Abstract
Two components of the germ-line-specific P granules of the nematode Caenorhabditis elegans have been identified using polyclonal antibodies specific for each. Both components are putative germ-line RNA helicases (GLHs) that contain CCHC zinc fingers of the type found in the RNA-binding nucleocapsid proteins of retroviruses. The predicted GLH-1 protein has four CCHC fingers; GLH-2 has six. Both GLH proteins localize in the P granules at all stages of germ-line development. However, the two glh genes display different patterns of RNA and protein accumulation in the germ lines of hermaphrodites and males. Injection of antisense glh-1 or glh-2 RNA into wild-type worms causes some offspring to develop into sterile adults, suggesting that either or both genes are required for normal germ-line development. As these very similar glh genes physically map within several hundred kilobases of one another, it seems likely that they represent a fairly recent gene duplication event.
Keywords: DEAD-box proteins, CCHC zinc fingers, glycine-rich repeats, gene duplications, chicken antibodies
Embryos of the free-living soil nematode Caenorhabditis elegans generate distinct founder cells via a series of asymmetric cell divisions. At each division, the germ-line daughter cell inherits distinctive non-membrane-bound particles, called P granules (1, 2, 3). P granules are partitioned to the primordial germ cell P4 of the 16- to 24-cell embryo and become perinuclear. P granules persist around the nuclei of all germ cells, until gametogenesis, at which point they are excluded from mature sperm and become dispersed within the cytoplasm of mature oocytes in preparation for cytoplasmic localization in the embryo. Although the distribution pattern of nematode P granules has been well-studied, the identity and function of P-granule components have yet to be determined.
Germ granules are found in many species (4, 5). The germ-line-specific polar granules of Drosophila melanogaster have been well-studied, with a number of different genes identified that are required for polar granule assembly and germ-cell formation, including vasa, staufen, valois, oskar, tudor, mago nashi, and germ-cell-less (6, 7, 8, 9, 10, 11, 12, 13, 14, 15). With the exception of vasa, these genes encode novel proteins. Vasa, however, is a member of a family of proteins with recognizable motifs and predictable function. Vasa is an RNA helicase of the DEAD-box family (8, 9) whose ATP-dependent RNA helicase activity has been demonstrated in vitro (16). As polar granules contain RNA as well as protein (11, 12, 15, 17), a germ-line-specific RNA helicase may function to bind and unwind RNAs necessary for germ-line development. Several potential vasa homologues have been cloned, including glh-1 (germ-line helicase 1) from Caenorhabditis, Xvh (Xenopus vasa homologue), mvh (mouse vasa homologue), and rvh (rat vasa homologue) (18, 19, 20, 21). glh-1 in C. elegans is unique among RNA helicase genes reported, including vasa, in that its predicted product contains four retroviral-like zinc fingers (18). We have identified a second C. elegans germ-line RNA helicase gene, glh-2, that also encodes zinc fingers. Immunolocalization with GLH-1 and GLH-2-specific antibodies demonstrates that both GLH-1 and GLH-2 localize to P granules. Thus, to our knowledge, GLH-1 and GLH-2 are the first components of C. elegans germ granules to be identified. glh-1 and glh-2 differ in their patterns of RNA and protein accumulation in the germ line, suggesting that these genes may have distinct functions. Injection of either glh-1 or glh-2 antisense RNA into the germ line of wild-type hermaphrodites results in sterile progeny, leading us to predict that mutations in glh-1 and glh-2 will result in a germ-line-defective mutant phenotype.
MATERIALS AND METHODS
Strains.
Wild-type worms were C. elegans strain N2 variety Bristol. Worms were grown using standard methods (22).
Sequence Analysis.
Both strands of genomic glh-1 and glh-2 clones and the glh-2 cDNA were sequenced using the chain-termination method with either Sequenase II (United States Biochemical) or with SequiTherm polymerase (Epicentre Technologies, Madison, WI). In addition, preliminary genomic sequence was obtained courtesy of the C. elegans sequencing consortium at Washington University (23). The glh-2 cDNA sequence is derived from a partial 2.3-kb cDNA isolated from a mixed-stage cDNA library made by S. Kim (Stanford University) and from a 927-bp PCR product generated with an upstream primer corresponding to the putative translation start site in the glh-2 genomic sequence (5′-CGAAGATGTCTGACGATTGG-3′) and a downstream primer corresponding to the 5′ end of the 2.3-kb cDNA (5′-CGCGGGATCCTTTCGGCCTTCACCCGGT-3′). Several different cDNA libraries yielded the same-sized PCR product.
In Situ Hybridization.
Whole-mount embryos permeabilized by freeze cracking were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) as described (24). Splayed adult worms were fixed in 4% paraformaldehyde in PBS. Fixed specimens were acetylated with acetic anhydride, dehydrated through an alcohol series, and stored at −80°C until needed. All probes were prepared by in vitro transcription of linearized templates of 5′- and 3′- gene-specific cloned fragments in the presence of 35S-labeled rCTP (New England Nuclear). Hybridizations were done as described by S. Petersen (25). Hybridizations were carried out overnight at 55°C in a moist chamber. The slides were washed four times in 4× SSPE (0.18 M NaCl/10 mM sodium phosphate, pH 7.4/1 mM EDTA), treated for 30 min at 37°C with RNase A (20 μg/ml), and then washed with increasing stringency to a final concentration of 0.1× SSPE/10 mM DTT at 55°C. Slides were dipped in NTB-2 emulsion (Kodak) and developed with Dektol developer (Kodak).
Generation and Purification of Anti-GLH Antibodies.
Mouse antisera were raised against the predicted GLH-1 protein from sequence between Gly-137 and Glu-572. The fragment was cloned into the pMALcRI vector (New England Biolabs), and the reading frame was verified by DNA sequencing. Fusion proteins were induced and isolated as suggested by the manufacturer. Three mice were injected subcutaneously with approximately 20 μg of the fusion protein after mixing 1:1 with Freund’s complete adjuvant. The animals were given three booster injections at 2-week intervals. Chicken antibodies were raised against the N-terminal 22 amino acids of both GLH-1 and GLH-2, peptides not shared between these proteins (Fig. 1, underlined sequence). Peptides were synthesized on a RaMPS multiple peptide synthesis system (DuPont) and conjugated to keyhole limpet hemocyanin according to the manufacturer’s directions (Pierce). Two laying hens were immunized subcutaneously with 100 μg of the conjugate with Freund’s complete adjuvant, followed by two biweekly booster injections, each of 100 μg. Anti-GLH chicken antibodies were concentrated from egg yolk with polyethylene glycol as described (27). For affinity purification, mouse anti-GLH-1 antibody was reacted with a nitrocellulose strip containing a fusion peptide from Gly-137 to Lys-489 fused to glutathione S-transferase. After washing the strips, the antibodies were eluted with 0.2 M glycine hydrochloride, pH 2.8/0.5 M NaCl and dialyzed against PBS (28). Affinity purification of the chicken anti-peptide antibodies was similarly performed against each peptide conjugated to BSA, using 5 M KI to elute anti-GLH-2.
Figure 1.

Alignment of GLH-1 and GLH-2. N-terminal glycine-rich imperfect repeats are shaded in grey. CCHC zinc fingers are indicated with black backing. The zinc finger consensus in the two GLH proteins is PxxCFNCxxxGHRSxxCPEP; these amino acids are found in at least 7/10 fingers. Comparisons of specific zinc fingers are shown at the bottom with differences in black. The conserved motifs found in all DEAD-box RNA helicases (26) are boxed. The 22-amino acid peptides used to produce GLH-1 and GLH-2-specific peptides are underlined (N termini). Sites of introns in the corresponding glh-1 and glh-2 genomic sequences are indicated with arrowheads. The 58 amino acids encoded by a 168-nt internal BamHI–BamHI fragment were absent in the original report of the glh-1 cDNA (18); the corrected sequence is GenBank accession no. L19948L19948.
Western Blot Analysis.
Western blot analysis was done essentially as described by Towbin (29). C. elegans protein extracts were prepared from animals grown in liquid culture (30), pelleted, washed twice in PBS, and suspended in an equal volume of a proteinase inhibitor mixture [1 mM phenylmethylsulfonyl fluoride/1 mM EDTA/5 mM N-ethylmaleimide/leupeptin (2 μg/ml)/aprotinin (4 μg/ml)/pepstatin (2 μg/ml) in PBS]. An equal volume of 2× SDS/PAGE sample buffer was added, and aliquots were boiled for 5 min. The undissolved carcasses were pelleted, and the supernatant was used for SDS/PAGE. Mouse antisera and PEG-concentrated chicken yolk were diluted 1:50 for anti-GLH-2 and 1:100 for anti-GLH-1. Secondary antibodies (Fisher for anti-chicken and Cappel/Worthington for anti-mouse) conjugated to horseradish peroxidase were diluted 1:1000.
Immunocytochemistry.
Worms and embryos were fixed and antibody staining was performed essentially as described (3). For double labeling, samples were incubated in a 1:1 mixture of affinity-purified chicken anti-GLH-1 and hybridoma supernatant containing mouse antibody OICID4 (31) or with a 1:1 mixture of affinity-purified mouse anti-GLH-1 and chicken anti-GLH-2, followed by a mixture of 1:100 rhodamine anti-mouse and 1:100 fluorescein anti-chicken secondary antibody. Samples were mounted in Elvanol (DuPont) and examined by epifluorescence microscopy.
Antisense RNA Injections.
Regions of glh-1 and glh-2 cDNAs were used as templates to produce capped antisense RNA in vitro, essentially as described (32) using Ambion megascript T3 and T7 kits. The glh-1 antisense RNA was 2.0 kb in length and extended from the end of the 3′ untranslated region (UTR) to Gly-137, while the glh-2 antisense RNA was 2.3 kb and extended from the end of the 3′ UTR to Asp-270. Wild-type animals were injected in the distal region of both gonad arms with either glh antisense RNA (1 μg/μl) or unc-54 antisense RNA (1 kb in length) (1 μg/μl) as a control. The injected worms were allowed to recover on individual plates at 20°C for 12 h and then were placed on new plates every 24 h. The progeny of the injected worms were scored for fertility (worms containing embryos) versus sterility (worms lacking embryos) 3 days after each transfer.
RESULTS AND DISCUSSION
Studies in Drosophila have identified several germ-granule components that are essential for granule assembly and have demonstrated that germ granules are required for formation of the primordial germ cells during early embryogenesis. To study germ-granule composition and function in Caenorhabditis, an organism with a very different mode of development than Drosophila, we began by identifying a potential vasa homolog, glh-1 (18). A second germ-line helicase gene, glh-2, has also been isolated. We report herein that both genes encode components of nematode germ granules and contain additional motifs not found in vasa.
Sequence Comparisons.
The glh-2 gene is represented by a 3136-bp cDNA that encodes a predicted 974-amino acid product containing several domains, including the conserved motifs found in all DEAD-box RNA helicases, zinc fingers, and glycine-rich N-terminal repeats (Fig. 1). The size of the glh-2 cDNA corresponds with the single ≈3.2-kb RNA species seen on Northern blots probed with a glh-2-specific probe (data not shown). Since no additional exons are predicted in the genomic sequence and the putative GLH-2 protein begins like that of Drosophila Vasa for the first 5/7 amino acids, we conclude that glh-2 cDNA is likely full length. GLH-2 shows 88% identity to GLH-1 from the first zinc finger of GLH-1 to the C terminus (Fig. 1). GLH-2 has six CCHC retroviral-like zinc fingers, compared with four in GLH-1 (Fig. 1). Conservation of coding sequence and intron–exon boundaries between glh-1 and glh-2 suggest that the distinct glh-1 and glh-2 genes arose from a duplication event, followed by either an internal duplication of zinc fingers in GLH-2 or the loss of two fingers from GLH-1. The function of the GLH zinc fingers has yet to be determined. Similar CCHC fingers in the nucleic acid binding domains of HIV Gag and the hexamer-binding protein HEXBP of Leishmania mediate sequence-specific binding to RNA and DNA, respectively (33, 34, 35); the two zinc fingers in HIV are not equivalent in their binding specificities (36).
Tandem imperfect glycine-rich repeats are a conserved feature in the N termini of GLH-1, GLH-2, and Vasa. The 17 repeats of GLH-1 and the 32 repeats of GLH-2 average 10 amino acids in length; those in Vasa are 7 amino acids long (8). The GLH-1 glycine repeat consensus is FGGG(N/K)(N/T)GG(S/T)G; the GLH-2 repeats are similar but more degenerate, with only the 2nd and 10th glycine invariant. One feature that distinguishes GLH glycine repeats from those in Vasa is a difference in charge; while the amino acids in the GLH-1 and GLH-2 repeats are generally neutral, the repeats in Vasa contain an arginine. The basic arginine/glycine/glycine repeats (RGG box) found in Vasa, Xvh, Mvh, and Rvh are predicted to function in RNA binding (37). The zinc fingers of the GLH proteins may functionally replace the RGG repeats, with the GLH glycine regions providing an entirely different function in P granules. Glycine-rich regions are predicted to favor the formation of globular structures and, therefore, may participate in GLH-mediated protein aggregation, as has been proposed for the glycine-rich N termini of loricins, plant cell wall proteins (38). Alternatively, the GLH glycine repeats, even though uncharged, may facilitate RNA binding, perhaps in combination with zinc fingers. The expanded glycine repeats and zinc fingers in GLH-2 could contribute, subtly or dramatically, to distinct roles for these two proteins.
RNA Analyses: In Situ Hybridizations. glh-1 RNA was previously shown to be highly enriched in the germ line (18).
Northern blot analyses of glh-2 with RNAs from several germ-line-defective strains yield very similar results (data not shown), with the exception of the fem-3(gf) strain, discussed below. To determine the tissue distribution of glh-1 and glh-2 transcripts, in situ hybridizations were performed to adult hermaphrodites, males, and embryos. In adults, glh-1 and glh-2 RNAs are restricted to germ-line tissue (Fig. 2). glh-1 RNA is present at all stages of germ-line development in the hermaphrodite gonad, from the distal region where germ cells divide mitotically through the proximal region where gametes mature (Fig. 2A). A similar pattern of strong glh-1 hybridization to all regions of the germ line is observed in males (Fig. 2B). The sense strand of glh-1 is shown as a negative control (Fig. 2C).
Figure 2.
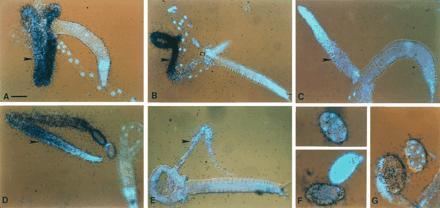
In situ hybridization to C. elegans adults and embryos. glh-1 hybridization to a splayed hermaphrodite (A) and male (B) using a 253-nt antisense probe from the glh-1-specific 5′ EcoRI–BamHI fragment (18). (C) Sense strand of the same glh-1 probe as a negative control. The gonads of the splayed worms are indicated with arrowheads. The 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei are blue. All glh-1 slides were hybridized with 5 × 105 dpm and exposed for 7 days. (D and E) Antisense glh-2 RNA hybridization to a hermaphrodite (D) and male (E). The probe used was 340 bp long, including 130 bp of the 3′-most coding region and the entire 210-bp glh-2 3′ UTR, minus the poly(A) tail. This probe was determined to be specific for glh-2 by both Northern and Southern blot analyses (data not shown). Exposures for glh-2 were 14 days, using 106 dpm. This glh-2 signal results from use of 2-fold higher probe concentration and exposure relative to the glh-1 conditions. (F) Antisense glh-1 hybridization to whole-mount embryos. From top to bottom, the embryo stages are: 8-cell stage, >60-cell stage, and 1-cell stage. (G) Antisense glh-2 hybridization to a whole mount 1-cell embryo (Left) and a 12- to 14-cell embryo (Right). Embryos were exposed for 7 days with 106 dpms. (Bars: A–E, 50 μm; F and G, 20 μm.)
Several aspects of the distribution of glh-2 RNA differ from glh-1. The glh-2 message is at least 3-fold less abundant than glh-1 mRNA in the hermaphrodite germ line. The glh-2 signal is weakest in the distal mitotic region and most concentrated in the central meiotic region of the gonad (Fig. 2D). In addition, glh-2 RNA is barely detectable in males (Fig. 2E). This result is consistent with findings in Northern blot analyses of fem-3(gf) mutant hermaphrodites, which over-produce sperm and produce no oocytes (39); in fem-3(gf) worms, the level of glh-2 RNA is lower than in wild-type (K.A.K. and K.L.B., unpublished results). The role of germ-line RNA helicases in spermatogenesis is unclear. The mouse germ-line RNA helicase PL10 is expressed only during spermatogenesis (41), although its function has yet to be determined. And while vasa is expressed in both male and female germ lines, male flies carrying a vasa deletion are fertile (40). Therefore, the dramatic differences between glh-1 and glh-2 mRNA levels in males (Fig. 2 B and E) may or may not be significant.
Both glh-1 and glh-2 RNAs are detected in all cells of young embryos, with the level of hybridization much reduced after the 8- to 10-cell stage (Fig. 2 F and G). Thus, while glh-1 and glh-2 RNA differ in their levels and their patterns of accumulation in hermaphrodite and male germ lines, both glh RNAs are found throughout the early embryo.
Protein Immunolocalization in Embryos and Adults.
To detect the localization of GLH-1 and GLH-2, antibodies were raised in mice against a fusion protein derived from GLH-1 and in chickens against synthetic peptides corresponding to each N terminus, the region that is least conserved between GLH-1 and GLH-2 (Fig. 1). By Western blot analysis, the anti-GLH-1 chicken antibodies detect a single band of ≈78 kDa, consistent with the predicted mass of the 763-amino acid GLH-1 protein (Fig. 3, lane 2, and data not shown); the sera from each of the mice detect the same size protein (Fig. 3, lane 6, and data not shown). The anti-GLH-2 chicken antibodies detect a single ≈100-kDa band (Fig. 3, lane 4, and data not shown), consistent with the predicted 974-amino acid GLH-2 protein. The preimmune chicken yolk and mouse sera are not reactive against worm proteins (Fig. 3, lanes 1, 3, and 5, and data not shown).
Figure 3.
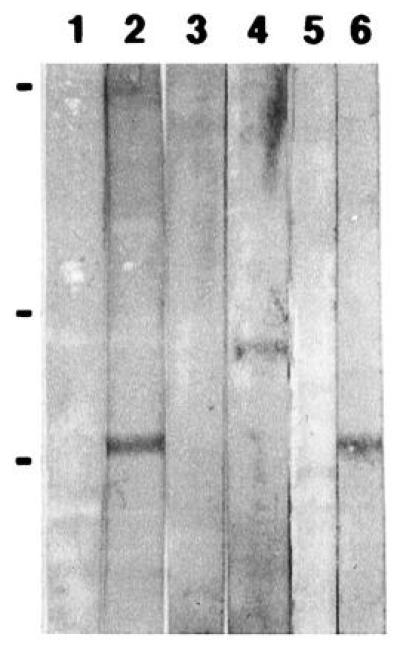
Western blot analysis using chicken yolk and mouse serum antibodies. Total C. elegans protein homogenate was resolved for 1100 V·h on an SDS/8% polyacrylamide gel and transferred to nylon-supported nitrocellulose membrane (Optibind-NC, Schleicher & Schuell). Strips were cut in half and incubated with anti-GLH antibodies as follows. Lanes: 1, preimmune yolk from one of the chickens immunized with GLH-1 peptide; 2, anti-GLH-1 chicken yolk; 3, preimmune yolk from one of the chickens immunized with GLH-2 peptide; 4, anti-GLH-2 yolk; 5, preimmune serum from one of the mice immunized with GLH-1 fusion protein; 6, anti-GLH-1 mouse serum. Prestained high molecular weight markers (GIBCO/BRL) of 202, 103, and 68 kDa are indicated.
Affinity-purified antibodies to either GLH-1 or GLH-2 proteins react with germ-line-specific P granules. P granules, as visualized by monoclonal antibodies directed against unidentified epitopes (3, 31), are present in germ cells of all developmental stages with the exception of mature sperm (see Introduction). Anti-GLH-1 and anti-GLH-2 stain the same granules recognized by the anti-P-granule monoclonal antibodies (Fig. 4A); the granules are cytoplasmic in the oocyte and early embryo and perinuclear in later stage embryos (Fig. 4). In adult worms, anti-GLH-1 brightly stains perinuclear P granules throughout the germ line of hermaphrodites and males (Fig. 5 A and B, Left). Anti-GLH-2 shows a more restricted pattern of staining that corresponds to the RNA distribution seen in situ; staining is less intense in the distal region than in the meiotic region of hermaphrodites (Fig. 5A Center) and is barely detectable in the male germ line (Fig. 5B Center). P-granule staining is not detected with any preimmune sera or yolk (data not shown). The results presented here demonstrate that anti-GLH-1 and anti-GLH-2 antibodies recognize distinct proteins that colocalize with P granules in the germ-line cells of embryos and adult worms.
Figure 4.
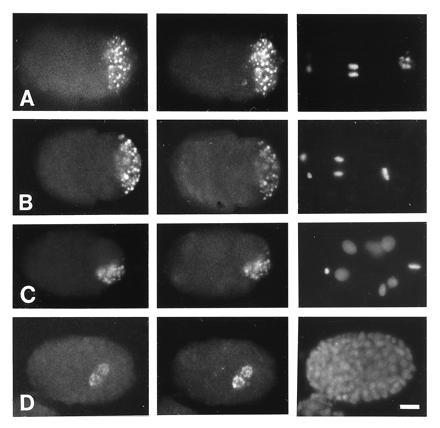
Immunofluorescence staining of embryos with OIC1D4 and anti-GLH-1 and anti-GLH-2 antibodies. Embryos are oriented with anterior left and ventral down. (A). Embryo stained with affinity-purified chicken anti-GLH-1 antibodies (Left), mouse monoclonal antibody OIC1D4 (Center), and DAPI (Right). (B–D). Embryos stained with affinity-purified mouse anti-GLH-1 (Left), chicken anti-GLH-2 (Center), and DAPI (Right). (A and B) Two-cell embryos. P1 is in mitosis, and P granules are segregated to the posterior cortex destined for P2. (C) Seven-cell embryo. P2 is in mitosis, and P granules are segregated to the ventral region destined for P3. (D) Late-stage embryo showing P granules in Z2 and Z3. (Bar = 10 μm.)
Figure 5.
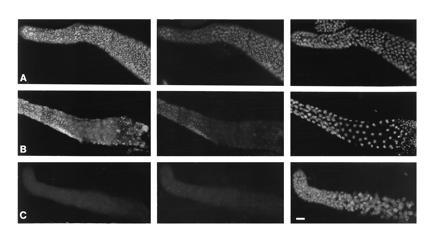
Immunofluorescence staining of adult gonads using anti-GLH-1 and anti-GLH-2 antibodies. Each row shows a sample stained with affinity-purified mouse anti-GLH-1 antibodies (Left), chicken anti-GLH-2 (Center), and DAPI (Right). Gonad arms are oriented with distal left. (A) Distal gonad arm from a wild-type hermaphrodite. (B) Gonad arm from a wild-type male. Sperm present at the far right of each panel fail to stain with anti-GLH-1 and anti-GLH-2. (C) Gonad arm from a sterile hermaphrodite worm produced by a mother injected with antisense RNA to glh-1. (Bar = 10 μm.)
Localization of glh gene products to P granules appears to occur at the protein level, since glh RNAs do not localize specifically to P granules and in embryos are not even restricted to the germ-line blastomeres. It is possible that translation of the glh RNAs is repressed in embryos by the multiple potential adenylation control elements or the nos response elements found in the 3′ UTRs of both glh mRNAs (42, 43, 44). Hyperadenylylated glh RNAs have been observed in Northern blot analysis of RNAs from several germ-line-defective C. elegans strains (ref. 18; K.A.K. and K.L.B., unpublished results) and nos-like elements are reported to spatially restrict the translation of glp-1 RNA in the C. elegans embryo (45). Alternatively, the glh mRNAs may be translated in the embryo, but the GLH proteins may be unstable in the somatic blastomeres due to the absence of other P-granule components.
Antisense RNA.
To address the function(s) of GLH-1 and GLH-2 in the C. elegans germ line, we determined whether injected antisense RNA produced against glh-1 or glh-2 affects germ-line development. Microinjection of antisense RNA into the syncytial germ line of wild-type hermaphrodites has been found to produce a gene-specific “phenocopy” of the mutant phenotype in the offspring of the injected worms, with the effects being most penetrant for maternal-effect genes but also observable for zygotic genes (refs. 32 and 46; C. Mello, personal commununication; L. Berkowitz, I. Kawasaki, R. Holdeman, I. Korf, and S.S., unpublished results). Injection of antisense RNAs complementary to glh-1 or glh-2 mRNAs resulted in sterility (and no other observable phenotype) in 10–11% of the progeny; a 2.0-kb glh-1 antisense RNA resulted in 63 sterile worms from 663 total offspring, and a 2.3-kb glh-2 antisense RNA resulted in 83 sterile worms from 790 total offspring. The germ lines in sterile worms were underproliferated and contained germ nuclei with altered morphology. Interestingly, staining with both anti-GLH-1 and anti-GLH-2 was absent in sterile offspring produced by either glh-1 or glh-2 antisense RNAs (n > 25 worms; Fig. 5C). Thus, injection of glh-1 or glh-2 antisense RNA reduces expression of both proteins to below detectable levels. The sterile worms also failed to stain with four other monoclonal antibodies directed against unknown P-granule epitopes (K76, OICID4, L416, and PIF4; refs. 3 and 31; data not shown), raising the exciting possibility that injection of antisense glh RNA prevents the assembly of P granules. Thus, antisense RNA against the glh genes can reduce the level of multiple P-granule components below detectability and prevent normal germ-line development. As a control, worms were injected with a 1.0-kb antisense RNA to the unc-54 gene, which encodes myosin heavy chain (47, 48). The injected worms produced some uncoordinated but all fertile offspring (31 Unc and no sterile worms out of 371 total progeny) whose germ lines stained positively for GLH protein (data not shown).
Since antisense RNA made to the coding region of either glh gene may cross-hybridize with transcripts from the other gene in vivo, we attempted to specifically inhibit production of each GLH protein by injecting antisense RNAs produced against 5′ and 3′ regions unique to each gene. These gene-specific RNAs, ranging in size from 140 to 900 nt, did not result in sterile offspring. This result could be due to inefficient antisense inhibition by the smaller RNAs. Alternatively, GLH-1 and GLH-2 may functionally compensate for one another, as in the case of LIN-12 and GLP-1 (49, 50, 51), and it may be necessary to target both glh genes by a cross-hybridizing antisense RNA to affect germ-line development.
Our results suggest that either wild-type glh-1 or glh-2 or both genes are required for normal germ-line development. Based on differences in the glycine repeats and zinc fingers of their predicted proteins, as well as differences in RNA abundance and patterns of accumulation, we predict that GLH-1 and GLH-2 have individual, although perhaps partially overlapping, roles in germ-line development. Generation and analysis of mutations in the genes should resolve this issue. Mutants should also reveal whether the sterile phenotype is maternal-effect or zygotic and enable determination of the cause of sterility.
A Third GLH?
The C. elegans genome sequencing project has identified a third potential RNA helicase gene located in the dpy-5–unc-13 interval on chromosome 1. glh-1 and glh-2 have been physically mapped to this same region. The novel helicase gene encodes an open reading frame with 69% and 72% identity to GLH-1 and GLH-2, respectively, when compared from the start of the zinc fingers to the potential C terminus. The predicted GLH-3 has two CCHC-type zinc fingers that are quite diverged from those in GLH-1 and GLH-2 and an N-terminal region that is not glycine-rich. In preliminary Northern blot analysis of germ-line-defective strains, this RNA helicase appears to be germ-line-specific (data not shown). Thus, the C. elegans germ line may utilize the activity of three germ-line RNA helicases, two quite similar to each other and the third more diverged.
GLHs: Multiple Genes, Multiple Motifs.
This report has established that the germ granules of both Drosophila and Caenorhabditis contain germ-line RNA helicases, with differences between these components in these two model organisms. Although vasa is a single copy gene, there are at least two glh genes that produce components of the P granules. The GLH helicases possess several motifs, including zinc fingers, uncharged glycine repeats, and potential 3′ regulatory elements, not found in Vasa. Therefore, the molecular mechanisms of nematode P-granule assembly and function may be found to differ in many respects from those currently being elucidated in Drosophila.
Note Added in Proof.
PIE-1 and MEX-3 were recently shown to transiently associate with P granules in early embryos (52, 53).
Acknowledgments
We are grateful to Dr. J. Im (University of Missouri, Columbia) for synthesizing and conjugating the peptides used in antibody production. Dr. C. Gunther (University of Missouri, Columbia) provided useful comments on the manuscript. The DNA Core facilities at University of Missouri, Columbia, and Indiana University and the University of Missouri, Columbia, Cytology core provided valuable services. M.E.G., D.L.R., and K.A.K. were supported by National Institutes of Health Training Grant T32 AI07276 to the Department of Molecular Microbiology and Immunology. A grant from the University of Missouri, Columbia, Research Board provided funds to M.E.G. and K.L.B. J.S.M. was partially supported by funds from the University of Missouri, Columbia, Medical Research Council, and K.L.B. was also supported with funds from the Council for Tobacco Research, Grant 2414AR2. S.S. acknowledges support from National Institutes of Health Grant GM34059.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
Abbreviations: GLH, germ-line helicase; UTR, untranslated region; DAPI, 4′,6-diamidino-2-phenylindole.
Data deposition: The sequences reported in this paper have been deposited in the GenBank data base (accession nos. glh-2 cDNA, U60194U60194; glh-2 genomic, U60449U60449; glh-1 cDNA, L19948L19948; glh-1 genomic, U62772U62772).
References
- 1.Wolf N, Priess J, Hirsh D. J Embryol Exp Morphol. 1983;73:297–306. [PubMed] [Google Scholar]
- 2.Strome S, Wood W B. Proc Natl Acad Sci USA. 1982;79:1558–1562. doi: 10.1073/pnas.79.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strome S, Wood W B. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- 4.Eddy E M. Int Rev Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- 5.Illmensee K, Mahowald A P. Exp Cell Res. 1976;97:127–140. doi: 10.1016/0014-4827(76)90662-5. [DOI] [PubMed] [Google Scholar]
- 6.Schubach T, Wieschaus E F. Roux Arch Dev Biol. 1986;195:302–317. doi: 10.1007/BF00376063. [DOI] [PubMed] [Google Scholar]
- 7.Schupbach T, Wieschaus E. Dev Biol. 1986;113:443–448. doi: 10.1016/0012-1606(86)90179-x. [DOI] [PubMed] [Google Scholar]
- 8.Hay B, Jan L Y, Jan Y N. Cell. 1988;55:577–587. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- 9.Lasko P F, Ashburner M. Nature (London) 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- 10.St. Johnston D, Beuchle D, Nusslein-Volhard C. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- 11.Kim-Ha J, Smith J L, Macdonald P M. Cell. 1991;66:23–35. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- 12.Ephrussi A, Dickinson L K, Lehmann R. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 13.Golumbeski G S, Bardsley A, Tax F, Boswell R E. Genes Dev. 1991;5:2060–2070. doi: 10.1101/gad.5.11.2060. [DOI] [PubMed] [Google Scholar]
- 14.Boswell R E, Prout M E, Steichen J C. Development (Cambridge, UK) 1991;113:373–384. doi: 10.1242/dev.113.1.373. [DOI] [PubMed] [Google Scholar]
- 15.Jongens T A, Hay B, Jan L Y, Jan Y N. Cell. 1992;70:569–584. doi: 10.1016/0092-8674(92)90427-e. [DOI] [PubMed] [Google Scholar]
- 16.Liang L, Diehl-Jones W, Lasko P. Development (Cambridge, UK) 1994;120:1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- 17.Whitfield W G F, Gonzalez C, Sanchez-Herrero E, Glover D M. Nature (London) 1989;338:337–340. doi: 10.1038/338337a0. [DOI] [PubMed] [Google Scholar]
- 18.Roussell D L, Bennett K L. Proc Natl Acad Sci USA. 1993;90:9300–9304. doi: 10.1073/pnas.90.20.9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komiya T, Itoh K, Ikenishi K, Furusawa M. Dev Biol. 1994;162:354–363. doi: 10.1006/dbio.1994.1093. [DOI] [PubMed] [Google Scholar]
- 20.Fujiwara Y, Komiya T, Kawabata H, Sato M, Fujimoto H, Furusawa M, Noce T. Proc Natl Acad Sci USA. 1994;91:12258–12262. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komiya T, Tanigawa Y. Biochem Biophys Res Commun. 1995;207:405–410. doi: 10.1006/bbrc.1995.1202. [DOI] [PubMed] [Google Scholar]
- 22.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson R, Ainscough R, Anderson K, Baynes C, Barks M, Bonfield J, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 24.Seydoux G, Fire A. Development (Cambridge, UK) 1994;120:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- 25.Petersen S L, McCrone S. In Situ Hybridization Applications to Neurobiology. New York: Oxford Univ. Press; 1993. pp. 78–95. [Google Scholar]
- 26.Pause A, Méthot N, Sonenberg N. Mol Cell Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gassmann M, Thömmes P, Weiser T, Hübscher U. FASEB J. 1990;4:2528–2532. doi: 10.1096/fasebj.4.8.1970792. [DOI] [PubMed] [Google Scholar]
- 28.Olmsted J B. Methods Enzymol. 1986;134:467–472. doi: 10.1016/0076-6879(86)34112-0. [DOI] [PubMed] [Google Scholar]
- 29.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood W B. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 31.Strome S. In: Gametogenesis and the Early Embryo. Gall J G, editor. New York: Liss; 1986. pp. 77–95. [Google Scholar]
- 32.Guo S, Kemphues K J. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 33.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Barklis E. J Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webb J R, McMaster W R. J Biol Chem. 1993;268:13994–14002. [PubMed] [Google Scholar]
- 36.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiledijian M, Dreyfuss G. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinert P M, Mack J W, Korge B P, Gan S Q, Haynes S R, Steven A C. Int J Biol Macromol. 1991;13:130–139. doi: 10.1016/0141-8130(91)90037-u. [DOI] [PubMed] [Google Scholar]
- 39.Barton M K, Schedl T B, Kimble J. Genetics. 1987;115:107–119. doi: 10.1093/genetics/115.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leroy P, Alzari P, Sassoon D, Wolgemuth D, Fellous M. Cell. 1989;57:549–559. doi: 10.1016/0092-8674(89)90125-6. [DOI] [PubMed] [Google Scholar]
- 41.Lasko P F, Ashburner M. Genes Dev. 1990;4:905–921. doi: 10.1101/gad.4.6.905. [DOI] [PubMed] [Google Scholar]
- 42.Sheets M D, Fox C A, Hunt T, VandeWoude G, Wickens M. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- 43.Wharton R P, Struhl G. Cell. 1989;59:881–892. doi: 10.1016/0092-8674(89)90611-9. [DOI] [PubMed] [Google Scholar]
- 44.Roussell, D. L., Gruidl, M. E., Kreutzer, M. A., Richards, J. P. & Bennett, K. L. (1995) in Molecular Approaches to Parasitology, MBL Lectures in Biology, eds. Boothroyd, J. C. & Komuniecki, R. (Wiley, New York), pp. 321–340.
- 45.Evans T C, Crittenden S, Kodoyianni V, Kimble J. Cell. 1994;77:183–194. doi: 10.1016/0092-8674(94)90311-5. [DOI] [PubMed] [Google Scholar]
- 46.Lin R, Thompson S, Priess J R. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 47.Epstein H F, Waterston R H, Brenner S. J Mol Biol. 1974;90:291–300. doi: 10.1016/0022-2836(74)90374-x. [DOI] [PubMed] [Google Scholar]
- 48.MacLeod A R, Waterston R H, Fishpool R M, Brenner S. J Mol Biol. 1977;114:133–140. doi: 10.1016/0022-2836(77)90287-x. [DOI] [PubMed] [Google Scholar]
- 49.Lambie E J, Kimble J. Development (Cambridge, UK) 1991;112:231–240. doi: 10.1242/dev.112.1.231. [DOI] [PubMed] [Google Scholar]
- 50.Fitzgerald K, Wilkinson H A, Greenwald I. Development (Cambridge, UK) 1993;119:1019–1027. doi: 10.1242/dev.119.4.1019. [DOI] [PubMed] [Google Scholar]
- 51.Fitzgerald K, Greenwald I. Development (Cambridge, UK) 1995;121:4275–4282. doi: 10.1242/dev.121.12.4275. [DOI] [PubMed] [Google Scholar]
- 52.Mello C C, Schubert C, Draper B, Zhang W, Lobel R, Priess J R. Nature (London) 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- 53.Draper B W, Mello C C, Bowerman B, Hardin J, Priess J R. Cell. 1996;87:205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]


