Summary
Genome stability requires that genomic DNA is replicated only once per cell cycle. The replication licensing system ensures that the formation of pre-replicative complexes is temporally separated from the initiation of DNA replication [1-4]. The replication licensing factors Cdc6 and Cdt1 are required for the assembly of pre-replicative complexes during G1 phase. During S phase, metazoan Cdt1 is targeted for degradation by the CUL4 ubiquitin ligase [5-8], and vertebrate Cdc6 is translocated from the nucleus to the cytoplasm [9,10]. However, because residual vertebrate Cdc6 remains in the nucleus throughout S phase [10-13], it has been unclear whether Cdc6 translocation to the cytoplasm prevents re-replication [1,2,14]. The inactivation of C. elegans CUL-4 is associated with dramatic levels of DNA re-replication [5]. Here, we show that C. elegans CDC-6 is exported from the nucleus during S phase in response to the phosphorylation of multiple CDK sites. CUL-4 promotes the phosphorylation of CDC-6 and its translocation via negative regulation of the CDK-inhibitor CKI-1. Re-replication can be induced by co-expressing non-exportable CDC-6 with non-degradable CDT-1, indicating that redundant regulation of CDC-6 and CDT-1 prevents re-replication. This demonstrates that Cdc6 translocation is critical for preventing re-replication, and that CUL-4 independently controls both replication licensing factors.
Keywords: CUL4, cullin, cell cycle, Cdt1
Results
CDC-6 is exported from the nucleus during S phase
The C. elegans ortholog of the DNA replication licensing factor Cdc6 is essential for DNA replication. C. elegans cdc-6(RNAi) embryos arrest with ∼100 cells, and contain only trace amounts of DNA, indicating a total failure of DNA replication (Figure S1).
We followed CDC-6 dynamics during the first cell division of the V1-V6 hypodermal seam cells using immunofluorescence with anti-CDC-6 antibody. The timing of S phase entry was determined by following ribonucleotide reductase promoter (Prnr-1) driven GFP expression [15]. Seam cells varied in the timing of S phase entry: 105-120 min post-hatch for V5, 120-135 min for V2-V4; 135-150 min for V6; and 180-195 min for V1 (data not shown).
At 10-20 min post-hatch, nuclear expression of CDC-6 was first observed in the seam cells, and by 100-115 min post-hatch, nuclear levels were high (Figure 1A; data not shown). As cells entered S phase, nuclear levels of CDC-6 dropped gradually with a concomitant increase in cytoplasmic CDC-6. Beyond 180 min post-hatch, endogenous CDC-6 remained at high levels in the cytoplasm, although residual nuclear CDC-6 was still present (Figure 1A).
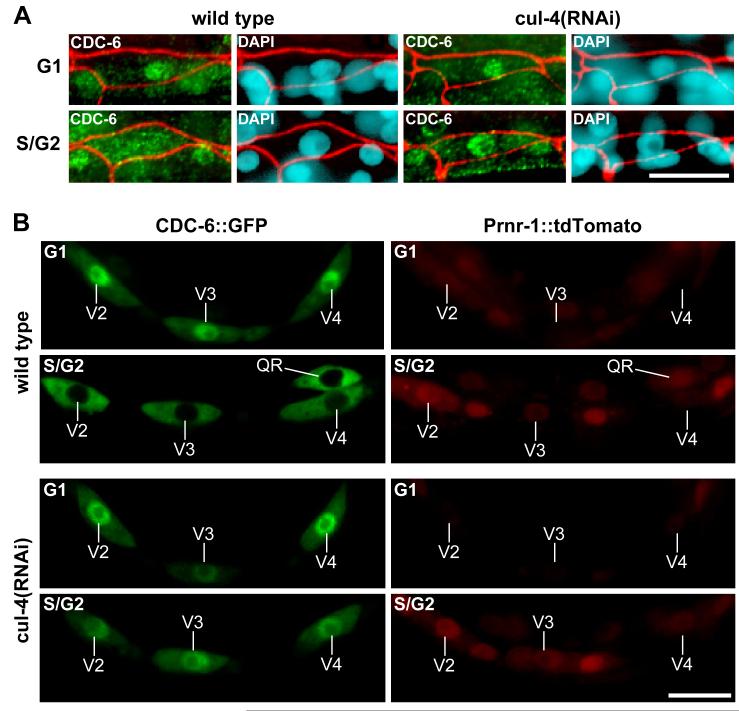
Figure 1. CDC-6 is exported from the nucleus during S phase in a CUL-4-dependent manner.
(A) Endogenous CDC-6 is exported to the cytoplasm during S phase in wild-type but not cul-4(RNAi) larvae. Wild-type and cul-4(RNAi) larvae were observed at 100-115 min post-hatch (G1 phase, upper panels) and at 180-195 min post-hatch (S/G2 phase, lower panels). Animals were stained with anti-CDC-6 (green), DAPI (blue), and anti-AJM-1 (red overlay, staining adhesion junctions to indicate seam cell boundaries [35]). (B) CDC-6::GFP translocates from the nucleus to the cytoplasm during S phase in wild-type larvae (top), but remains nuclear in cul-4(RNAi) larvae (bottom). CDC-6::GFP that was expressed from the wrt-2 promoter was observed at 90-105 min post-hatch (G1 phase; upper panels) and at 195-210 min post-hatch (S/G2 phase; lower panels). Prnr-1::tdTomato was used as an S phase marker. Scale bars, 10 μm.
We also followed CDC-6::GFP that was expressed in seam cells under the control of the wrt-2 promoter [16]. In contrast to the partial translocation of endogenous CDC-6, transgenic CDC-6::GFP appeared to localize completely to the cytoplasm during S phase (Figures 1B, S2D). CDC-6::GFP was present throughout the cell cycle, and localized to condensed chromosomes during mitosis (Figure S2A,B).
To determine the time course of CDC-6 nuclear export during S phase, we investigated CDC-6::GFP or CDC-6::tdTomato localization at set time points (Figure S2C). At 120-135 min post-hatch, CDC-6::GFP remained nuclear in most of the seam cells except V5. Cytoplasmic localization was first detected at 150-165 min post-hatch for V2-V4, at 165-180 min for V6 and at 195-185 min for V1. The timing of CDC-6 translocation for the seam cells correlated with the timing of S phase entry.
To determine if the exclusive cytoplasmic localization of CDC-6::GFP during S phase resulted from the degradation of nuclear CDC-6::GFP, we created a constitutively nuclear-localized CDC-6::GFP by: adding two SV40 large T-antigen nuclear localization signals (NLSs) to the C-terminus; and inactivating the predicted nuclear-export signal (NES) through L434A and L436A amino acid subsitutions [17,18]. This CDC-6mNES+2NLS protein had stable nuclear protein levels throughout the cell cycle, indicating that CDC-6 does not undergo appreciable degradation in the nucleus during S phase (Figure S3; data not shown).
CUL-4 is required for CDC-6 phosphorylation and nuclear export in S phase
In cul-4(RNAi) larvae, blast cells arrest in S phase and undergo extensive re-replication, and this is associated with a failure to degrade CDT-1 [5]. We sought to address whether CUL-4 also regulates the licensing factor CDC-6 during S phase. Endogenous CDC-6 localization was followed in cul-4(RNAi) larvae by immunofluorescence. Prior to S phase, nuclear-localized CDC-6 accumulated in cul-4(RNAi) seam cells with a time course similar to that of wild type (Figure 1A; data not shown). However, during S phase, CDC-6 in cul-4(RNAi) seam cells remained nuclear with no appreciable increase in cytoplasmic staining (Figure 1A). CUL-4 was similarly required for the nuclear export of exogenous CDC-6::GFP during S phase (Figure 1B). These results indicate that CDC-6 nuclear export requires the CUL-4 ubiquitin ligase.
In humans, Cdc6 nuclear export is triggered by the phosphorylation of three CDK phosphorylation sites that are located near two N-terminal NLSs [9,10] (Figure 2A). C. elegans CDC-6 has six sites similar to the CDK phosphorylation consensus that are located near three N-terminal NLSs (Figure 2A).
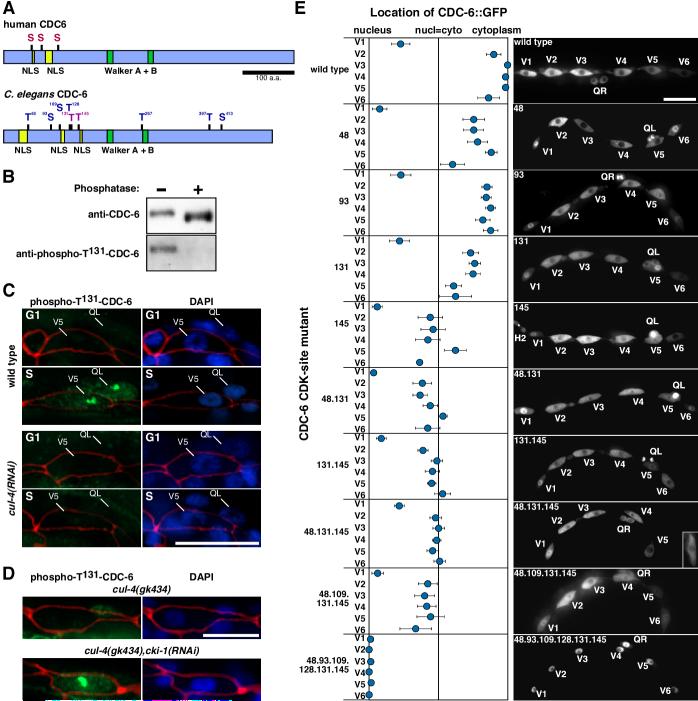
Figure 2. CDC-6 is phosphorylated in a CUL-4-dependent manner on CDK sites during S phase.
(A) Schematic representation of human and C. elegans CDC-6 orthologs. Potential CDK phosphorylation sites are shown as black bars labeled S (serine) or T (threonine); NLSs are shown as yellow boxes (the first C. elegans NLS is bi-partite and the next two are simple NLSs [36]); and Walker A and B domains as green boxes (these are required for the loading of Mcm2-7 onto chromatin [37]). In humans, the phosphorylation of S54, S74, and S106 (red) near the two N-terminal NLSs is required for Cdc6 nuclear export [9,10]. C. elegans CDC-6 has two consensus CDK sites (S/T-P-X-K/R; purple lettering) and seven minimum CDK phosphorylation sites (S/T-P; blue) [38]. Six of these potential CDK phosphorylation sites are located near the NLSs. Scale bar, 100 amino acids. (B) Characterization of affinity purified anti-phospho-T131-CDC-6 antibody. Wild-type C. elegans lysate, either untreated (left lane) or treated with λ phosphatase (right lane), was analyzed by western blot. Note that anti-phospho-T131-CDC-6 antibody recognized the endogenous CDC-6 protein but could not recognize CDC-6 after treatment with phosphatase, thereby demonstrating specificity. (C) CDC-6 T131-phosphorylation in wild-type and cul-4(RNAi) seam cells in G1 and S/G2 phases. Wild-type (top) and cul-4(RNAi) (bottom) larvae were observed in G1 phase (upper panels) and in S phase (lower panels). Animals were stained with anti-phospho-T131-CDC-6 (green), DAPI (blue), and anti-AJM-1 (red overlay). (D) RNAi depletion of cki-1 in cul-4(gk434) restores the phosphorylation of CDC-6 in S phase. Staining as in (C) for cul-4(gk434) (top) and cul-4(gk434), cki-1(RNAi) (bottom) seam cells in S phase (180-195 min post-hatch). (E) Subcellular localization of wild-type and CDK-site mutant CDC-6::GFP proteins expressed from the wrt-2 promoter. Serine and threonine residues in the predicted N-terminal CDK phosphorylation sites were replaced by alanines, making the sites unphosphorylatable. CDC-6::GFP localization in V1-V6 seam cells during S/G2 phase (180-225 min post hatch) is plotted on a continuum from nuclear localization to cytoplasmic localization. Numbers on the left of the graph indicate the serine or threonine residues replaced with alanine in the mutant proteins. The panels to the right show epifluorescence images of CDK-site mutant CDC-6::GFP proteins in V1-V6 seam cells at 195-225 min post-hatch. Numbers in the upper left corner indicate the serine or threonine residues replaced with alanine. In the CDC-6m48.131.145 image, an inset shows a longer exposure of the V6 seam cell, whose expression was too weak to view with the normal exposure. Error bars represent SEM. See Experimental Procedures for the number of cells analyzed. Scale bars, 10 μm.
To examine whether phosphorylation near the NLSs promotes CDC-6 nuclear export, we generated a phospho-specific antibody against the threonine-131 CDK phosphorylation site (Figure 2A,B). Anti-phospho-T131-CDC-6 signal was not detected in wild-type V1-V6 seam cells before S phase, but was observed in both the nucleoli and cytoplasm of S phase seam cells (Figure 2C). This indicates that at least a subset of CDC-6 that undergoes nuclear export is phosphorylated on position T131. Within the nucleus, the obvious nucleolar anti-phospho-T131-CDC-6 signal contrasted with the more uniform nuclear signal observed with anti-CDC-6 antibody, suggesting either that a higher percentage of nucleolar-localized CDC-6 is phosphorylated on the T131 site or that the phosphorylation of T131 induces nucleolar localization. In contrast to wild-type larvae, anti-phospho-T131-CDC-6 signal was not detected in cul-4(RNAi) larvae during S phase, indicating that CUL-4 is required for the phosphorylation of T131 (Figure 2C).
Phosphorylation of multiple CDK sites is required for CDC-6 nuclear export
To determine the functional relevance of CDC-6 phosphorylation, we expressed CDC-6::GFP with potential N-terminal CDK phosphorylation sites replaced by unphosphorylatable alanines. All of the CDC-6::GFP CDK-site mutants had normal nuclear localization in G1 phase (Figure S4; data not shown).
Single mutations of T48, S93, T131, or T145 each caused partial retention of CDC-6::GFP in the nucleus during S phase, with the T145 mutation having the greatest effect (Figure 2E). Combining T48, S109, T131, and T145 mutations gave approximately the same translocation rate as the T145 mutation alone (Figure 2E). However, mutation of all six N-terminal CDK phosphorylation sites (CDC-6mCDK) completely blocked translocation during S phase (Figures 2E, S4). This implies that the phosphorylation of multiple sites is required for CDC-6 nuclear export. No obvious defects in DNA replication or cell cycle progression resulted from expressing the constitutively nuclear localized CDC-6mCDK or CDC-6mNES+2NLS proteins, suggesting that continuous CDC-6 nuclear localization does not, by itself, deregulate DNA replication.
CDC-6 nuclear export is independent of CDT-1 degradation
Cdt1 and Cdc6 physically interact in fission yeast and mammals [19,20]. This raises the possibility that CDT-1 degradation is a necessary precedent for CDC-6 nuclear export, perhaps by allowing kinases to gain access to CDC-6 phosphorylation sites. To address whether the nuclear retention of CDC-6 in cul-4(RNAi) animals is a secondary consequence of CDT-1 perdurance, we asked if the expression of a non-degradable CDT-1 prevents CDC-6 nuclear export in a wild-type background.
Human Cdt1 can be stabilized during S phase by mutation of CDK-phosphorylation sites and the PCNA-binding PIP-box sequence [8, 11]. To stabilize C. elegans CDT-1, we substituted alanines for three conserved PIP-box residues and five N-terminal CDK phosphorylation residues to create the CDT-1mCDK+PIP3A mutant (Figure S5A). CDT-1mCDK+PIP3A::GFP was present in 83% of S/G2 phase seam cells (n=18), indicating that the mutant protein is partially stabilized during S phase (Figure S5B).
We observed that despite the presence of stabilized CDT-1mCDK+PIP3A::GFP, CDC-6::tdTomato was still exported to the cytoplasm during S phase, indicating that CDC-6 nuclear export occurs independently of CDT-1 degradation (Figure 3A). It should be noted that the stabilized CDT-1 protein is functional and can promote DNA replication (see below).
Figure 3. CUL-4 regulates CDC-6 nuclear export through a CKI-1-dependent pathway.
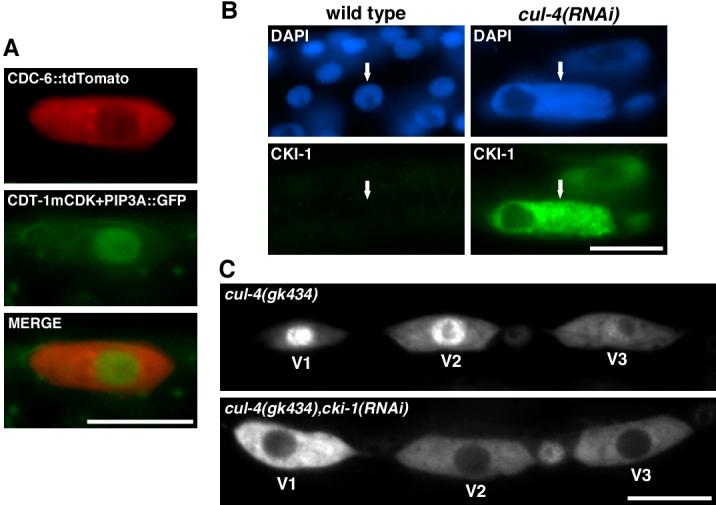
(A) CDT-1 perdurance does not disrupt CDC-6 nuclear export. CDC-6::tdTomato is exported in the presence of stabilized CDT-1::GFP at 195-210 min post hatch (S/G2 phase). Pwrt-2::CDC-6::tdTomato (red) and Pnhr-168::CDT-1mCDK+PIP3A::GFP (green) were expressed in a strain that has ajm-1::GFP (green) marking seam cell boundaries. (B) CKI-1 accumulates in enlarged cul-4(RNAi) seam cells. L2-stage wild-type and arrested L2-stage cul-4(RNAi) larvae were stained with anti-CKI-1 (green) and DAPI (blue). Arrows indicate seam cells. (C) cki-1(RNAi) restores CDC-6 nuclear export in cul-4(gk434) mutant seam cells. Pwrt-2::CDC-6::GFP was observed in cul-4(gk434) and cul-4(gk434), cki-1(RNAi) larvae at 195-210 min post-hatch (S/G2 phase). For both genetic backgrounds, the expression of Pwrt-2::CDC-6::GFP was nuclear during G1 phase (data not shown). Scale bars, 10 μm.
cki-1 inactivation rescues CDC-6 nuclear export in cul-4 mutants
We have found that CUL-4 negatively regulates the level of the CDK-inhibitor CKI-1, with CKI-1 accumulating in cul-4(RNAi) re-replicating cells (Figure 3B) [7]. cki-1 RNAi reduces the size of nuclei and DNA levels in cul-4(gk434) seam cells, indicating suppression of the cul-4 re-replication phenotype (Figure S6A) [7]. cki-1 RNAi does not affect the accumulation of CDT-1 in cul-4(gk434) mutants, indicating that the prevention of re-replication is independent of CDT-1 accumulation [7].
We hypothesized that CUL-4 promotes CDC-6 nuclear export by negatively regulating CKI-1 levels; and that CKI-1 accumulation in cul-4 mutants prevents CDK(s) from phosphorylating CDC-6 to induce translocation. To test this model, we investigated the localization of Pwrt-2::CDC-6::GFP in cki-1(RNAi), cul-4(gk434) mutants. CDC-6 is predominantly nuclear-localized during S phase in 73% of cul-4(gk434) mutant V2-V6 seam cells (n=30) (the failure to observe full penetrance for nuclear localization is likely the effect of cul-4 maternal product [5]). cki-1 RNAi in the cul-4(gk434) mutant significantly increases the percentage of seam cells with cytoplasmically-localized CDC-6::GFP (83%, n=29, vs. 27% with no RNAi, n=30), indicating that CKI-1 is required for CDC-6 nuclear retention in cul-4 mutants (Figure 3C).
A prediction of the model is that cki-1(RNAi) will restore the phosphorylation of CDC-6 in cul-4(gk434) mutants during S phase. The status of CDC-6 phosphorylation was assessed by immunofluorescence with anti-phospho-T131-CDC-6 antibody. In S phase, anti-phospho-T131-CDC-6 signal was detected in a relatively small percentage of cul-4(gk434) seam cells (33%, n=30 vs. 80% for wild-type, n=41) (Figure 2C,D). However, cki-1(RNAi) in cul-4(gk434) increased the percentage of seam cells with phospho-T131 signal to the wild-type level (82% n=45) (Figure 2D). If cki-1 RNAi induces CDC-6 nuclear export in cul-4 mutants by permitting CDC-6 phosphorylation, then non-phosphorylatable CDC-6mCDK should not undergo nuclear export in cki-1(RNAi), cul-4(gk434) seam cells; and this was observed (47/47 nuclear localized) (Figure S6B). These results indicate that CKI-1 is essential to prevent CDC-6 phosphorylation in cul-4 mutants.
Deregulation of both CDT-1 and CDC-6 can induce re-replication
We investigated the biological significance of CDT-1 degradation and CDC-6 nuclear export by overexpressing wild-type and deregulated proteins using the heat-shock promoters hsp16-2 and hsp16-41 [21]. Wild-type CDT-1 and CDC-6, or stabilized CDT-1 (CDT-1mCDK+PIP6A) and non-exportable CDC-6 (CDC-6m5CDK), were expressed individually or in combination (Figure 4A). Heatshock expression of individual deregulated CDT-1 or CDC-6 produced higher levels of lethality (embryonic- or L1-stage-arrest) relative to wild-type proteins (Figure 4A). Combinations of CDT-1 and CDC-6 produced more lethality than individually expressed proteins. However, ∼100% lethality was obtained only when deregulated CDC-6 was combined with either wild-type CDT-1 or deregulated CDT-1 (Figure 4A).
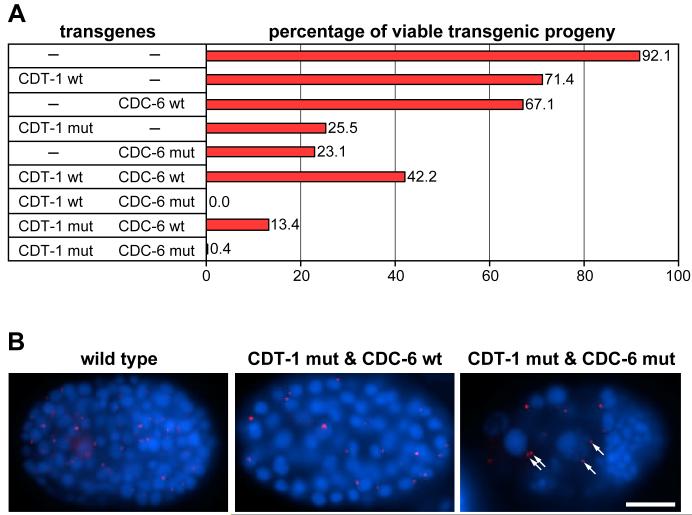
Figure 4. Expression of deregulated CDT-1 and CDC-6 produces re-replication.
(A) Graph of viability upon overexpression of wild-type or deregulated CDT-1 and CDC-6 transgenes. ‘CDT-1 wt’ and ‘CDC-6 wt’ are wild-type genes; ‘CDT-1 mut’ is CDT-1mCDK+PIP6A; and ‘CDC-6 mut’ is CDC-6m5CDK. Each gene was expressed under the control of both the hsp16-41 and hsp16-21 heat-shock promoters. Embryos from injected hermaphrodites were incubated at 25°C (a semi-permissive temperature for hsp expression [39]); and heat-shocked for 30 min at 33°C 12 hrs prior to harvest. The percentages of viable progeny are listed to the right of each bar in the graph. (B) Expression of non-degradable CDT-1 and non-exportable CDC-6 induces DNA re-replication. Epifluorescence images of a wild-type embryo and transgenic embryos expressing either CDT-1mCDK+PIP6A plus wild-type CDC-6 or CDT-1mCDK+PIP6A plus CDC-6m5CDK. Animals were stained with DAPI (blue) and anti-SPD-2 (red), which highlights centrosomes [23]. Arrows indicate pairs of centrosomes in cells with increased DNA content (the cell on the left has 13.9C DNA content, and the cell on the right has 11.1C). Scale bar, 10 μm.
Increased nuclear DNA levels were observed in 47% (n=36) of embryos expressing non-exportable CDC-6 with stabilized CDT-1 (21.5 ± 2.4C DNA content, n=63, vs. 2.4 ± 0.3C, n=13, for wild-type embryonic interphase cells) (Figure 4B). In contrast, other combinations of wild-type and/or deregulated CDT-1 and CDC-6 did not produce noticeably increased DNA levels (Figure 4B; data not shown). Increased DNA content can arise either from re-replication, in which DNA replication initiates continuously during S phase, or from failed mitosis, in which cells with duplicated genomes re-enter the cell cycle without DNA segregation. These two mechanisms can be distinguished by analyzing centrosome numbers. Cells that undergo failed mitosis have extra centrosomes because centrosomes will duplicate in the subsequent cell cycle, while re-replicating cells that undergo S phase arrest have only two centrosomes [22]. We analyzed centrosome numbers with anti-SPD-2 antibody, which stains centrioles [23]. We observed that cells with excessive DNA levels had only two centrosomes, implying that the increased ploidy arises from re-replication (Figure 4B). Therefore, the stabilized CDT-1 and non-exportable CDC-6 are functional, and can act in concert to induce DNA re-replication.
Discussion
CDC-6 phosphorylation-dependent nuclear export prevents DNA re-replication
We found that C. elegans CDC-6 is exported from the nucleus during S phase, similar to vertebrate Cdc6 [1,2,14], suggesting that this is an ancient regulatory mechanism. Further, our study indicates that the strategy to trigger Cdc6 nuclear export is also conserved: the phosphorylation of multiple CDK sites to inactivate N-terminal NLSs. All six N-terminal CDK-sites must be phosphorylated to promote CDC-6 nuclear export. Interestingly, the phosphorylation of T131 is associated with both nuclear export and nucleoli localization. This suggests the possibility that the phosphorylation of a subset of sites, while not sufficient to induce nuclear export, can direct CDC-6 to specific sub-nuclear locations.
In humans and Xenopus, ectopically-expressed Cdc6 is completely exported to the cytoplasm during S phase; in contrast, a substantial fraction of endogenous Cdc6 remains nuclear localized during S phase [9-13]. Strikingly, we observed a similar result in C. elegans with a substantial fraction of endogenous CDC-6 remaining in the nucleus during S phase, while ectopically-expressed CDC-6 appears exclusively cytoplasmic. The reason(s) for these differential localizations are not understood.
The presence of nuclear-localized Cdc6 during S phase in mammalian cells has led to the proposal that Cdc6 translocation is not important for restraining DNA replication licensing [1,2,14]. Further, there is currently no evidence for a functional role of Cdc6 translocation in preventing re-replication [1,2,14]. In this study, we observed that non-exportable CDC-6 can synergize with deregulated CDT-1 to induce re-replication. This implies that CDC-6 translocation is a redundant safeguard to prevent the re-initiation of DNA replication, and provides the first evidence in any organism of a functional role for phosphorylation-dependent CDC-6 nuclear export.
Deregulation of both CDC-6 and CDT-1 is required for re-replication
In S. pombe, the overexpression of the Cdc6 ortholog (Cdc18) is sufficient to induce significant re-replication [24]. In contrast, overexpression of Cdc6 does not induce re-replication in S. cerevisiae, Drosophila, and humans [25-27]. In humans, co-overexpression of wild-type Cdt1 and Cdc6 in cells that lack a cell cycle checkpoint produces only modest re-replication in a subset of cells [27].
We observed that co-expression of non-degradable CDT-1 and non-exportable CDC-6 produced significant re-replication in a subset of early-stage C. elegans embryos. In contrast, overexpression of combinations of deregulated and wild-type CDT-1 or CDC-6 did not induce re-replication. This indicates that redundant regulation of CDT-1 and CDC-6 prevents re-replication. The failure to observe re-replication in every embryonic cell expressing deregulated CDT-1 and CDC-6 suggests the presence of additional safeguards in the early embryo to prevent DNA re-replication. Expression of combinations of wild-type and deregulated CDT-1 and CDC-6 produced embryonic lethality that was not associated with increased DNA levels. The cause of this lethality is unclear, but may arise from changes in the timing of DNA replication, which is known to produce embryonic arrest [28].
CUL-4 regulates both CDC-6 and CDT-1 replication licensing factors
Inactivation of CUL-4 produces dramatic levels of re-replication that is associated with a failure to degrade CDT-1 [5]. However, overexpressing Cdt1 in fission yeast does not induce re-replication, and overexpressing human Cdt1 several log-fold higher than the endogenous protein produces only modest re-replication in a subset of cells [27,29,30]. Given the negligible or limited effects of greatly overexpressing Cdt1 in other organisms, it was hard to reconcile the substantial re-replication associated with merely failing to degrade CDT-1 during S phase in cul-4(RNAi) animals.
Our work reveals that the CDC-6 replication licensing factor is also deregulated in cul-4(RNAi) animals. CDC-6 remains nuclear throughout S phase in cul-4(RNAi) animals, and this is correlated with a failure to phosphorylate CDC-6 on CDK sites. CUL-4 negatively regulates the levels of the CDK-inhibitor CKI-1 ([7], this study). The negative regulation of CKIs of the CIP/KIP family by CUL4 is conserved in Drosophila and humans [31-33]. cki-1 RNAi suppresses re-replication in cul-4 mutants without affecting CDT-1 accumulation [7], indicating that CKI-1 is independently required for the induction of re-replication. Significantly, the presence of CKI-1 is required for the block on CDC-6 phosphorylation and nuclear export in cul-4(gk434) cells. Our results suggest that CUL-4 promotes CDC-6 nuclear export by negatively regulating CKI-1 levels, thereby allowing CDK(s) to phosphorylate CDC-6 and induce its nuclear export. The evidence that CDK(s) are the relevant kinases is that CDC-6 is phosphorylated on CDK-consensus sites and the phosphorylation is blocked by a CDK inhibitor. In yeast and mammals, CDK activity prevents re-replication, and siRNA co-depletion of CDK1 and CDK2 in human cells induces limited re-replication [1,3,34]. Our results suggest that in metazoa, Cdc6 is one of the critical targets of CDKs for preventing re-replication. Our work further indicates that CUL-4 is a master regulator that restrains DNA replication through two independent pathways: mediating CDT-1 degradation and promoting CDC-6 nuclear export via the negative regulation of CKI-1.
Supplementary Material
Acknowledgments
We are grateful to Yuji Kohara, Roger Tsien, Richard Roy, Andrew Fire, Kevin O'Connell, and Sudhir Nayak for clones, strains or reagents; Christopher Dowd for technical assistance; members of the Kipreos laboratory for critical reading of the manuscript; and the Caenorhabditis Genetics Center for strains. This work was supported by grant R01GM055297 from NIGMS (NIH) to ETK.
Footnotes
Supplemental Data
Supplemental Data include Experimental Procedures and six figures and are available with this article online at
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 2.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–2843. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- 5.Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–889. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- 6.Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol. 2006;8:84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y, Kipreos ET. The Caenorhabditis elegans replication licensing factor CDT-1 is targeted for degradation by the CUL-4/DDB-1 complex. Mol Cell Biol. 2007;27:1394–1406. doi: 10.1128/MCB.00736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A. PCNA is a co-factor for Cdt1 degradation by CUL4/DDB1 mediated N-terminal ubiquitination. J Biol Chem. 2006;281:6246–6252. doi: 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- 9.Jiang W, Wells NJ, Hunter T. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc Natl Acad Sci U S A. 1999;96:6193–6198. doi: 10.1073/pnas.96.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen BO, Lukas J, Sorensen CS, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. Embo J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coverley D, Pelizon C, Trewick S, Laskey RA. Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J Cell Sci. 2000;113:1929–1938. doi: 10.1242/jcs.113.11.1929. [DOI] [PubMed] [Google Scholar]
- 12.Fujita M, Yamada C, Goto H, Yokoyama N, Kuzushima K, Inagaki M, Tsurumi T. Cell cycle regulation of human CDC6 protein. Intracellular localization, interaction with the human mcm complex, and CDC2 kinase-mediated hyperphosphorylation. J Biol Chem. 1999;274:25927–25932. doi: 10.1074/jbc.274.36.25927. [DOI] [PubMed] [Google Scholar]
- 13.Alexandrow MG, Hamlin JL. Cdc6 chromatin affinity is unaffected by serine-54 phosphorylation, S-phase progression, and overexpression of cyclin A. Mol Cell Biol. 2004;24:1614–1627. doi: 10.1128/MCB.24.4.1614-1627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A. Regulating the licensing of DNA replication origins in metazoa. Curr Opin Cell Biol. 2006;18:231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Hong Y, Roy R, Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 1998;125:3585–3597. doi: 10.1242/dev.125.18.3585. [DOI] [PubMed] [Google Scholar]
- 16.Aspock G, Kagoshima H, Niklaus G, Burglin TR. Caenorhabditis elegans has scores of hedgehog-related genes: sequence and expression analysis. Genome Res. 1999;9:909–923. doi: 10.1101/gr.9.10.909. [DOI] [PubMed] [Google Scholar]
- 17.Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 19.Cook JG, Chasse DA, Nevins JR. The regulated association of Cdt1 with minichromosome maintenance proteins and Cdc6 in mammalian cells. J Biol Chem. 2004;279:9625–9633. doi: 10.1074/jbc.M311933200. [DOI] [PubMed] [Google Scholar]
- 20.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 21.Stringham EG, Dixon DK, Jones D, Candido EPM. Temporal and Spatial Expression Patterns of the Small Heat Shock (hsp16) Genes in Transgenic Caenorhabditis elegans. Mol Biol Cell. 1992;3:221–223. doi: 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng H, Zhong W, Punkosdy G, Gu S, Zhou L, Seabolt EK, Kipreos ET. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat Cell Biol. 1999;1:486–492. doi: 10.1038/70272. [DOI] [PubMed] [Google Scholar]
- 23.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell. 2004;6:511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- 24.Nishitani H, Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 25.Crevel G, Mathe E, Cotterill S. The Drosophila Cdc6/18 protein has functions in both early and late S phase in S2 cells. J Cell Sci. 2005;118:2451–2459. doi: 10.1242/jcs.02361. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA rereplication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 27.Vaziri C, Saxena S, Jeon Y, Lee C, Murata K, Machida Y, Wagle N, Hwang DS, Dutta A. A p53-dependent checkpoint pathway prevents rereplication. Mol Cell. 2003;11:997–1008. doi: 10.1016/s1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 28.Encalada SE, Martin PR, Phillips JB, Lyczak R, Hamill DR, Swan KA, Bowerman B. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev Biol. 2000;228:225–238. doi: 10.1006/dbio.2000.9965. [DOI] [PubMed] [Google Scholar]
- 29.Yanow SK, Lygerou Z, Nurse P. Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. Embo J. 2001;20:4648–4656. doi: 10.1093/emboj/20.17.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gopalakrishnan V, Simancek P, Houchens C, Snaith HA, Frattini MG, Sazer S, Kelly TJ. Redundant control of rereplication in fission yeast. Proc Natl Acad Sci USA. 2001;98:13114–13119. doi: 10.1073/pnas.221467598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banks D, Wu M, Higa LA, Gavrilova N, Quan J, Ye T, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 and PCNA interact with p53 and regulate p53 polyubiquitination and protein stability through MDM2 and CUL4A/DDB1 complexes. Cell Cycle. 2006;5:1719–1729. doi: 10.4161/cc.5.15.3150. [DOI] [PubMed] [Google Scholar]
- 32.Bondar T, Kalinina A, Khair L, Kopanja D, Nag A, Bagchi S, Raychaudhuri P. Cul4A and DDB1 associate with Skp2 to target p27Kip1 for proteolysis involving the COP9 signalosome. Mol Cell Biol. 2006;26:2531–2539. doi: 10.1128/MCB.26.7.2531-2539.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higa LA, Yang X, Zheng J, Banks D, Wu M, Ghosh P, Sun H, Zhang H. Involvement of CUL4 ubiquitin E3 ligases in regulating CDK inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle. 2006;5:71–77. doi: 10.4161/cc.5.1.2266. [DOI] [PubMed] [Google Scholar]
- 34.Machida YJ, Dutta A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007;21:184–194. doi: 10.1101/gad.1495007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C, Hardin JD. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol. 2001;3:983–991. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- 36.Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 37.Cook JG, Park CH, Burke TW, Leone G, DeGregori J, Engel A, Nevins JR. Analysis of Cdc6 function in the assembly of mammalian prereplication complexes. Proc Natl Acad Sci U S A. 2002;99:1347–1352. doi: 10.1073/pnas.032677499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson RB, Kemp BE. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- 39.Dawe AS, Smith B, Thomas DW, Greedy S, Vasic N, Gregory A, Loader B, de Pomerai DI. A small temperature rise may contribute towards the apparent induction by microwaves of heat-shock gene expression in the nematode Caenorhabditis elegans. Bioelectromagnetics. 2006;27:88–97. doi: 10.1002/bem.20192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.