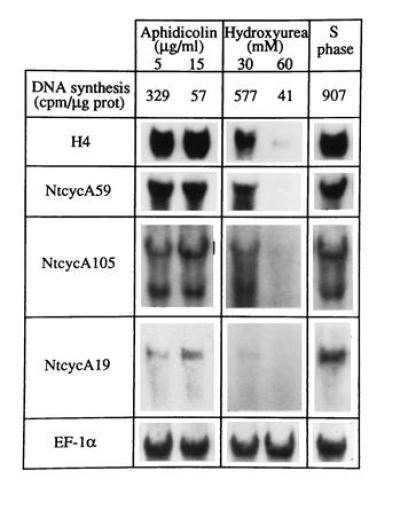
Response patterns of tobacco A-type cyclin mRNAs toward an arrest of the cell cycle. Stationary-phase cells were subcultured for 24 hr in presence of the indicated concentrations of aphidicolin or hydroxyurea. The DNA synthesis rate was then determined. Part of the cells were released from an aphidicolin block (5 μg/ml) and collected 1 hr after the release (S phase). RNA (25 μg) gel blots were hybridized with the indicated cDNA probes.
The effect of both inhibitors was also investigated when added at mid-S phase of the synchronized cells. No qualitative differences were observed between the two inhibitors and thus only the results obtained with aphidicolin are presented in Fig. 6. The addition of aphidicolin rapidly and efficiently blocked DNA synthesis. The increase of NtcycA19 mRNA level usually occurring from mid-S phase onward did not occur under these conditions, thus indicating that the signal directing mRNA accumulation takes place during the second part of S phase and is dependent on ongoing DNA replication. The decrease of H4 histone mRNA amount usually observed at the end of S phase in the unperturbed cell cycle no longer occurred in aphidicolin-blocked cells. Experiments extending further than that shown in Fig. 6 showed that the drop of NtcycA105 transcripts usually observed at the onset of mitosis in control cells was prevented by the aphidicolin block, like for the histone genes (data not shown). This suggests that the signals that govern the arrest of NtcycA105 and histone mRNA accumulation are impaired by a block of DNA synthesis. Interestingly, the amount of the early expressed cyclin NtcycA59 mRNA was enhanced. Thus, these results suggest the existence of feed-back mechanisms controlling the mRNA abundance of at least some of these cyclin genes.
Figure 6.
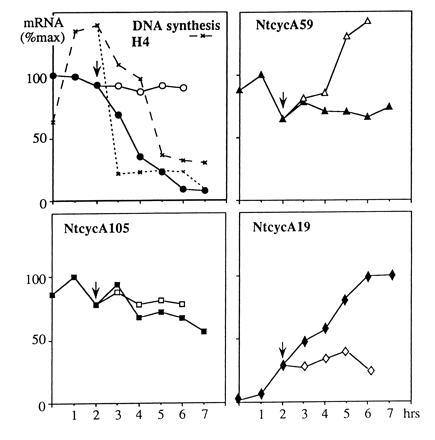
Response pattern of tobacco A-type cyclin mRNAs toward an arrest of ongoing DNA synthesis. Aphidicolin (20 mg/liter) was added in the middle of the first S phase (arrowhead) of synchronized cells. DNA synthesis is indicated for control (dashed line) and blocked (dotted line) cells. mRNA levels were monitored by quantification of RNA gel blots in control cells (solid symbols) and after the addition of the inhibitor (open symbols) and plotted relative to their maximal level in the control cells.
DISCUSSION
The cyclins described herein fall into the class II of plant cyclins that has been renamed recently class CycA for their close phylogenic relationship with the animal A-type cyclins (15, 16). From the four full-length and one partial cDNAs characterized (Fig. 1), two of them (NtcycA19 and NtcycA30) represented allelic variants originating from the two ancestors of N. tabacum and encoded highly homologous proteins. The NtcycA19 cDNA was found to be the equivalent in the tobacco BY2 line of the clone Ntcyc25 previously isolated from another tobacco line (17). The other cDNAs, NtcycA13, NtcycA59, and NtcycA105 encoded different and novel cyclins. Thus, with another cyclin (Ntcyc27) previously characterized in tobacco (17), five distinct A-type cyclins have been found so far in N. tabacum. This number may be even an underestimate of the reality as indicated by additional related genes cross-hybridizing with the cyclin probes on DNA gel blots (Fig. 3). Multiple A-type cyclins have also been identified in other plant species, although less extensively than in tobacco (15, 18, 22, 23). Thus, the presence of multiple A-type cyclins in one single species seems to be a general feature in plants. This is different from animals where only one A-type cyclin exists in a given species.
Phylogenetic analysis indicated that the plant A-type cyclins thus far isolated could be classified into three groups named CycA1, CycA2, and CycA3 (Fig. 2). The destruction boxes are clearly different from one group to another, at both the level of the consensus sequences and their positions relative to the cyclin-box (see Results for details), suggesting that distinct mechanisms of protein degradation may exist for each group. The tobacco cyclins cover all three groups: CycA1 (Ntcyc25 = NtcycA19 = NtcycA30), CycA2 (Ntcyc27), and CycA3 (NtcycA59, NtcycA105, and NtcycA13). Members of the three groups have also been isolated from glycine max (Fig. 2; ref. 18), thus, suggesting that these three structurally distinct groups might be represented in all the plant species and, consequently, might play different functions.
This suggestion is partially supported by the expression profile of the different cyclins during the cell cycle, as judged from mRNA accumulation. In the CycA3 group, the genes NtcycA59 and NtcycA105 were found to be both induced at the G1/S transition and highly expressed during S and G2 phases (Fig. 4B). The gene NtcycA19 (Ntcyc25) belonging to the CycA1 group was found to be induced later, at mid-S phase, peaked in G2 phase and the mRNA disappeared at the end of mitosis (Fig. 4B; ref. 17). In agreement with these results, in situ hybridization in G. max also revealed that the CycA3 group cyclin cyc1Gm is expressed earlier than the CycA1 group cyclin cyc3Gm (18). The expression profile of the tobacco cyclin gene Ntcyc27 belonging to the CycA2 group looked very similar to that of the CycA1 group (17). This expression profile seems to result mainly from a transcriptional regulation, since the promoter of an Arabidopsis cyclin belonging to the CycA2 group directed accumulation of GUS transcripts with similar kinetics (24). To better understand the mechanisms controlling plant cyclin expression, it would be necessary to study other putative levels of regulation such as control of mRNA stability, mRNA translation and protein stability. Nevertheless, if one assumes that the cyclin mRNA abundance may be taken as a rough estimate of the amount of the corresponding protein, our results indicate that CycA1 and CycA2 groups may act at the same step of the cell cycle, while CycA3 group acts at a much earlier step. The expression profile of the CycA3 group during the cell cycle appears very similar to that described for the mammalian A cyclin and consequently cyclins of this group might fulfill the dual functions in both DNA replication and G2/M transition, as described in animal cells. Surprisingly, the expression profile of CycA1 and CycA2 cyclins is intermediate between that of the animal A and B cyclins and the corresponding cyclins might thus play functions at S/G2 and G2/M transitions in plants. On the other hand, a more precise analysis indicated that, although belonging to the same group CycA3, the accumulation of the transcripts of NtcycA59 and NtcycA105 were not induced exactly at the same time point during the G1/S transition (Fig. 4C). Ntcyc105 mRNAs increased in parallel to histone transcripts and DNA replication whereas NtcycA59 mRNA did earlier, suggesting that these two cyclins may also fulfill different functions, NtcycA59 being possibly involved in the control of the entry in S phase and Ntcyc105 in the progression through S phase.
The hypothesis attributing different functions to these A-type cyclins according to the accumulation profiles of the corresponding mRNAs was further supported by the differential responses obtained upon an arrest of S phase by treatment with inhibitors of DNA synthesis. These responses revealed that different feed-back control mechanisms exist for the different cyclin genes, the steady-state level of NtcycA59 mRNAs increasing in response to the arrest whereas those of NtcycA105 and NtcycA19 remained constant. These different responses may be interpreted accordingly with the above suggested functions in that (i) enhanced expression of cyclin NtcycA59, which is assumed to play a function at the entry of S phase, would allow cells to overcome the S phase blockage; (ii) maintenance of NtcycA105 (as well as histones), which may play a role in the progression through S phase (like histones), would ensure the requirements for the completion of S phase; and (iii) prevention of the accumulation of NtcycA19, which may play a function at S/G2 and G2/M transitions, would allow the completion of S phase before initiating premature mitosis.
In our study, we have used two PCR fragments as B-type cyclin probes, which are representative of the two groups of plant cyclins B, CycB1 and CycB2 (16). One of them corresponds to the Ntcyc1 cDNA already described in tobacco (25). Thus, with the other isolated cycB1 clone Ntcyc29 (17), at least three different B-type cyclins exist in tobacco. In contrast to the sequential expression pattern observed for the different A-type cyclins, all three B-type cyclin mRNAs were found to accumulate exclusively during mitosis (Fig. 4B; ref. 17). Intriguingly, the transcriptional rate directed by a Arabidopsis B-type cyclin promoter increased earlier, at the S/G2 transition (24), which might indicate an involvement of posttranscriptional mechanisms in regulating B-type cyclin mRNA stability during G2 phase or a differential regulation of some cyclin genes within the CycB groups. The narrow window of expression we observe at the mRNA level clearly differs from the expression pattern of animal B-type cyclins that are synthesized in late S phase but whose association in the complex with the Cdc2 kinase is rendered inactive by phosphorylation throughout G2 phase until G2/M transition (2). This result suggests that, rather than being regulated by phosphorylation as in animal cells, the activity of some cycB/Cdc2 complexes might be directly regulated by the transcriptional timing of cycB gene expression in plants. This hypothesis requires further support from studies at the protein level, but it is in agreement with the finding that mutations introduced in the potential phosphorylation site of cdc2a do not affect plant development nor entry into mitosis (26). Alternatively, one might wonder if some of the multiple A-type cyclins in plants could not functionally play a role similar to that of the animal B-type cyclins at particular stages of the cell cycle, since their expression profile, especially for the late expressed A-type plant cyclins (Cyc A1 and A2 groups) shows similarities to that of the animal B cyclins. Moreover, it has been well documented at the sequence level that the plant A-type cyclins carry not only some consensus motifs of the animal A cyclins but also some motifs specific to the B cyclins (Fig. 1; refs. 15, 16, and 22).
Our findings and the fact that only a few cdks have been found in a given plant species (7) suggest that plants may have taken a different way than animals to diversify their cdk–cyclin complexes. In animals, only one A-type cyclin has been found in a given species and this cyclin has been shown to exert different functions upon association with different cdks (2) whereas in plants, different A-type cyclins might associate with a same cdk to form different cdk–cyclin complexes. Studies will be necessary at the level of the proteins and the cdk–cyclin kinase activities at different steps of the cell cycle to ascertain correspondence between mRNA and protein accumulation and to acquire a better insight into the differential functions of the multiple A-type cyclins in plants.
Acknowledgments
We thank Bernadette Clément and Martine Flénet for skillful technical assistance. J.P.R. was supported by Rhône-Poulenc. We thank Tobacco Science Research Laboratory, Japan Tobacco, Inc., for allowing us to use the BY2 cell suspension.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
References
- 1.Jacobs T. Dev Biol. 1992;153:1–15. doi: 10.1016/0012-1606(92)90087-w. [DOI] [PubMed] [Google Scholar]
- 2.Pines J. Trends Biol Sci. 1993;18:195–197. doi: 10.1016/0968-0004(93)90185-p. [DOI] [PubMed] [Google Scholar]
- 3.King R W, Jackson P K, Kirschner M W. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 4.Girard F, Strausfeld U, Fernandez A, Lamb N J C. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- 5.Pagano M, Pepperkik R, Verde F, Ansorge W, Draetta G. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zindy F, Lamas E, Chenivesse X, Scobcza K J, Wang J, Fesquet D, Henglein B, Brechot C. Biochem Biophys Res Commun. 1992;182:1144–1154. doi: 10.1016/0006-291x(92)91851-g. [DOI] [PubMed] [Google Scholar]
- 7.Hirt H, Heberle-Bors E. Dev Biol. 1994;5:147–154. [Google Scholar]
- 8.Jacobs T W. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:317–339. [Google Scholar]
- 9.Soni R, Carmichael J R, Shah Z H, Murray J A H. Plant Cell. 1995;7:85–103. doi: 10.1105/tpc.7.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meskiene I, Bögre L, Dahl M, Pirck M, Ha D T C, Swoboda I, Heberle-Bors E, Ammerer G, Hirt H. Plant Cell. 1995;7:759–771. doi: 10.1105/tpc.7.6.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Nagata T, Nemoto Y, Hasezawa S. Int Rev Cytol. 1992;132:1–30. [Google Scholar]
- 13.Reichheld J-P, Sonobe S, Clément B, Chaubet N, Gigot C. Plant J. 1995;7:245–252. [Google Scholar]
- 14.Joshi C P. Nucleic Acids Res. 1987;15:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renaudin J P, Colasanti J, Rime H, Juan Z, Sundaresan V. Proc Natl Acad Sci USA. 1994;91:7375–7379. doi: 10.1073/pnas.91.15.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renaudin J P, Savouré A, Philippe H, Van Montagu M, Inzé D, Rouzé P. In: Plant Cell Division. Francis D, Dudits D, Inzé D, editors. Colchester, U.K.: Portland; 1997. in press. [Google Scholar]
- 17.Setiady Y Y, Sekine M, Hariguchi N, Yamamoto T, Kouchi H, Shinmyo A. Plant J. 1995;8:949–957. doi: 10.1046/j.1365-313x.1995.8060949.x. [DOI] [PubMed] [Google Scholar]
- 18.Kouchi H, Sekine M, Hata S. Plant Cell. 1995;7:1143–1155. doi: 10.1105/tpc.7.8.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebwohl D E, Muise-Helmericks R, Sepp-Lorenzino L, Serve S, Timaul M, Bol R, Borgen P, Rosen N. Oncogene. 1994;9:1925–1929. [PubMed] [Google Scholar]
- 20.Regad F, Hervé C, Marinx O, Bergounioux C, Tremoussaygue D, Lescure B. Mol Gen Genet. 1995;248:703–711. doi: 10.1007/BF02191710. [DOI] [PubMed] [Google Scholar]
- 21.Slater M L. J Bacteriol. 1973;113:263–270. doi: 10.1128/jb.113.1.263-270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira P, Hemerly A, de Almeida J, Bergounioux C, Burssens S, Van Montagu M, Engler G, Inzé D. Proc Natl Acad Sci USA. 1994;91:11313–11317. doi: 10.1073/pnas.91.24.11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szarka S, Fitch M, Schaerer S, Moloney M. Plant Mol Biol. 1995;27:263–275. doi: 10.1007/BF00020182. [DOI] [PubMed] [Google Scholar]
- 24.Shaul O, Mironov V, Burssens S, Van Montagu M, Inzé D. Proc Natl Acad Sci USA. 1995;93:4868–4872. doi: 10.1073/pnas.93.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin L-X, Richard L, Perennes C, Gadal P, Bergounioux C. Plant Physiol. 1995;108:425–426. doi: 10.1104/pp.108.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemerly A, de Almeida Engler J, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P. EMBO J. 1995;14:3925–3936. doi: 10.1002/j.1460-2075.1995.tb00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


