Abstract
“Centralized” (ancestral and consensus) HIV-1 envelope immunogens induce broadly cross-reactive T cell responses in laboratory animals; however, their potential to elicit cross-reactive neutralizing antibodies has not been fully explored. Here, we report the construction of a panel of consensus subtype B (ConB) envelopes and compare their biologic, antigenic and immunogenic properties to those of two wildtype Env controls from individuals with early and acute HIV-1 infection. Glycoprotein expressed from full-length (gp160), uncleaved (gp160-UNC), truncated (gp145) and N-linked glycosylation site deleted (gp160-201N/S) versions of the ConB env gene were packaged into virions and, except for the fusion defective gp160-UNC, mediated infection via the CCR5 co-receptor. Pseudovirions containing ConB Envs were sensitive to neutralization by patient plasma and monoclonal antibodies, indicating the preservation of neutralizing epitopes found in contemporary subtype B viruses. When used as DNA vaccines in guinea pigs, ConB and wildtype env immunogens induced appreciable binding, but overall only low level neutralizing antibodies. However, all four ConB immunogens were significantly more potent than one wildtype vaccine at eliciting neutralizing antibodies against a panel of tier 1 and tier 2 viruses, and ConB gp145 and gp160 were significantly more potent than both wildtype vaccines at inducing neutralizing antibodies against tier 1 viruses. Thus, consensus subtype B env immunogens appear to be at least as good as, and in some instances better than, wildtype B env immunogens at inducing a neutralizing antibody response, and are amenable to further improvement by specific gene modifications.
Introduction
Genetic variation is a hallmark of human immunodeficiency virus type 1 (HIV-1) infection and a major obstacle to AIDS vaccine development (Korber et al., 2001; Mullins and Jensen, 2006, Worobey, in press). Since its introduction into the human population almost a century ago (Korber et al., 2000; Sharp et al., 2000), pandemic HIV-1 (HIV-1 group M) has continued to diversify and today comprises a spectrum of viral variants of unprecedented genetic complexity. Viruses belonging to this “main” group of HIV-1 have been classified into “subtypes” and “circulating recombinant forms” (CRFs) based on their phylogenetic relationships (Leitner et al., 2005). Subtypes represent major clades that resulted from the expansion of founder viruses early in the group M epidemic (Vidal et al., 2000; Rambaut et al., 2001; Worobey, in press); CRFs represent descendants of complex recombinants of two or more group M subtypes (Robertson et al., 1995; Leitner et al., 2005). Among all known subtypes and CRFs, subtype C is the most prevalent, accounting for more than 50% of group M infections worldwide and representing the predominant HIV-1 lineage in southern Africa, China and India (Osmanov et al. 2002). Subtype A and related CRFs account for roughly 30% of group M infections, and are primarily found in west and central Africa. Subtype B comprises about 15% of group M infections and is the predominant subtype in Europe, Australia and the Americas (subtype B and related recombinants are also common in Asia). Since all other subtypes and CRFs are less prevalent (Osmanov et al., 2002), candidate vaccines have historically been selected from members of subtypes A, B and C (Douek, et al., 2006, IAVI, 2006; HVTN, 2006). However, with envelope protein sequence distances as high as 38%, selecting a single contemporary virus as a vaccine strain is unlikely to provide sufficient global, or even regional, coverage of HIV-1 diversity.
An inherent problem associated with selecting a contemporary HIV-1 strain as a candidate immunogen is that this virus is as distant from other contemporary viruses as these are from each other. To reduce this distance, we and others have proposed the use of “centralized” HIV-1 immunogens, expressed from consensus or reconstructed ancestor gene sequences (Korber et al., 2001; Gaschen et al., 2002; Ellenberger et al., 2002; Mullins et al., 2004; Nickle et al., 2003; Novitsky et al., 2002). Because of their “central” position within an evolutionary tree, these inferred sequences are almost half as distant from contemporary HIV-1 strains as the latter are from each other and should thus contain a greater number of conserved epitopes. However, since centralized sequences encode artificial gene products, their antigenicity and immunogenicity cannot be predicted. Moreover, their biological properties may vary since their exact sequence depends on the input data, the alignment, and the particular algorithm used for reconstruction. For example, ancestral sequences which represent an attempt to reconstruct the common ancestor of a given viral lineage, tend to be artificially enriched for certain nucleotides, may include recently fixed escape mutations, and are vulnerable to sampling bias (Gaschen et al., 2002). Consensus sequences which represent the most common amino acid residue at any one position in a protein alignment are also vulnerable to sampling bias and may bring together polymorphisms not linked in natural infections (Doria-Rose et al., 2005). Finally, genomic regions that evolve by frequent insertions and deletions, like the variable loop regions in the envelope glycoprotein, have to be reconstructed manually, using conserved elements (e.g., glycosylation sites) spanning these regions as a guide. Thus, centralized sequences represent only imperfect approximations of HIV-1’s evolutionary history and require extensive experimental validation to ensure that they represent suitable immunogens.
To date, five centralized Env immunogens, derived from M group (Con6, ConS) and subtype specific consensus (ConC) and ancestral (An1-EnvB, AncC) env genes, have been generated and tested as DNA and/or protein vaccines (Doria-Rose et al., 2005; Gao et al., 2005; Weaver et al., 2006; Kothe et al., 2006; Liao et al., 2006). All of these genes expressed functional Env glycoproteins and elicited humoral and/or cellular immune responses in small animal models (Doria-Rose et al., 2005; Gao et al., 2005; Weaver et al., 2006; Kothe et al., 2006; Liao et al., 2006). However, only the group M specific ConS Env immunogen (delivered as an oligomeric gp140 protein) elicited high titer antibodies that neutralized primary isolates from three different group M clades (Liao et al., 2006). The other four immunogens elicited either no or only negligible neutralizing antibody responses. Interestingly, the ConS vaccine used expressed a secreted Env glycoprotein that lacked the gp120/gp41 cleavage site (C), the fusion peptide (F), and an immunodominant (I) region in the transmembrane gp41 domain (ConSΔCFIgp140). The improved immunogenicity of the ConS Env protein may thus be due, at least in part, to these gene modifications (Chakrabarti et al., 2002), although its ability to induce cross-clade responses is likely a consequence of its central nature.
To examine directly whether centralized env immunogens can be improved by targeted gene modifications, we constructed a panel of consensus subtype B (ConB) env genes that expressed glycoproteins with changes in functional domains previously shown to influence antigenic and/or immunogenic properties of wildtype Env proteins (Edwards et al., 2002, Barnett et al., 2001; Bower et al., 2004; Grundner et al., 2005; Yang et al., 2001; Hu et al., 2005). The rationale for generating such a panel was several-fold. First, subtype B is the most extensively studied group M subtype (Leitner et al., 2005), and thus an obvious target for immunogen design. Second, despite a growing number of centralized gene products, consensus subtype B env immunogens have not yet been reported. Third, a subtype B env ancestor vaccine elicited only weak neutralizing antibody responses in laboratory animals (Doria-Rose et al, 2005). Finally, a formal comparison of consensus and wildtype subtype B env vaccines has not been conducted. Here, we describe the antigenic properties of native and modified ConB Env proteins and compare the immunogenicity of ConB env DNA vaccines to those of two contemporary (wildtype) env controls.
Results
Design of native and modified consensus subtype B (ConB) env genes
The ConB Env sequence was generated by selecting the most common amino acid at each position of a full-length Env protein alignment derived from 137 subtype B viruses deposited in the 2001 HIV Sequence Database (Gaschen et al., 2002; Kothe et al., 2006). Hypervariable loop regions were reconstructed as described (Gaschen et al., 2002, Kothe et al., 2006). Fig. 1A depicts an alignment of ConB gp160 and the deduced protein sequences of two contemporary wildtype env controls derived from patients during the acute (WITO4160.27) and early (CAAN5342.A2) phase of HIV-1 infection (Li et al., 2005). Of note, ConB Env is considerably shorter (850 amino acids) and less extensively glycosylated (24 N-linked glycosylation sites in gp120) than a previously reported subtype B Env ancestor (884 amino acids; 30 N-linked glycosylation sites in gp120) (Doria-Rose et al., 2005); this is because we opted to create hypervariable regions that contain a minimum number of residues, reasoning that shorter loops might give better accessibility to neutralizing epitopes.
Fig. 1.
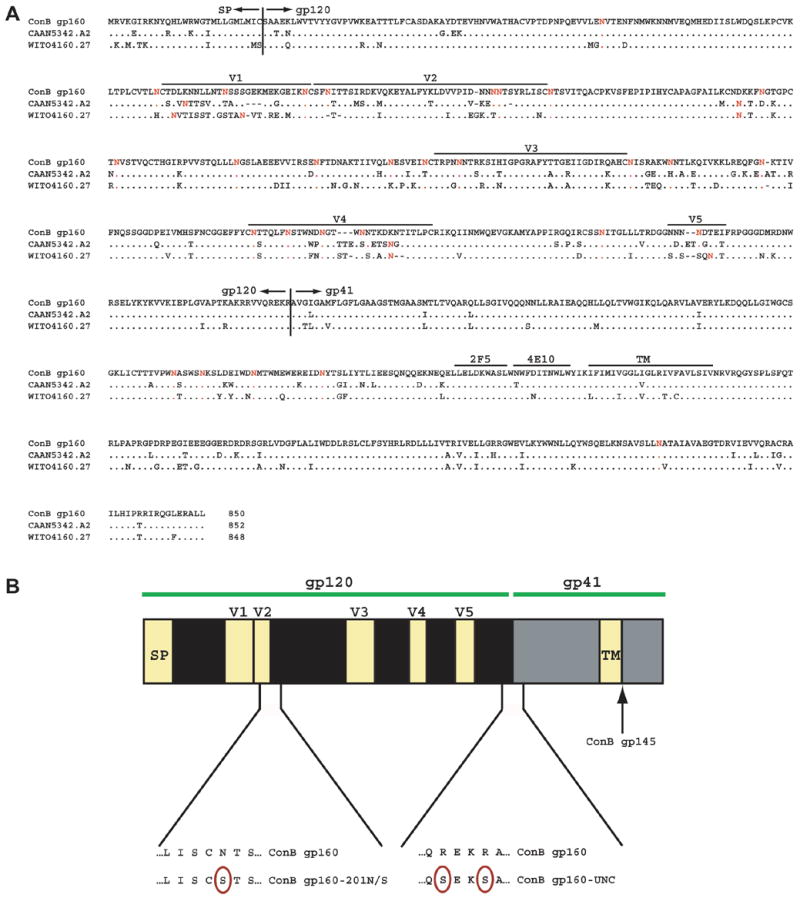
Generation of consensus subtype B (ConB) envelopes. (A) Alignment of the deduced protein sequence of full length (unmodified) ConB Env with those of recently transmitted, contemporary subtype B Env controls (CAAN5342.A2, WITO4160.27). Sequences are compared to ConB gp160, with dots indicating sequence identity, and dashes indicating gaps introduced for optimal alignment. Potential N-linked glycosylation sites are highlighted in red. The locations of signal peptide (SP) and gp120/gp41 cleavage sites are indicated, as are the positions of the variable loops (V1–V5) and the transmembrane (TM) domain. (B) Location of specific mutations within the ConB Env sequence. Amino acid substitutions are highlighted (see text for design and construction details of ConB gp160-201N/S, ConB gp160-UNC and ConB gp145).
To investigate whether synthetic Env proteins could be modified to improve their immunogenicity, we generated three ConB variants. Cleavage site mutations can alter the antigenicity of wildtype Env proteins (Schulke et al., 2002; Si et al., 2003; Herrera et al., 2005; Pancera and Wyatt, 2005) and are thought to be responsible (at least in part) for the improved immunogenicity of soluble gp140 oligomers compared to monomeric gp120 (Barnett et al., 2001; Bower et al., 2004; Grundner et al., 2005; VanCott et al., 1997; Yang et al., 2001). We thus generated a fusion-defective version of ConB (ConB gp160-UNC) by replacing two amino acid residues in the gp120/gp41 cleavage site (REKR→SEKS). Truncations of the gp41 cytoplasmic domain have also been reported to influence the conformation of the gp120 surface subunit and to expose conserved neutralizing determinants in some wildtype Env proteins (Edwards et al., 2002). We therefore introduced a stop codon immediately following the membrane-spanning domain to generate a truncated, yet still anchored, version of ConB Env (ConB gp145). Finally, in wildtype subtype B viruses, the V1V2 loop is believed to occlude the CCR5 binding site, but the position of this loop can be altered by removal or repositioning of a N-linked glycosylation site at its base (Kolchinsky et al., 2001a,b). To examine whether the removal of an analogous glycosylation site would expose ConB neutralization epitopes that are normally masked by the V1V2 loop, we replaced an asparagine residue at position 201 with a serine residue, generating ConB gp160-201N/S. Fig. 1B depicts a schematic representation of these modifications.
Functional analysis of ConB Env glycoproteins
Codon-usage optimization of HIV/SIV gene sequences enhances protein expression in vitro (Haas et al., 1996; Andre et al., 1998; Gao et al., 2003; Kothe et al., 2006) and increases humoral and cellular immune responses to DNA vaccines in laboratory animals in vivo (Robinson and Torres, 1997; Torres et al., 1997; Letvin et al., 2004; Mascola et al., 2005; Rao et al., 2006). To generate constructs suitable for DNA vaccination, we synthesized codon-usage optimized ConB, CAAN5342.A2 and WITO4160.27 env genes using an algorithm first described by Seed and colleagues (Haas et al., 1996; Andre et al., 1998). All env gene modifications were introduced into the codon-usage optimized sequence of ConB. The integrity of the newly derived constructs was tested following transfection into 293T cells. Consistent with previous reports (Doria-Rose et al., 2005; Gao et al., 2005; Kothe et al., 2006; Liao et al., 2006), consensus and codon optimized wildtype env genes expressed high levels of Env glycoprotein. Moreover, all constructs expressed the same amounts of Env glycoprotein as determined by Western blot analysis of transfected cell lysates (data not shown).
To determine whether the ConB-derived envelope glycoproteins were packaged into HIV-1 particles, expression plasmids were co-transfected with an env-deficient HIV-1 (SG3Δenv) backbone vector. Optimized CAAN5342.A2 and WITO4160.27 env constructs, as well as the env-minus backbone vector alone, were included as controls. Culture supernatants were harvested, centrifuged through a 20% sucrose cushion, normalized by p24 Gag content, and examined by Western Blot analysis using antibodies specific for HIV-1 Gag (p24) and Env (gp120) proteins. As shown in Fig. 2, the ConB-derived Envs incorporated efficiently into virus particles, although most of the virion-associated glycoprotein was only incompletely processed. Cleaved gp120 was detectable in all preparations, except for the cleavage defective ConB gp160-UNC protein. However, based on Western blot intensities, the relative amounts of gp120 were lower than those of the corresponding precursors (Fig. 2). Importantly, incomplete processing was not only observed for the synthetic ConB Envs, but also for the contemporary CAAN5342.A2 and WITO4160.27 Envs, suggesting saturation of furin and other cellular proteases involved in gp160 processing in 293T cells over-expressing Env proteins (Binley et al., 2002; Gao et al., 2003; Kothe et al., 2006).
Fig. 2.
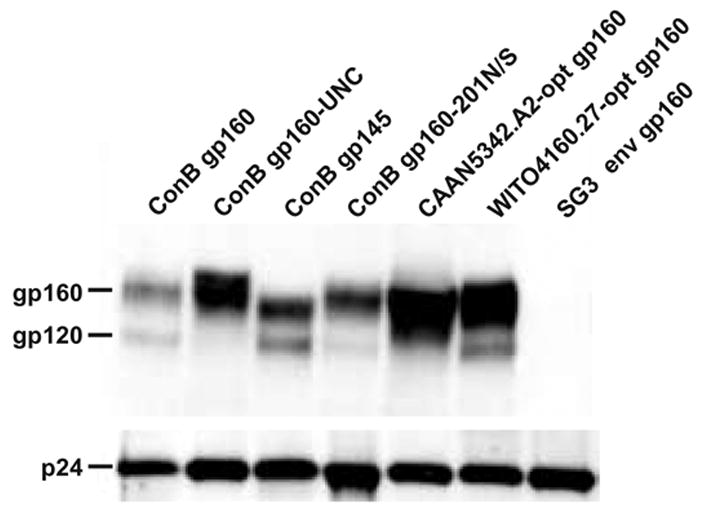
Processing and virion incorporation of ConB Envs. Consensus (ConB gp160, ConB gp160-UNC, ConB gp145, and ConB gp160-201N/S) and wildtype (CAAN5342.A2 and WITO41260.27) env genes were co-transfected with the HIV-1/SG3Δenv backbone vector. Purified particles were examined by Western blot analysis using antibodies specific for HIV-1 Env (polyclonal anti-gp120; upper panel) and Gag (monoclonal anti-p24; lower panel) proteins (the SG3Δenv backbone vector was included for control). The position of the uncleaved gp160 precursor, the mature gp120 protein, and the p24 Gag protein are indicated.
To determine whether the particle-associated ConB envelope glycoproteins were capable of mediating fusion and entry into appropriate target cells, purified pseudovirion preparations were analyzed in a single round infectivity assay (Derdeyn et al., 2000; Platt et al., 1998; Wei at al., 2002). JC53-BL cells express high levels of CD4, CCR5 and CXCR4 receptor molecules and are stably transfected with β-galactosidase and luciferase reporter genes under the control of the HIV-1 long terminal repeat (LTR). The infectious titer of any given virus stock can thus be determined by staining cultures for β-galactosidase expression and counting the number of blue cells. Fig. 3 depicts the infectious titer of ConB Env containing pseudovirions normalized for Gag p24 protein content. Except for the cleavage-defective gp160-UNC variant (which as expected was fusion defective), all other ConB glycoproteins conferred infectivity to HIV-1/SG3Δenv, albeit with varying efficiencies. In three different experiments, ConB gp160 Env-containing particles were the most infectious, reaching titers of up to 8,078 IU/ng p24. ConB gp145 Env containing pseudovirions were slightly less infectious, but this difference was not significant (Fig. 3). However, markedly reduced levels of β-galactosidase expressing JC53-BL cells were observed for the glycosylation site mutant of ConB. In three independent experiments, the relative infectivity of ConB gp160-201N/S Env containing pseudovirions was approximately 10-fold lower than that of ConB gp160 containing particles, and this difference was statistically significant (p<0.0005). The contemporary control envelopes also varied in their ability to mediate cell entry. Pseudovirions containing the CAAN5342.A2 Env infected JC53-BL cells efficiently, exhibiting titers well within the range of standard laboratory controls, YU-2 and NL4.3. In contrast, particles containing WITO4160.27 Env were only poorly infectious (23 IU/ng p24), suggesting a partially impaired envelope structure and/or function (Fig. 3). Of note, all consensus and wildtype Envs used CCR5 as the co-receptor for cell entry (Fig. 4).
Fig. 3.
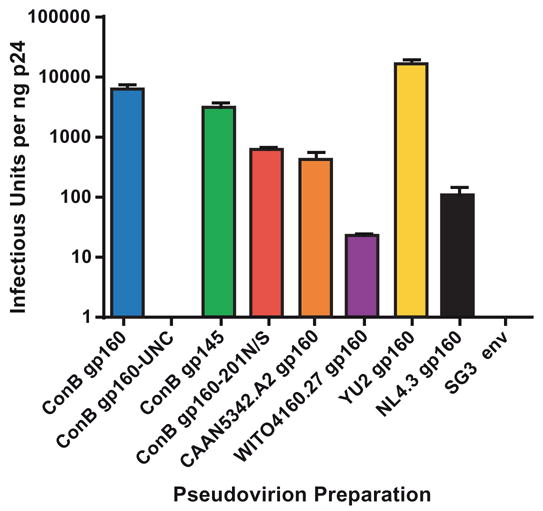
Infectivity of virions containing ConB Envs. Virus stocks were generated by co-transfection of ConB (ConB gp160, ConB gp160-UNC, ConB gp145, ConB gp160-201N/S) and contemporary (CAAN5342.A2, WITO41260.27) env genes with HIV-1/SG3Δenv. Infectivity was determined in JC53-BL cells by determining the number of blue cells (infectious units, IU) per nanogram of p24. Bars indicated standard errors (values are averaged from 3 independent experiments). Infectivity values of virions containing YU-2 and NL4.3 Envs are shown for control.
Fig. 4.
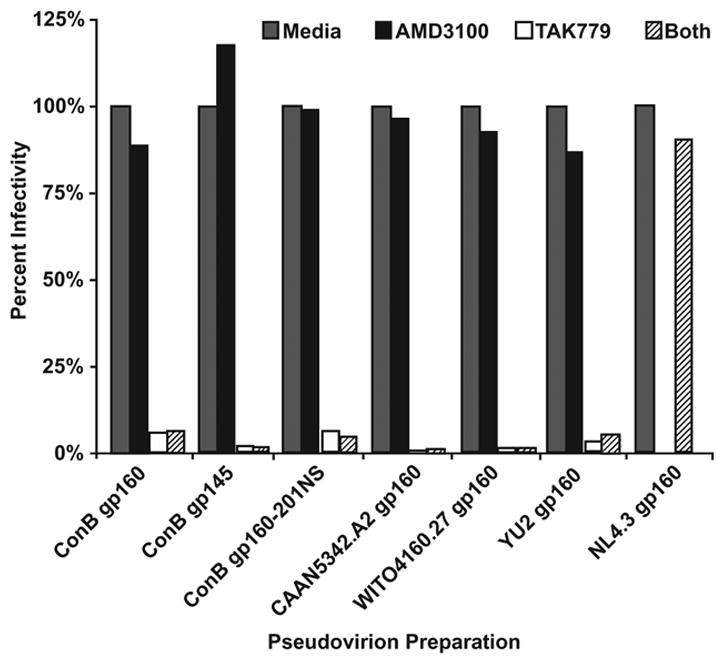
Co-receptor preference of ConB Envs. JC53-BL cells were pretreated with AMD3100 (inhibitor of CXCR4), TAK779 (inhibitor of CCR5), both or neither (media only) prior to addition of virions containing the Env proteins indicated. Virus infectivity is plotted on the vertical axis as a percentage of the untreated control. The CCR5-tropic YU-2 and the CXCR4-tropic NL4.3 Envs were included as controls.
Sensitivity of ConB Env glycoproteins to neutralization by patient plasma
To determine whether native and modified ConB envelope glycoproteins retained neutralizing epitopes also found in contemporary subtype B viruses, ConB gp160, gp145 and gp160-201N/S Env containing virions were tested for sensitivity to neutralization by “high reactive” and “low reactive” plasma pools (derived from HIV-1 infected individuals previously determined to have high and low titers of neutralizing antibodies against subtype B viruses, respectively). As shown in Figs. 5A and 5B, ConB gp160 and gp145 Envs were more sensitive to neutralization by patient plasma than YU2 Env, but less sensitive than NL4.3 Env. The neutralization profile of CAAN5342.A2 Env was comparable to that of YU-2, while WITO4160.27 Env exhibited greater sensitivity similar to NL4.3 Env. Interestingly, ConB gp160-201N/S represented the most sensitive Env, yielding IC50 values of 1: 2891 and 1:288 for the high and low reactive pools, respectively. Thus, a single amino acid substitution and associated glycosylation site change at the base of the V1V2 loop resulted in a greater than 5-fold increase in neutralization sensitivity of ConB Env to pooled patient plasma.
Fig. 5.
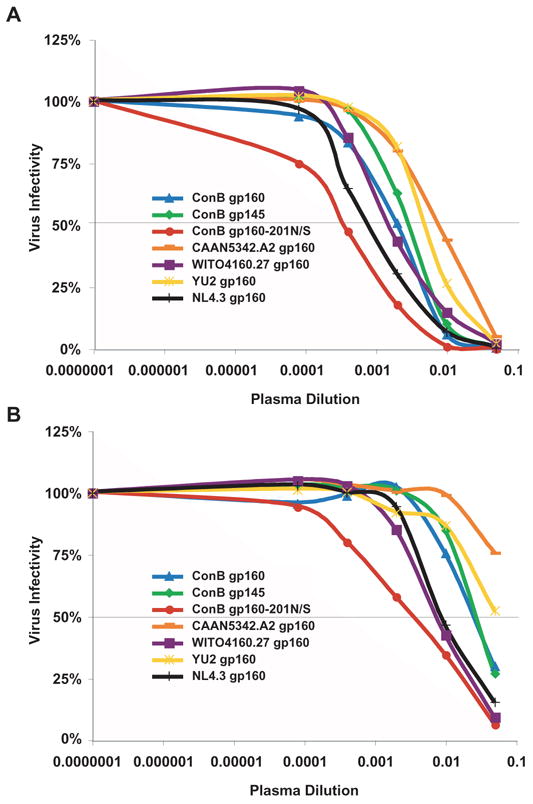
Neutralization of ConB Env containing virions by “high reactive” (A) and “low reactive” (B) plasma pools. Neutralization of viral infectivity in JC53-BL cells (y-axis) was scored as the plasma dilution (x-axis) required to reduce virus infectivity by 50% (IC50) (grey line).
To follow-up on these results, the same panel of ConB and wildtype Envs was tested for neutralization by plasma samples from seven individuals chronically infected with HIV-1 subtype B. As shown in Fig. 6, ConB gp160 and gp145 Envs were equally sensitive to neutralization by patient plasma, with IC50 titers of up to 1:700 for ConB gp160 and 1:500 for ConB gp145. CAAN5342.A2 Env was similar to YU2 Env (median titers were 7 and 15, respectively), while WITO4160.27 Env was similar to NL4.3 Env (median titers were 209 and 296, respectively). Again, the most sensitive Env was the glycosylation site mutant ConB gp160-201N/S, exhibiting IC50 titers of 1:402 to 1:7485. Thus, both ConB gp160 and gp145 envelope glycoproteins retained neutralizing epitopes common to contemporary (primary) subtype B viruses. Moreover, an Env modification believed to alter the position of the V1V2 loop increased the sensitivity of the ConB gp160 to neutralization by both pooled and individual patient plasma by up to 12-fold.
Fig. 6.
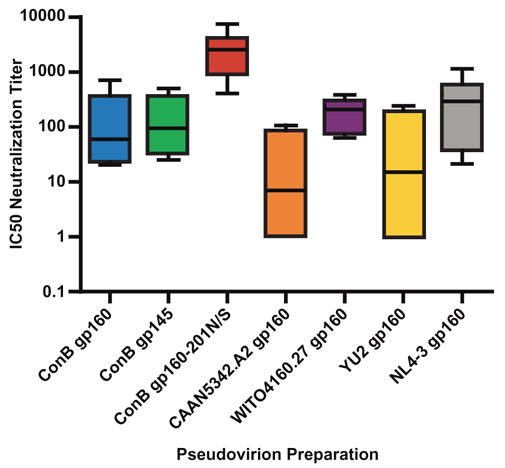
Neutralization of ConB Env containing virions by patient plasma. Plasma samples from seven subtype B infected individuals were tested for their ability to neutralize ConB or contemporary Env containing virions. Neutralization was scored as the plasma dilution required to reduce virus infectivity by 50% (IC50). Vertical boxes represent the 25th and 75th percentiles of the IC50 values, the line in the box the median, and the lines emerging from the box the highest and lowest serum dilutions observed for the group, respectively.
Sensitivity of ConB Env glycoproteins to neutralization by soluble CD4 and monoclonal antibodies
Primary isolates of HIV-1 display varying degrees of sensitivity to neutralization by soluble CD4 (sCD4) and monoclonal antibodies (e.g., IgG1b12, 2G12, 2F5, and 4E10) (Binley et al., 2004; Li et al., 2005). We thus tested the sensitivity of the ConB Env proteins to neutralization by these same reagents. As shown in Fig. 7, ConB gp160, ConB gp145 and CAAN5342.A2 Envs were completely resistant to neutralization by sCD4 when assayed at a maximum concentration of 100nM. ConB gp160-201N/S Env was significantly more sensitive, yielding an IC50 value of 54nM. Finally, NL4.3, YU2 and WITO4160.27 Envs were most sensitive, yielding IC50 values of 1.7 nM, 9.2 nM and 9.5 nM, respectively.
Fig. 7.
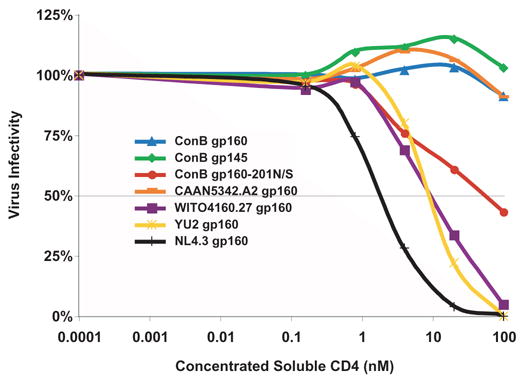
Neutralization of ConB Env containing virions by soluble CD4. Neutralization of viral infectivity in JC53-BL cells (y-axis) was scored as the concentration of sCD4 (x-axis) required to reduce virus infectivity by 50% (IC50) (grey line).
The monoclonal antibody IgG1b12 recognizes a neutralizing epitope on gp120 that overlaps the CD4-binding site (Pantophlet and Burton, 2006). As shown in Fig. 8A, all envelopes tested were sensitive to neutralization by IgG1b12, except ConB gp160-201N/S and CAAN5342.A2. ConB gp160 and WITO4160.27 Envs yielded IC50 values of 0.6 μg/ml; ConB gp145 Env was about 4-fold more sensitive, yielding an IC50 of 0.16 μg/ml. As reported previously (Wei et al., 2003), YU2 Env was quite resistant (IC50 2.5 μg/ml), while NL4.3 was highly sensitive (IC50 0.03 μg/ml). Thus, in the context of the ConB envelope glycoprotein, truncation of the gp41 cytoplasmic domain caused a conformational change in gp120 that resulted in increased accessibility of the IgG1b12 epitope, while an asparagine to serine substitution at position 201 in gp120 had the opposite effect.
Fig. 8.
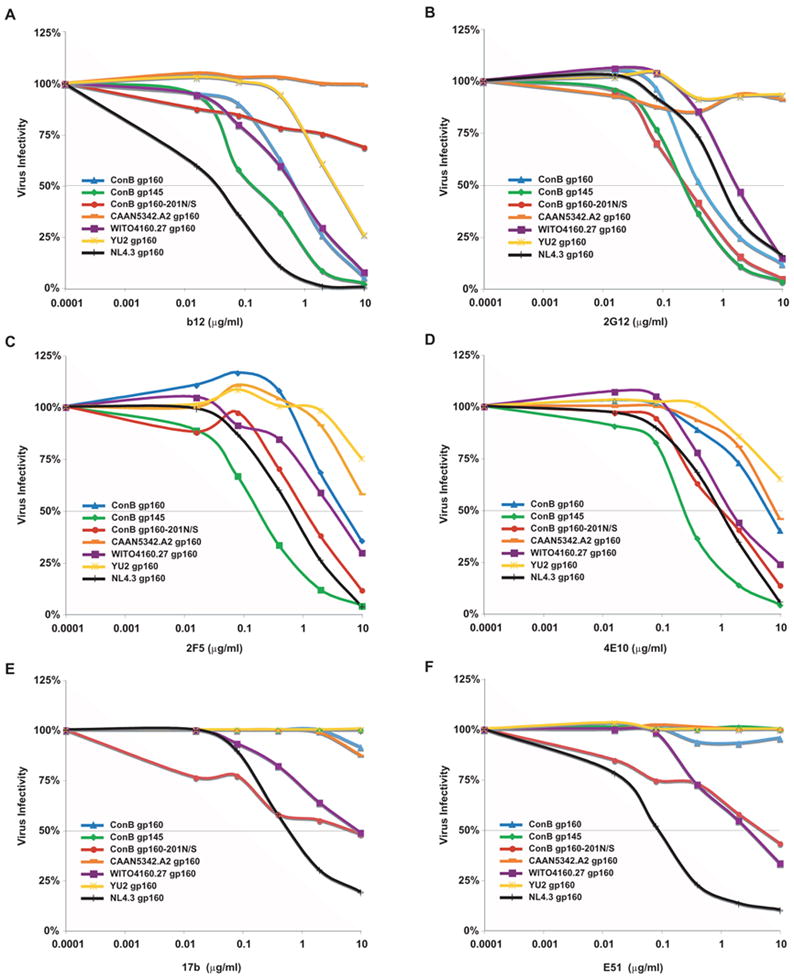
Neutralization of ConB Env containing virions by monoclonal antibodies. Neutralization of viral infectivity in JC53-BL cells (y-axis) by monoclonal antibodies IgGb12 (A), 2G12 (B), 2F5 (C), 4E10 (D), 17b (E) and E51 (F) was scored as the antibody concentration (μg/ml; x-axis) required to reduce virus infectivity by 50% (IC50) (grey line).
The various ConB and control Envs were also tested for neutralization by the monoclonal antibody 2G12 (Fig. 8B) which recognizes clustered mannose residues on the silent face of the gp120 outer domain (Scanlan et al., 2002; Pantophlet and Burton, 2006). CAAN5342.A2 and YU-2 Envs were completely resistant to neutralization by 2G12 (IC50 values > 10 μg/ml), while WITO4160.27 and NL4.3 Envs were somewhat sensitive (IC50 values 1.3 μg/ml and 1.0 μg/ml, respectively). The three ConB Envs were most sensitive, with very similar IC50 values ranging from 0.2 μg/ml to 0.3 μg/ml. Thus, the 2G12 epitope is present on the consensus subtype B envelope glycoprotein and not influenced by the Env modifications present in ConB gp145 and ConB gp160-201N/S.
The monoclonal antibodies 2F5 and 4E10 recognize epitopes in the membrane proximal external region (MPER), and 4E10 has broad neutralizing activity (Binley et al., 2004). CAAN5342.A2 and YU-2 Envs were resistant to neutralization by both 2F5 and 4E10, while WITO4160.27 and NL4.3 Envs were relatively more sensitive (Fig. 8C and D). ConB gp160 was quite resistant (5.2 μg/ml and 3.7 μg/ml for 2F5 and 4E10, respectively); ConB gp160-201N/S was more sensitive (1.1 μg/ml and 0.9 μg/ml); however, surprisingly, ConB gp145 Env was by far the most sensitive envelope, yielding IC50 values of 0.18 μg/ml and 0.27 μg/ml for 2F5 and 4E10, respectively. Since the 2F5 and 4E10 epitopes were not altered in ConB gp145, it seems clear that truncation of the gp41 cytoplasmic domain facilitates greater accessibility of the MPER region, at least within the context of the ConB Env protein.
Finally, to determine whether removal of an N-linked glycosylation site at position 201 in the ConB Env resulted in increased exposure of the CCR5-binding site, we performed neutralization assays using the co-receptor binding site antibodies, 17b and E51 (Wyatt et al., 1995; Trkola et al., 1996a; Sullivan et al., 1998; Wyatt et al., 1998; Kwong et al., 1998; Xiang et al., 2003). As expected, in the absence of sCD4, neither antibody was able to neutralize ConB gp160, ConB gp145, CAAN5342.A2 or YU2 Envs (Fig. 8E and F); however, ConB gp160-201N/S Env reached IC50 values of approximately 10 μg/ml for both antibodies, suggesting increased formation and/or exposure of the coreceptor binding site in this particular mutant. Of note, the poorly infectious WITO4160.27 Env was also relatively more sensitive to neutralization by 17b and E51 (6.0 μg/ml and 2.0 μg/ml, respectively), as was the control NL4.3 Env as previously described (Wei et al., 2003).
Humoral immune responses to ConB Envs in DNA immunized guinea pigs
To explore the immunogenicity of synthetic subtype B env constructs in vivo, guinea pigs (six or twelve animals per group) were immunized three times at four-week intervals with 400μg of ConB gp160, ConB gp160-UNC, ConB gp145, and ConB gp160-201N/S env DNA, as well as 400μg of CAAN5342.A2-opt and WITO4160.27-opt env DNA for wildtype control. Two weeks following the second and third immunization, serum samples were collected from each animal and assayed for the presence of binding antibodies to the respective (cognate) gp120 glycoprotein using an ELISA assay. As shown in Fig. 9, all ConB DNA vaccines elicited binding antibodies that reached endpoint titers of up to 1:160,000 after the third immunization. Surprisingly, the binding antibody titers of guinea pigs vaccinated with the two contemporary env genes, WITO4160.27-opt and CAAN5342.A2-opt, were 10 to 40-fold lower. This was not due to type specific immune responses since the cognate gp120 proteins were used for their detection. Moreover, the lower binding antibody titers did not reflect an overall reduced immunogenicity, since CAAN5342.A2 env was as potent as ConB gp160 with respect to inducing neutralizing antibodies (see below).
Fig. 9.
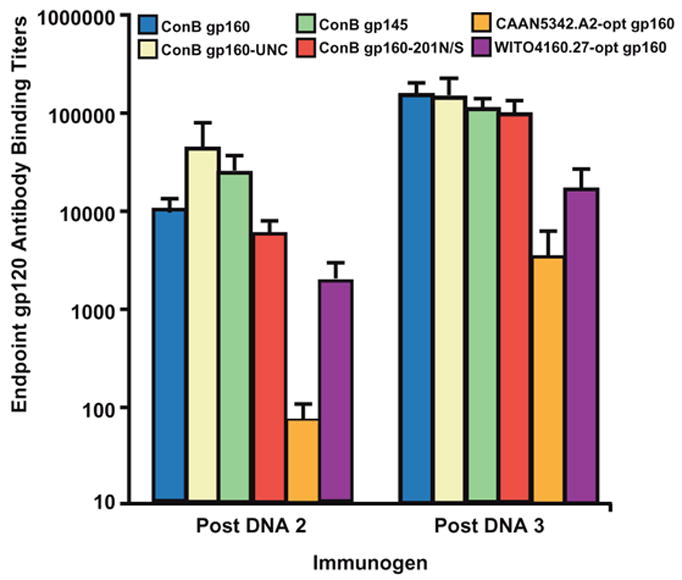
Humoral immune responses to DNA vaccination. Guinea pigs were vaccinated three times at four-week intervals with plasmid DNA containing ConB (ConB gp160, ConB gp160-UNC, ConB gp145, ConB gp160-201N/S) or contemporary env genes (CAAN5342.A2-opt, WITO4160.27-opt). Two-weeks following the third vaccination, sera were assayed for the presence of binding antibodies to their respective (cognate) gp120 proteins. Sera were serially diluted and the last dilution giving absorbance values greater than twice the optical density (OD) value of the negative control was identified as the endpoint titer. Vertical boxes indicate the mean endpoint titer ± SEM for each of the groups indicated.
Sera from immunized guinea pigs were next tested for neutralizing antibodies using a single-round infectivity assay as described (Wei et al., 2003; Decker et al., 2005; Li et al., 2005). Each serum was analyzed at a 1:10 dilution and its neutralization activity determined relative to the baseline activity of the pre-immune serum from the same animal. In addition, each pre- and post-immunization serum was tested for neutralization of MuLV envelope-containing virions. Thus, all neutralization values generated were quality controlled not only for non-specific anti-viral but also anti-cellular serum activities (Table 1). We developed this approach specifically for DNA vaccination studies because of the expected low titer neutralizing responses. We reasoned that after correction of non-specific serum activity both at the pre- and post immune level, even a modest reduction of infectivity at a 1:10 serum dilution would indicate HIV-1 Env specific neutralization. Using this approach, we were thus able to conduct meaningful comparisons even of very low level neutralization responses.
Table 1 summarizes the results of the neutralization studies. All guinea pigs except one (animal 229) developed antibodies that neutralized at least one of the viruses listed, albeit at very low levels. The viruses most commonly neutralized were those containing globally sensitive envelopes, including WITO4160.27 and ConB gp160-201N/S (also see Fig. 6), as well as the tier 1 viruses, SS1196.1 and SF162 (Mascola et al., 2005; Li et al., 2005). Interestingly, 81% of guinea pig sera also had neutralizing activity against the relative more resistant CAAN5342.A2 Env. However, less than half of the immunized animals developed a neutralizing antibody response against tier 2 viruses 3988.25, SC422661.8, THRO4156.18, and REJO4541.67 which represent relative resistant (subtype matched) primary isolates (Li et al., 2005). Importantly, there were no significant differences between autologous and heterologous responses for any of the immunogens tested, indicating that immunization with ConB env vaccines did not confer significantly greater neutralizing power against consensus B glycoproteins.
We next asked whether statistically significant differences existed between consensus and wildtype immunogens with respect to breadth and magnitude of the elicited response. In a first step, we combined heterologous neutralization data for the four ConB and the two wildtype env immunogens, and compared these as two different groups. Using a Wilcoxon rank sum test with continuity correction, we found no significant differences in the breadth of the neutralizing antibody response, i.e., antibodies from both immunogen groups neutralized a similar number of viruses (p = 0.1482); however, compared to the contemporary immunogens, ConB env vaccines elicited an antibody response of greater magnitude (p=0.0121), as reflected by the number of sera that reduced virus infectivity by 50% or more (highlighted in dark blue in Table 1).
In a second step, we wished to compare individual immunogens with respect to their relative potency (defined as a combination of both breadth and magnitude). For this analysis, all neutralization data listed Table 1 were subjected to multiple pairwise comparisons using Tukey’s Honestly Significant Difference procedure as described in Materials and Methods. Fig. 10 depicts the estimated differences between neutralization coefficients (solid circles) determined for all possible combinations of immunogens, as well as a 95% confidence interval for those differences (brackets). A negative difference (circle to the left of the vertical line) indicates that the first named immunogen in the pair is more potent at eliciting a neutralizing antibody response than the second named immunogen. Conversely, a positive difference (circle to the right of the vertical line) indicates that the first named immunogen in the pair is less potent than the second named immunogen. Confidence intervals that do not cross the dotted vertical line indicate statistically significant differences in potency. When responses against all 12 viruses listed in Table 1 were included (Fig. 10A), ConB gp145 env emerged as the best and WITO4160.27 env as the worst immunogen, with no significant potency differences observed for the remainder of the immunogens. Analyzing only responses to tier 1 viruses (SF162 and SS1196.1) yielded similar results (Fig. 10B): ConB gp145, and ConB gp160 env vaccines, were found to be significantly more potent than the two wildtype immunogens WITO4160.27 and CAAN5342.A2. However, restricting the analysis to tier 2 viruses (3988.25, QH0692.42, SC422661.8, THRO4156.18, REJO4541.67) resulted in the loss of all statistically significant differences between consensus and wildtype immunogens (Fig. 10C).
Fig. 10.
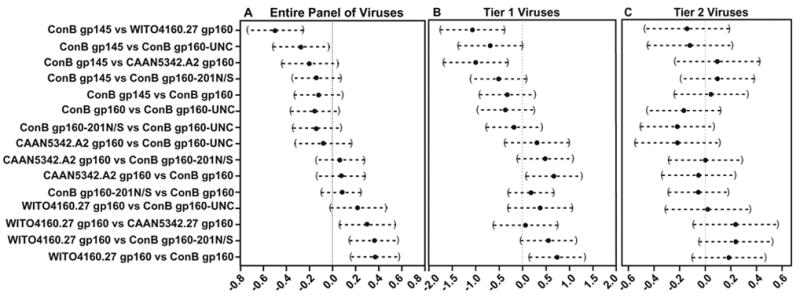
Relative potency of ConB and wildtype env vaccines as determined by comparison of neutralization responses against the entire panel of viruses (A), tier 1 viruses (B), and tier 2 viruses (C), as indicated in Table 1. Neutralization data were analyzed by performing multiple pairwise comparisons (Tukey’s Honestly Significant Difference procedure). An estimated difference between neutralization coefficients (solid circles) as well as a 95% confidence interval for that difference (brackets) are shown for all pairwise comparisons of immunogens (as indicated on the left). A negative difference (circle to the left of the vertical dotted line) indicates the first named immunogen of the pair was better at eliciting a neutralizing antibody response than the second named immunogen. Pairwise comparison differences with 95% confidence intervals that do not cross the dotted vertical line indicate statistically significant differences in potency.
Finally, to test for the presence of CD4i antibodies, guinea pig sera were incubated with virions containing an HIV-2 envelope glycoprotein (HIV-2/7312A) in the presence and absence of sCD4 as described (Decker et al., 2005). No reduction of infectivity was observed indicating the absence of neutralizing antibodies directed against the CCR5 binding site (not shown). Guinea pig sera were also tested using virions that contained a chimeric HIV-2 envelope expressing the entire 23 amino acid HIV-1 MPER region. Such HIV-2 Env scaffolds have previously been shown to detect 2F5 and 4E10 neutralizing antibodies as well as antibodies directed against other epitopes in the MPER region (Bibollet-Ruche et al., 2006). Again, no reduction in infectivity was observed, indicating the absence of MPER neutralizing antibodies in all vaccinated guinea pigs (not shown).
Discussion
In this study, we characterized the biologic and antigenic properties of four synthetic HIV-1 subtype B env gene products and compared their ability to elicit neutralizing antibodies when used as DNA vaccines in guinea pigs. The purpose of this study was to address three questions: (i) can consensus subtype B env genes be generated that encode functional glycoproteins; (ii) is there an advantage for the use of consensus over contemporary subtype B env immunogens for eliciting neutralizing antibodies; and (iii) can consensus glycoproteins be modified to improve their immunogenicity.
Biologic and antigenic properties ConB envelope glycoproteins
Despite their artificial nature, full length and truncated subtype B consensus env genes encode functional glycoproteins. Both ConB gp160 and ConB gp145 Envs conferred infectivity to pseudotyped virions at levels comparable to wildtype envelope glycoproteins. ConB gp160-UNC was cleavage defective and thus non-infectious. Finally, the ConB gp160201N/S mutant was processed and packaged, but particles containing this envelope were less infectious. Since a similar phenotype has recently been described for HIV-1 and SHIV Env proteins with glycosylation site changes at analogous sites (Kolchinsky et al., 2001a,b; Hu et al., 2005), these data indicate that consensus subtype B envelopes, like their wildtype counterparts, can be modified in an overall predictable fashion.
Truncation of the cytoplasmic domain of gp41 has been reported to expose otherwise hidden neutralizing epitopes in wildtype HIV-1 envelopes (Edwards et al., 2002). Although increased sensitivity to neutralization by certain monoclonal antibodies was observed (Fig. 8), ConB gp145 did not exhibit the globally sensitive phenotype of the previously described cytoplasmic domain truncated HIV-1 envelopes, JRFL and ADA (Edwards et al., 2002). The latter were highly sensitive to neutralization by pooled patient sera as well as monoclonal antibodies directed to the CD4-binding site (IgG1b12) and the bridging sheet (17b, 48d) (Edwards et al., 2002). ConB gp145 was not particularly sensitive to neutralization by patient plasma and resistant to CD4i monoclonal antibodies (Figs. 5, 6 and 8). However, ConB gp145 was 5 to 10-fold more sensitive than ConB gp160 to neutralization by IgG1b12, 2F5 and 4E10. Of note, the truncated JRFL and ADA mutants each retained a cytoplasmic domain of 43 amino acids, while ConB gp145 was truncated immediately after the membrane spanning domain. It is thus possible that the only modest increase in neutralization sensitivity of ConB gp145 Env is due to these length differences and/or is context dependent. Nonetheless, our results show that gp41 truncations do alter the presentation of neutralizing epitopes within the ConB envelope.
Removal of a single N-linked glycosylation site at the base of the V1V2 loop has been reported to render wildtype HIV-1 envelopes more neutralization sensitive (Kolchinsky et al., 2001a,b; Hu et al., 2005), and in the case of one envelope (HIV-1/ADA) to result in a CD4 independent phenotype (Kolchinsky et al., 2001b). Alteration of an N-linked glycosylation site at the analogous position in ConB gp160-201N/S also resulted in an increased sensitivity to neutralization by patient plasma, sCD4, and monoclonal antibodies recognizing the MPER region and the bridging sheet (Figs. 5–8). However, this increase was only modest compared to the corresponding ADA mutant (197N/K). Moreover, the ConB gp160-201N/S Env remained CD4 dependent (not shown) and was resistant to neutralization by IgG1b12. It is believed that the CD4 binding site of the HIV-1 Env is partially occluded by the V1V2 loop and that the position of this loop (and thus access to the CD4 binding site) can be altered by the removal of a carbohydrate at its base (Kolchinsky et al., 2001a). We found that removal of this glycosylation site in ConB Env was not sufficient to increase neutralization by the CD4 binding site antibody IgG1b12 (Fig. 8A). However, introduction of a lysine rather than a serine at position 201 in ConB Env increased IgG1b12 neutralization sensitivity by 30-fold (ConB gp160 – 201N/K; IC50 = 0.02 μg/ml; data not shown). Thus, in addition to the glycosylation site, the particular amino acid at the base of the V1V2 loop (position 201 in ConB and 197 in ADA) appears to represent an important determinant of IgG1b12 sensitivity, at least in the context of the ConB envelope.
Immunogenic properties ConB envelope glycoproteins
A major goal of AIDS vaccine development is the induction of potent and broadly cross-reactive antibodies that neutralize the majority of currently circulating HIV-1 variants. In this study, we compared four ConB and two wildtype env immunogens following DNA immunization of guinea pigs. Sera were tested against a panel of 12 viruses (Table 1) which included the vaccine strains as well as subtype matched tier 1 and tier 2 viruses (Mascola et al., 2005; Li et al., 2005). Neutralizing antibodies were detected in all but one immunized guinea pig (Table 1), although the magnitude of the response was overall very low. At a 1:10 dilution, only 30% of all antibody positive sera reduced viral infectivity by more than 50% and none of these exceeded 85% neutralization. Given that env DNA vaccines rarely elicit neutralizing antibodies, these results are not unexpected. In fact, compared to neutralizing responses induced by wildtype and centralized subtype C env DNA vaccines, the subtype B responses are considerably more potent (Kothe et al., 2006).
A key question in current AIDS vaccine development is whether there is an advantage of using consensus rather than contemporary env immunogens for eliciting neutralizing antibodies. The results in Table 1 were rigorously controlled for both non-specific anti-viral and anti-cellular serum activities. Thus, it was possible to examine the relative utility of consensus versus wildtype env immunogens even when comparing only low level antibody responses. Conducting multiple pairwise comparisons, we found that ConB env DNA vaccines were at least as potent as wildtype subtype B env DNA vaccines (Fig. 10). Thus, there certainly was no disadvantage associated with using consensus env vaccines. Moreover, depending on the viruses selected for analysis, ConB gp145, and to a lesser extent ConB gp160, were significantly more potent than either wildtype env vaccine (Fig. 10A and B). Between the two wildtype controls, the neutralization resistant CAAN5342.A2 env elicited a significantly more potent neutralizing antibody response than the globally neutralization sensitive WITO4160.27 env. (Fig. 10A). When analyses were restricted to tier 1 or tier 2 viruses, most of the significant relationships were lost (Fig. 10B and C); however, in these instances, WITO4160.27 env still tended to rank at the bottom of all immunogens. These results highlight the difficulty of randomly selecting a contemporary envelope as a suitable vaccine candidate.
Numerous env modifications including the removal of N-linked glycosylation sites, deletion of variable loops, and cytoplasmic tail truncations have been tested for their ability to improve the neutralizing antibody response to HIV-1 env vaccines (Barnett et al., 2001; Hu et al., 2005; Kim et al., 2005; Liao et al., 2006; Quinones-Kochs et al., 2002). Collectively, these studies have shown that improvements are possible, but that successful modifications are frequently strain and/or context dependent (Barnett et al., 2001; Chakrabarti et al, 2002). We found that in the context of the ConB envelope, truncation of the cytoplasmic domain of gp41 was the most effective means to improve immunogenicity. ConB gp145 was significantly more potent than all other immunogens when tested against the entire virus panel (Fig. 10A) and remained the best immunogen when only tier 1 virus responses were analyzed (Fig. 10B). Even when analyses were restricted to tier 2 virus responses, ConB gp145 still ranked among the most potent immunogens, although this was no longer statistically significant (Fig. 10C). Interestingly, a recent study of patients with unusually broad neutralizing antibody responses revealed that these individuals harbor viruses whose envelopes have increased sensitivity to neutralization by 2F5 and 4E10 (Cham et al., 2006). ConB gp145 was also markedly more sensitive to neutralization by 2F5 and 4E10 (Fig. 8); yet, it did not elicit detectable MPER antibodies in guinea pigs. Thus, both in the patients with broadly cross-reactive neutralizing antibodies (Cham et al., 2006) and in the ConB gp145 immunized guinea pigs, the enhanced 4E10 and 2F5 sensitivity may be a surrogate of increased exposure of other neutralizing epitopes. Additional studies will be necessary to test this hypothesis.
ConB gp160-201N/S was constructed with the expectation that the asparagine to serine change at position 201 would increase antibody access to conserved epitopes normally occluded by the V1V2 loop. Although the sensitivity of ConB gp160-201N/S to neutralization by patient sera was markedly enhanced, the removal of the glycosylation site did not generate a superior immunogen. ConB gp160-201N/S elicited a neutralizing antibody response comparable to ConB gp160 and ConB gp160-UNC, but not as good as ConB gp145 (Fig. 10). Interestingly, a glycosylation change at the analogous site in a vaccinia based 89.6 Env immunogen induced neutralizing antibodies in macaques that were protective against an 89.6 Env containing SHIV challenge (Hu et al., 2005). These findings reemphasize the context dependent nature of Env modifications. Finally, the uncleaved version of ConB gp160 was not improved relative to cleaved ConB gp160 version as an immunogen.
Conclusions
In summary, we report here that consensus subtype B env genes express glycoproteins that resemble wildtype Envs in their overall structure and function, and that they elicit low level neutralizing antibody responses when administered as DNA to guinea pigs. Comparisons of individual immunogens indicate that ConB env vaccines are at least as potent, and in some instances more potent than wildtype env vaccines. Moreover, like their wildtype counterparts, consensus env immunogens are amenable to improvement by specific gene modifications. Given their documented utility for eliciting broadly cross-reactive T cell responses, consensus envelope immunogens should continue to be evaluated as potential components of future AIDS vaccines. In this context, it will be important to establish the relative merits of vaccines designed to be central to the entire M group versus those that are central to a single clade.
Materials and methods
Gene synthesis
The HIV-1 subtype B consensus full-length env gene sequence was codon-usage optimized as described (Haas et al., 1996; Andre et al., 1998; Gao et al., 2003; Kothe et al., 2006) and is available at Genbank under accession number DQ667594. All ConB env gene modifications were generated using the QuikChange Site-Directed Mutagenesis Kit (Invitrogen, Carlsbad, CA). The ConB gp160-UNC mutant was made by altering the primary cleavage site REKR to SEKS. ConB gp145 was generated by introduction of a premature stop codon (TAA) that truncates the envelope glycoprotein immediately after the membrane spanning domain. Lastly, ConB gp160-201N/S was generated by removing a potential N-linked glycosylation site at position 201 (201N/S). Individual env variants were sequence confirmed and cloned into pcDNA3.1 (Invitrogen, Carlsbad, CA).
Contemporary subtype B env genes were cloned by reverse transcriptase polymerase chain reaction (RT-PCR) amplification from the plasma of individuals with acute (WITO4160) and early (CAAN5342) HIV-1 infection (Li et al., 2005). CAAN5342 clone A2 has been characterized extensively and represents one of twelve subtype B env clones (SVPB19) included in a standard virus panel to assess neutralizing antibody responses (Li et al., 2005); WITO4160 clone 27 has not previously been described, but was derived from the same plasma sample as WITO4160 clone 33, also included in the standard subtype B env panel (SVPB18) (Li et al., 2005). The nucleotide sequences of the codon-usage optimized CAAN5342.A2 and WITO4160.27 env genes are available under accession numbers DQ821487 and DQ667595 (the corresponding wildtype sequences are available under AY835452 and DQ824742, respectively).
Western blot analysis of pseudovirion stocks
To test whether full-length and modified subtype B consensus Envs were capable of incorporating into virus particles, each gene was transfected with an env-minus HIV-1 backbone vector (SG3Δenv). Transfections were performed in 100mm-diameter dishes containing a 60% confluent layer of 293T cells in complete Dulbecco’s Modified Eagle Media (DMEM) using FuGene 6 (Roche Applied Science, Indianapolis, IN) as specified by the manufacturer. Forty-eight hours post-transfection, virus-containing supernatants were collected, clarified by low-speed centrifugation, passed through a 0.2 μm filter, and pelleted through a 20% sucrose cushion. Pelleted virus was normalized for p24 Gag content and then subjected to SDS-PAGE and Western blot analysis. Blots were probed with antibodies specific for HIV-1 Env glycoproteins (USB 6001-15, US Biologicals, Swampscott, MA) and p24 Gag (AG3.0, NIH AIDS Research & Reference Reagent Program, Bethesda, MD) and developed using HRP-labeled antibodies (SouthernBiotech, Birmingham, AL) and an enhanced chemiluminescence (ECL) based detection system (GE Healthcare Life Sciences, Piscataway, NJ).
Infectivity and co-receptor usage of ConB Env containing pseudovirions
Infectivity assays were performed as described (Derdeyn et al., 2000). Briefly, JC53-BL cells were seeded in 24-well plates at 50,000 cells per well in DMEM supplemented with 10% fetal bovine serum, and incubated overnight. Pseudotyped virus stocks containing ConB gp160, ConB gp160-UNC, ConB gp145, or ConB gp160-201NS Envs were added to each well in the presence of DEAE-Dextran hydrochloride (80 μg/ml) (Sigma-Aldrich, St. Louis, MO) in a final volume of 250μl. Following a 48-hour incubation, plates were washed, stained, and the number of blue cells counted to determine the infectious virus titer.
A modification of the infectivity assay was used to determine the co-receptor usage of the various consensus envelopes using antagonists to CXCR4 (AMD3100) and CCR5 (TAK-779) (Zhang et al., 2000; Spenlehauer et al., 2001). Briefly, JC53-BL cells were seeded overnight and then treated for 1 hour with AMD3100 (1.2 μM/well), TAK-779 (10 μM/well), a combination of these chemokine receptor blocking agents, or media. 2,000 infectious units of pseudotyped virions were added to each well in the presence of 80 μg/ml DEAE-Dextran hydrochloride and incubated at 37°C. Following a two-day incubation, supernatant was removed, and cells were lysed using a luciferase assay system kit (Promega, Madison, WI). The light intensity of each cell lysate was measured on a Tropix luminometer using Tropix WinGlow version 1.24 software. A reduction in infectivity in the presence of a specific inhibitor reflects a requirement of the targeted co-receptor for entry.
Neutralization sensitivity of ConB Env containing virions to patient plasma and neutralizing monoclonal antibodies
Pseudovirions containing the ConB gp160, ConB gp145, ConB gp160-201N/S, CAAN5342.A2, or WITO4160.27 envelope glycoproteins were examined for sensitivity to neutralization by patient plasma and monoclonal antibodies using a single round infectivity assay. Pseudovirions containing the standard laboratory Envs, YU-2 and NL4.3, were included as controls. JC53-BL cells were seeded at 8,000 cells per well in a 96-well plate in 10% DMEM media overnight at 37°C with 5% CO2. Plasma from HIV-1 subtype B infected individuals were serially diluted and incubated with 2,000 units of infectious virus per well for 1 hour at 37°C. Pre-incubated virus/plasma dilutions were added to the cells in the presence of DEAE-Dextran and incubated for 2 days at 37°C. Control wells containing pseudovirions that had not been pre-incubated with antibody were included for each virus tested. Additionally, cell-only wells were included on each plate as a measure of background. Cells were lysed and analyzed for luciferase activity by measuring relative light units (RLU) in a Tropix luminometer using Tropix WinGlow version 1.24 software. Neutralization was measured as the percent reduction of viral infectivity in comparison to control wells infected with virus alone. A similar assay was used to assess pseudovirion sensitivity to neutralization by soluble CD4 (sCD4) (R&D Systems, Minneapolis, MN) and a panel of human monoclonal antibodies. The following monoclonal antibodies were obtained through the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health: IgG1b12 (Roben et al., 1994), 2G12 (Trkola et al., 1996b), 2F5 (Muster et al., 1993), 4E10 (Buchacher et al., 1994), and 17b (Sullivan et al., 1998). The E51 monoclonal antibody was obtained from J. Robinson (Tulane University School of Medicine, New Orleans, LA). For these assays, infectious virus (2,000 IU) was pre-incubated with serial dilutions of sCD4 (using concentration ranging from 0.0001 μM to 100 μM) and monoclonal antibodies (using concentration ranging from 0.0001 μg/ml to 10 μg/ml) and then added to JC53-BL cells.
Guinea pig immunization and serum collection
Guinea pigs were housed according to Accreditation of Laboratory Animal Care (AALAC) guidelines. All protocols were approved by the Institutional Animal Care and Use Committee. Female Hartley guinea pigs (Harlan Sprague, Indianapolis, IN) (n = 12/group for ConB gp160 and ConB gp160-201N/S DNA vaccines; n = 6/group for ConB gp160-UNC, ConB gp145, CAAN5342.A2-opt, and WITO4160.27-opt DNA vaccines) were immunized intramuscularly three times at 4-week intervals with 400μg plasmid DNA. Two weeks following the last immunization, 5ml of blood was collected from each animal via the cranial vena cava. Sera were obtained by tabletop centrifugation using Becton Dickinson SST Tubes (BD, Franklin Lakes, NJ). Samples were stored at −20°C until analysis.
Endpoint binding titer ELISA for anti-Env antibody detection in immunized guinea pigs
Guinea pig sera were tested for binding antibodies to their cognate gp120 protein in an enzyme linked immunosorbent assay (ELISA). ConB, CAAN5342.A2 and WITO4160.27 gp120 glycoproteins were produced by transfecting 293T cells with the corresponding plasmids (engineered to contain a premature stop codon immediately prior to the gp160 cleavage site). Supernatants were harvested 72 hours post-transfection, clarified by centrifugation, and passed through a 0.2μM filter. Recombinant gp120 was purified using Galanthus Nivalis Lectin (GNL) (Vector Laboratories, Burlingame, CA), eluted with 500mM alpha-methyl mannoside (Vector Laboratories, Burlingame, CA), dialyzed overnight in PBS and quantified using a BCA Protein Assay (Pierce Biotechnology, Rockford, IL). Microtiter plates were coated with recombinant gp120 (0.5μg/ml in PBS), washed, and blocked with 200μl per well 5% nonfat milk in PBS-T. Serial five-fold dilutions were made of each guinea pig serum, added to individual wells, and set to incubate for 1 hour at 37°C. Following a wash, 100μl of HRP-conjugated goat anti-guinea pig antibody (ICN Pharmaceuticals, Costa Mesa, CA), diluted to 1:50,000 in blocking buffer was added to each well. After an additional 1-hour incubation at 37°C, 100μl of liquid TMB (3,3′,5,5′-tetramethylbenzidine) was added to each well. Reactions were stopped by the addition of 100μl of 4N sulfuric acid. Absorbances were read at 405nm on an MRX Microplate reader (DYNEX Technologies, West Sussex, UK). Endpoint titers were determined as the serum titer at which the absorbance value was 2x the mean OD of the negative serum control.
Neutralization analysis of sera from immunized guinea pigs
Sera from immunized guinea pigs were tested for neutralizing antibodies in duplicate. Pre- and post-immunization sera from the same animal were diluted 1:10, incubated with 100,000 relative light units (RLUs) of pseudovirions containing autologous or heterologous envelope glycoproteins in a total volume of 150 μl, and added to JC53-BL cells on a 96-well plate. To control for non-specific antiviral activity, these same sera were also tested for neutralization of viruses containing the amphotropic MuLV Env. Background RLUs were measured in wells containing only JC53-BL cells and media. Following a 48-hour incubation of cells and serum dilutions, cells were lysed and the corresponding RLUs measured using a Wallac 1420 luminometer (Perkin Elmer, Boston, MA) using Wallac 1420 version 3.00 software. Percent neutralization (as shown in Table 1) was calculated for each serum using the following equation (“ENV” represents autologous or heterologous HIV-1 Env glycoproteins):
Statistical analyses
To determine the relative potency of each immunogen, detailed statistical modeling of the respective neutralization data was undertaken. A survey of the raw neutralization results suggested that they were log-normally distributed (Shapiro-Wilk test on all available data, p > 0.5; also supported by separate Shapiro-Wilk tests on each microtiter plate). Thus, log-transformed neutralization ratios were used in all subsequent statistical analyses. All statistical modeling was performed with the statistical package R (R Development Core Team, 2006). Models were developed that predicted the log of the specific neutralization ratio: that is, the ratio of pre- and post-immune HIV-1 Env specific neutralization divided by the ratio of pre- and post-immune MuLV Env specific neutralization. Using a weighted least-squares approach to account for variations in the pattern of data replication, models were fit (Crawley, 2002) that predicted the log specific neutralization ratio as a function of the immunogen and the target strain. There were no significant interactions between these parameters (so, for example, immunization with a specific env vaccine did not confer significantly greater neutralizing power against that envelope). To account for multiple testing, a recent implementation of Tukey’s Honestly Significant Difference procedure (Bretz et al., 2001, Westfall 1997) was applied, yielding the simultaneous 95% confidence intervals plotted in Fig. 10. This procedure is appropriate for experimental designs that are statistically unbalanced such as the one described here.
Acknowledgments
We thank Larry Liao for unpublished information, the NIAID-sponsored Reagent Resource Support Program for AIDS Vaccine Development for providing the monoclonal antibodies IgG1b12, 2G12, 2F5, 4E10, Wendy J. Abbott and Jamie C. White for artwork and manuscript preparation. This work was supported in part by grants from the National Institutes of Health (NO1 AI85338, U19 AI 028147, P01 AI 061734, P30 AI27767, P30 CA13148, R21 AI 055386), an internal directed research (DR) grant for vaccine design at Los Alamos National Laboratory, the Bill and Melinda Gates Foundation (ID #37874), and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andre S, Seed B, Eberle J, Schraut W, Bultmann A, Haas J. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72:1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett SW, Lu S, Srivastava I, Cherpelis S, Gettie A, Blanchard J, Wang S, Mboudjeka I, Leung L, Lian Y, Fong A, Buckner C, Ly A, Hilt S, Ulmer J, Wild CT, Mascola JR, Stamatatos L. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J Virol. 2001;75:5526–5540. doi: 10.1128/JVI.75.12.5526-5540.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibollet-Ruche F, Li H, Decker JM, Goepfert PA, Hahn BH, Delaporte E, Peeters M, Allen S, Hunter E, Robinson JE, Kwong PD, Shaw GM. Keystone Symposia 2006: HIV Pathogenesis/ HIV Vaccines. 2006. Detection Of novel neutralizing antibody responses to the membrane proximal external region (MPER) of GP41 following infection by HIV-1 subtypes A, B, C, D, F, G, H, CRF01, CRF02, or CRF11; p. abstr 110. Abstr. [Google Scholar]
- Binley JM, Sanders RW, Master A, Cayanan CS, Wiley CL, Schiffner L, Travis B, Kuhmann S, Burton DR, Hu SL, Olson WC, Moore JP. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2002;76:2606–2616. doi: 10.1128/JVI.76.6.2606-2616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JF, Yang X, Sodroski J, Ross TM. Elicitation of neutralizing antibodies with DNA vaccines expressing soluble stabilized human immunodeficiency virus type 1 envelope glycoprotein trimers conjugated to C3d. J Virol. 2004;78:4710–4719. doi: 10.1128/JVI.78.9.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretz F, Genz A, Hothorn LA. On the numerical availability of multiple comparison procedures. Biom J. 2001;43:645–656. [Google Scholar]
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- Chakrabarti BK, Kong WP, Wu BY, Yang ZY, Friborg J, Ling X, King SR, Montefiori DC, Nabel GJ. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J Virol. 2002;76:5357–5368. doi: 10.1128/JVI.76.11.5357-5368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cham F, Zhang PF, Heyndrickx L, Bouma P, Zhong P, Katinger H, Robinson J, van der Groen G, Quinnan GV., Jr Neutralization and infectivity characteristics of envelope glycoproteins from human immunodeficiency virus type 1 infected donors whose sera exhibit broadly cross-reactive neutralizing activity. Virology. 2006;347:36–51. doi: 10.1016/j.virol.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Crawley MJ. Statistical Computing: An Introduction to Data Analysis Using S-Plus. John Wiley & Sons, Ltd; Chichester, West Sussex, England: 2002. [Google Scholar]
- Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med. 2005;201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Learn GH, Rodrigo AG, Nickle DC, Li F, Mahalanabis M, Hensel MT, McLaughlin S, Edmonson PF, Montefiori D, Barnett SW, Haigwood NL, Mullins JI. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J Virol. 2005;79:11214–11224. doi: 10.1128/JVI.79.17.11214-11224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douek DC, Kwong PD, Nabel GJ. The rational design of an AIDS vaccine. Cell. 2006;124:677–681. doi: 10.1016/j.cell.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Edwards TG, Wyss S, Reeves JD, Zolla-Pazner S, Hoxie JA, Doms RW, Baribaud F. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J Virol. 2002;76:2683–2691. doi: 10.1128/JVI.76.6.2683-2691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger DL, Li B, Lupo LD, Owen SM, Nkengasong J, Kadio-Morokro MS, Smith J, Robinson H, Ackers M, Greenberg A, Folks T, Butera S. Generation of a consensus sequence from prevalent and incident HIV-1 infections in West Africa to guide AIDS vaccine development. Virology. 2002;302:155–163. doi: 10.1006/viro.2002.1577. [DOI] [PubMed] [Google Scholar]
- Gao F, Li Y, Decker JM, Peyerl FW, Bibollet-Ruche F, Rodenburg CM, Chen Y, Shaw DR, Allen S, Musonda R, Shaw GM, Zajac AJ, Letvin N, Hahn BH. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res Hum Retroviruses. 2003;19:817–823. doi: 10.1089/088922203769232610. [DOI] [PubMed] [Google Scholar]
- Gao F, Weaver EA, Lu Z, Li Y, Liao HX, Ma B, Alam SM, Scearce RM, Sutherland LL, Yu JS, Decker JM, Shaw GM, Montefiori DC, Korber BT, Hahn BH, Haynes BF. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J Virol. 2005;79:1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, Korber B. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- Grundner C, Li Y, Louder M, Mascola J, Yang X, Sodroski J, Wyatt R. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology. 2005;331:33–46. doi: 10.1016/j.virol.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- Herrera C, Klasse PJ, Michael E, Kake S, Barnes K, Kibler CW, Campbell-Gardener L, Si Z, Sodroski J, Moore JP, Beddows S. The impact of envelope glycoprotein cleavage on the antigenicity, infectivity, and neutralization sensitivity of Env-pseudotyped human immunodeficiency virus type 1 particles. Virology. 2005;338:154–172. doi: 10.1016/j.virol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- HIV Vaccine Trials Network. [accessed March 1, 2006]; http://www.hvtn.org.
- Hu SL, Klots I, Cleveland B, Polacino P, Richardson B, Anderson D, Montefiori D. Abstr. Enhanced neutralizing antibody response elicited by a “Prime-Boost” immunization with HIV-1 Env Proteins with a single N-linked Glycosylation site mutation in the V2 Loop. AIDS Vaccine. Int Conf Prgm, abstr. 2005;73:2005. [Google Scholar]
- IAVI database of AIDS vaccines in human trials. IAVI Report. [accessed March 1, 2006]; http://www.iavi.org/trialsdb.
- Kim M, Qiao ZS, Montefiori DC, Haynes BF, Reinherz EL, Liao HX. Comparison of HIV Type 1 ADA gp120 monomers versus gp140 trimers as immunogens for the induction of neutralizing antibodies. AIDS Res Hum Retroviruses. 2005;21:58–67. doi: 10.1089/aid.2005.21.58. [DOI] [PubMed] [Google Scholar]
- Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J Virol. 2001a;75:3435–3443. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolchinsky P, Kiprilov E, Sodroski J. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol. 2001b;75:2041–2050. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- Kothe DL, Li Y, Decker JM, Bibollet-Ruche F, Zammit K, Salazar M, Chen Y, Weng Z, Weaver E, Gao F, Haynes BF, Shaw GM, Korber BTM, Hahn BH. Ancestral and Consensus Envelope Immunogens for HIV-1 Subtype C. Virology. 2006;352:438–449. doi: 10.1016/j.virol.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S, Korber B. HIV Sequence Compendium. Theoretical Biology and Biophysics Group; Los Alamos National Laboratory, NM, LA-UR 06-0680: 2005. [Google Scholar]
- Letvin NL, Huang Y, Chakrabarti BK, Xu L, Seaman MS, Beaudry K, Korioth-Schmitz B, Yu F, Rohne D, Martin KL, Miura A, Kong WP, Yang ZY, Gelman RS, Golubeva OG, Montefiori DC, Mascola JR, Nabel GJ. Heterologous envelope immunogens contribute to AIDS vaccine protection in rhesus monkeys. J Virol. 2004;78:7490–7497. doi: 10.1128/JVI.78.14.7490-7497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-X, Sutherland LL, Xia S-M, Brock ME, Scearce RM, Vanleeuwen S, Alam SM, McAdams M, Weaver EA, Ma B-J, Li Y, Decker JM, Nabel GJ, Montefiori DC, Hahn BH, Korber BT, Gao F, Haynes BF. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. doi: 10.1016/j.virol.2006.04.043. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Sambor A, Beaudry K, Santra S, Welcher B, Louder MK, Vancott TC, Huang Y, Chakrabarti BK, Kong WP, Yang ZY, Xu L, Montefiori DC, Nabel GJ, Letvin NL. Neutralizing antibodies elicited by immunization of monkeys with DNA plasmids and recombinant adenoviral vectors expressing human immunodeficiency virus type 1 proteins. J Virol. 2005;79:771–779. doi: 10.1128/JVI.79.2.771-779.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JI, Nickle DC, Heath L, Rodrigo AG, Learn GH. Immunogen sequence: the fourth tier of AIDS vaccine design. Expert Rev Vaccines. 2004;3:S151–S159. doi: 10.1586/14760584.3.4.s151. [DOI] [PubMed] [Google Scholar]
- Mullins JI, Jensen MA. Evolutionary dynamics of HIV-1 and the control of AIDS. Curr Top Microbiol Immunol. 2006;299:171–192. doi: 10.1007/3-540-26397-7_6. [DOI] [PubMed] [Google Scholar]
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J, Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickle DC, Jensen MA, Gottlieb GS, Shriner D, Learn GH, Rodrigo AG, Mullins JI. Consensus and ancestral state HIV vaccines. Science. 2003;299:1515–1518. doi: 10.1126/science.299.5612.1515c. author reply 1515–1518. [DOI] [PubMed] [Google Scholar]
- Novitsky V, Smith UR, Gilbert P, McLane MF, Chigwedere P, Williamson C, Ndung'u T, Klein I, Chang SY, Peter T, Thior I, Foley BT, Gaolekwe S, Rybak N, Gaseitsiwe S, Vannberg F, Marlink R, Lee TH, Essex M. Human immunodeficiency virus type 1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J Virol. 2002;76:5435–5451. doi: 10.1128/JVI.76.11.5435-5451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanov S, Pattou C, Walker N, Schwardlander B, Esparza J. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J Acquir Immune Defic Syndr. 2002;29:184–190. doi: 10.1097/00042560-200202010-00013. [DOI] [PubMed] [Google Scholar]
- Pantophlet R, Burton DR. GP120: Target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- Pancera M, Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology. 2005;332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones-Kochs MI, Buonocore L, Rose JK. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J Virol. 2002;76:4199–4211. doi: 10.1128/JVI.76.9.4199-4211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2006. [Google Scholar]
- Rambaut A, Robertson DL, Pybus OG, Peeters M, Holmes EC. Human immunodeficiency virus. Phylogeny and the origin of HIV-1 Nature. 2001;410:1047–1048. doi: 10.1038/35074179. [DOI] [PubMed] [Google Scholar]
- Rao SS, Gomez P, Mascola JR, Dang V, Krivulka GR, Yu F, Lord CI, Shen L, Bailer R, Nabel GJ, Letvin NL. Comparative evaluation of three different intramuscular delivery methods for DNA immunization in a nonhuman primate animal model. Vaccine. 2006;24:367–373. doi: 10.1016/j.vaccine.2005.07.072. [DOI] [PubMed] [Google Scholar]
- Roben P, Moore JP, Thali M, Sodroski J, Barbas CF, 3rd, Burton DR. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DL, Sharp PM, McCutchan FE, Hahn BH. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- Robinson HL, Torres CA. DNA vaccines. Semin Immunol. 1997;9:271–283. doi: 10.1006/smim.1997.0083. [DOI] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulke N, Vesanen MS, Sanders RW, Zhu P, Lu M, Anselma DJ, Villa AR, Parren PW, Binley JM, Roux KH, Maddon PJ, Moore JP, Olson WC. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol. 2002;76:7760–7776. doi: 10.1128/JVI.76.15.7760-7776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Bailes E, Gao F, Beer BE, Hirsch VM, Hahn BH. Origins and evolution of AIDS viruses: estimating the time-scale. Biochem Soc Trans. 2000;28 :275–282. doi: 10.1042/bst0280275. [DOI] [PubMed] [Google Scholar]
- Si Z, Phan N, Kiprilov E, Sodroski J. Effects of HIV type 1 envelope glycoprotein proteolytic processing on antigenicity. AIDS Res Hum Retroviruses. 2003;19 :217–226. doi: 10.1089/088922203763315722. [DOI] [PubMed] [Google Scholar]
- Spenlehauer C, Kirn A, Aubertin AM, Moog C. Antibody-mediated neutralization of primary human immunodeficiency virus type 1 isolates: investigation of the mechanism of inhibition. J, Virol. 2001;75:2235–2245. doi: 10.1128/JVI.75.5.2235-2245.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres CA, Iwasaki A, Barber BH, Robinson HL. Differential dependence on target site tissue for gene gun and intramuscular DNA immunizations. J Immunol. 1997;158:4529–4532. [PubMed] [Google Scholar]
- Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996a;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996b;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanCott TC, Mascola JR, Kaminski RW, Kalyanaraman V, Hallberg PL, Burnett PR, Ulrich JT, Rechtman DJ, Birx DL. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J Virol. 1997;71:4319–4330. doi: 10.1128/jvi.71.6.4319-4330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal N, Peeters M, Mulanga-Kabeya C, Nzilambi N, Robertson D, Ilunga W, Sema H, Tshimanga K, Bongo B, Delaporte E. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol. 2000;74:10498–10507. doi: 10.1128/jvi.74.22.10498-10507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver EA, Lu Z, Camacho ZT, Moukdar F, Liao HX, Ma BJ, Muldoon M, Theiler J, Nabel GJ, Letvin NL, Korber BT, Hahn BH, Haynes BF, Gao F. Cross-subtype T cell immune responses induced by an HIV-1 group M Consensus Env immunogen. J Virol. 2006;80:6745–6756. doi: 10.1128/JVI.02484-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Westfall P. Multiple testing of general contrasts using logical constraints and correlation. J Am Stat Assoc. 1997;92:299–306. [Google Scholar]
- Worobey M. In: The origins and diversity of HIV. Sande M, Lange J, Volberding P, Greene WC, editors. Global HIV/AIDS Medicine; Elsevier, Philadelphia: in press. [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang SH, Wang L, Abreu M, Huang CC, Kwong PD, Rosenberg E, Robinson JE, Sodroski J. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology. 2003;315:124–134. doi: 10.1016/s0042-6822(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Yang X, Wyatt R, Sodroski J. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol. 2001;75:1165–1171. doi: 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lou B, Lal RB, Gettie A, Marx PA, Moore JP. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J Virol. 2000;74:6893–68910. doi: 10.1128/jvi.74.15.6893-6910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


