Abstract
Background
As part of our investigation into the genetic basis of tumor cell radioresponse, we have isolated several clones with a wide range of responses to X-radiation (XR) from an unirradiated human colorectal tumor cell line, HCT116. Using human cDNA microarrays, we recently identified a novel gene that was down-regulated by two-fold in an XR-resistant cell clone, HCT116Clone2_XRR. We have named this gene as X-ray radiation resistance associated 1 (XRRA1) (GenBank BK000541). Here, we present the first report on the molecular cloning, genomic characterization and over-expression of the XRRA1 gene.
Results
We found that XRRA1 was expressed predominantly in testis of both human and macaque. cDNA microarray analysis showed three-fold higher expression of XRRA1 in macaque testis relative to other tissues. We further cloned the macaque XRRA1 cDNA (GenBank AB072776) and a human XRRA1 splice variant from HCT116Clone2_XRR (GenBank AY163836). In silico analysis revealed the full-length human XRRA1, mouse, rat and bovine Xrra1 cDNAs. The XRRA1 gene comprises 11 exons and spans 64 kb on chromosome 11q13.3. Human and macaque cDNAs share 96% homology. Human XRRA1 cDNA is 1987 nt long and encodes a protein of 559 aa. XRRA1 protein is highly conserved in human, macaque, mouse, rat, pig, and bovine. GFP-XRRA1 fusion protein was detected in both the nucleus and cytoplasm of HCT116 clones and COS-7 cells. Interestingly, we found evidence that COS-7 cells which over-expressed XRRA1 lacked Ku86 (Ku80, XRCC5), a non-homologous end joining (NHEJ) DNA repair molecule, in the nucleus. RT-PCR analysis showed differential expression of XRRA1 after XR in HCT116 clones manifesting significantly different XR responses. Further, we found that XRRA1 was expressed in most tumor cell types. Surprisingly, mouse Xrra1 was detected in mouse embryonic stem cells R1.
Conclusions
Both XRRA1 cDNA and protein are highly conserved among mammals, suggesting that XRRA1 may have similar functions. Our results also suggest that the genetic modulation of XRRA1 may affect the XR responses of HCT116 clones and that XRRA1 may have a role in the response of human tumor and normal cells to XR. XRRA1 might be correlated with cancer development and might also be an early expressed gene.
Background
Treatment with X-radiation (XR) remains a major modality for eradicating cancer and it has been estimated that half of all cancer patients undergo radiotherapy at some point during their treatment [1]. At least partly because of the relevance of radiotherapy to cancer treatment, the genetic control of the expression of resistance, or conversely, sensitivity to XR has been investigated for several decades. It is now generally accepted that there are several components to the cellular response to XR stress: sensing of the DNA damage, cell signaling pathways, and repair of the DNA damage [2]. However, the molecular mechanisms of how cells detect, communicate, and cope with XR-induced DNA damage are not well characterized. There is strong evidence that ataxia telangiectasia mutated (ATM) protein act as damage sensors and that their activation initiates mobilization and/or activation of repair complexes [3]. In addition, the ATM kinase activity mediates the prompt induction of various signaling pathways with some of these events leading to either cellular apoptosis or cell cycle arrest [4]. There is also evidence that non-homologous end joining involving Ku86/Ku70 is likely the major DNA double strand breaks rejoining pathway following XR with a smaller role for homologous recombination in post replicated DNA [5].
Many of the studies on the genetic basis of cellular response to XR have benefited either from mutants that have increased ionizing radiation (IR) sensitivity compared to the parental cell lines or from the study of human genetic syndromes that are associated with increased IR sensitivity (i.e. Fanconi's anemia, Bloom's, and Werner's syndromes) [6,7]. By contrast, mutants or human genetic syndromes associated with increased IR resistance (as exemplified by Li Fraumeni) are rare [8]. Furthermore, Bennett et al [9] recently identified 107 new loci that are associated with response to γ-radiation in yeast. While over 50% of these newly identified putative yeast genes shared homology with human genes, it remains to be established that the human counterparts of these genes are actually associated with similar responses to γ-radiation.
In order to further investigate the genetic basis of tumor cell radioresponse, we have isolated several clones with a wide range of responses to X-radiation from an unirradiated human colorectal tumor cell line, HCT116. Interestingly, we found a cell clone, HCT116Clone2_XRR that was resistant to both fractionated and single dose X-radiation when compared to the parental cells. In addition, we also isolated two other clones: (a) HCT116CloneK_XRS that was more sensitive than the parental cells, and (b) HCT116Clone10 that possessed similar X-radiation responses as the parental cells, to both of these types of X-radiation (Qutob, S.S., Ng, C.E., to be published elsewhere). Global gene analysis between cells HCT116Clone2_XRR and HCT116Clone10 using cDNA microarray identified an unknown GenBank expression sequence tag (EST) accession no. R40588, as one of the spots on the 19,200 human cDNA microarray (University Health Network, Ontario Cancer Institute, Toronto), that was down regulated by approximately two-fold in HCT116Clone2_XRR in comparison with HCT116Clone10 cells (Qutob, S.S., Ng, C.E., to be published elsewhere).
Here we describe the in silico analysis of R40588 that resulted in the identification of a novel gene, XRRA1. We cloned the XRRA1 full-length cDNA from macaque testis and an XRRA1 splice variant from HCT116Clone2_XRR cells. We then extended our analysis to identify XRRA1 in several mammals (i.e. mouse, rat, pig and bovine). This paper also reports the experiments to evaluate XRRA1 expression in human and macaque normal tissue/organs and in various human cancer and normal cell lines, as well as over-expression of GFP-XRRA1 fusion protein in HCT116 clones and COS-7 cells. We did immunostaining of Ku86 in the presence of over-expressed GFP-XRRA1 24 hours after XR treatment at 4 and 10 Gy of COS-7 cells. This is the first report on the molecular cloning, genomic characterization and over-expression of the novel XRRA1 gene.
Results
Genomic approaches revealed a novel XRRA1 gene
Using the EST sequence of R40588 as template, we performed ESTs "walking" either towards the 5' or 3' direction on both the sense and anti-sense strands that we refer to as ESTs-based ORF assembling (EBOA). By EBOA, we acquired a cDNA candidate that putatively contained start-, stop-codon and poly-A signal. ESTs that overlapped each other sequentially were as follows: GenBank accession nos R40588, BQ648278, BU187847, BE782795, BI819448, BQ925455, and BM563944. Out of this ORF candidate, we found 31 ESTs as primary sequences that could be constructed into one potential novel gene in silico (Table 1, Fig. 1). We have named this gene as X-ray radiation resistance associated 1 (XRRA1) (GenBank accession no. BK000541). BLAST [10] analysis demonstrated that it was a novel gene.
Table 1.
ESTs that were used to build Hs XRRA1 and Mm Xrra1.
| Organism | EST | Nucleotides | Exons | Genomic Clone | Exons |
| Homo sapiens XRRA1 (1987 nt) | 1. BM563944.1 | 1 – 822 | 1 – 7 | AP001992.4 | 1 – 5 |
| 2. BE780143.1 | 1 – 233 | 1 – 2 | AP000560.4 | 3 – 11 | |
| 3. AW962535.1 | 1 – 175 | 1 – 2 | AP001324.4 | 5 – 11 | |
| 4. BM921116.1 | 1 – 107 | 1 | |||
| 5. BQ925455.1 | 1 – 348, 513 – 1066 | 1 – 3, 4 – 7 | |||
| 6. AI651142.1 (+/-) | 225 – 348, 513 – 726 | 2 – 3, 4 – 6 | |||
| 7. AW448937.1 (+/-) | 234 – 348, 513 – 726 | 2 – 3, 4 – 6 | |||
| 8. AI651143.1 (+/-) | 245 – 348, 513 – 726 | 2 – 3, 4 – 6 | |||
| 9. AW197451.1 (+/-) | 270 – 348, 513 – 726 | 2 – 3, 4 – 6 | |||
| 10. AW895676.1 (+/-) | 513–609 | 4 – 5 | |||
| 11. BE836017.1 (+/-) | 513 – 609 | 4 – 5 | |||
| 12. BF810439.1 | 513 – 589 | 4 – 5 | |||
| 13. AI652186.1 (+/-) | 526 – 726 | 5 – 6 | |||
| 14. BE313013.1 | 583 – 1190 | 5 – 8 | |||
| 15. BF891539.1 (+/-) | 607 – 694 | 5 – 6 | |||
| 16. BI819118.1 | 897 – 1755 | 7 – 11 | |||
| 17. BQ319609.1 | 1271 – 1388 | 8 – 10 | |||
| 18. BU620818.1 (+/-) | 1275 – 1976 | 9 – 11 | |||
| 19. BU620757.1 (+/-) | 1312 – 1911 | 9 – 11 | |||
| 20. AI057634.1 | 1474 – 1981 | 10 – 11 | |||
| 21. BQ051594.1 | 1492 – 1979 | 10 – 11 | |||
| 22. BE782795.1 | 1500 – 1898, 1939–1987 | 11 | |||
| 23. AW189295.1 | 1502 – 1979 | 11 | |||
| 24. AI655334.1 | 1546 – 1976 | 11 | |||
| 25. AA854636.1 | 1556 – 1981 | 11 | |||
| 26. AI218588.1 | 1593 – 1979 | 11 | |||
| 27. AI457916.1 | 1594 – 1981 | 11 | |||
| 28. AW513519.1 (+/-) | 1614 – 1758 | 11 | |||
| 29. AA961245.1 (+/-) | 1680 – 1977 | 11 | |||
| 30. BU187847.1 | 1796 – 1987 | 11 | |||
| 31. AA953084.1 (+/-) | 1845 – 1977 | 11 | |||
| Mus musculus Xrra1 (1903 nt) | 1. BI990168.1 | 277 – 870 | 2 – 7 | NW_000328 | 1 – 11 |
| 2. AV046823.1 | 715 – 978 | 6 – 7 | |||
| 3. BE650319.1 | 857 – 1380 | 7 – 9 | |||
| 4. BU610007.1 | 1134 – 1459 | 8 – 10 | |||
| 5. AA144723.1 | 1144 – 1396 | 8 – 10 | |||
| 6. AI647383.1 | 1184 – 1372 | 8 – 9 | |||
| 7. BE956083.1 (+/-) | 1381 – 1903 | 9 – 11 | |||
| 8. BQ840215.1 | 1398 – 1649 | 10 – 11 | |||
| 9. BE631695.1 | 1458 – 1604 | 10 – 11 | |||
| 10. BQ175761.1 (+/-) | 1458 – 1903 | 10 – 11 | |||
| 11. BE956870.1 (+/-) | 1471 – 1903 | 10 – 11 | |||
| 12. AI449753.1 (+/-) | 1600 – 1901 | 11 | |||
| 13. AW125246.1 (+/-) | 1704 – 1903 | 11 |
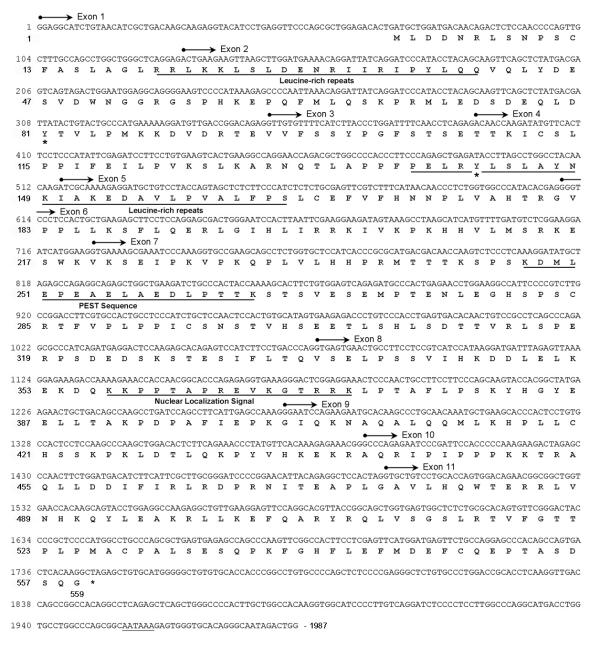
Figure 1.
Human XRRA1 cDNA sequences and its translation. The gene comprises 11 exons, and produces 559 aa protein. XRRA1 protein consists several motifs i.e. leucine rich repeats, PEST sequence, nuclear localization signal, and two sites of tyrosine phosphorylation (*). Poly A-signal, AATAA is underlined.
We used human genomic clones RP11-147I3 and CMB9-8M21 (GenBank accession nos AP001992 and AP000560) of chromosome 11q13.3 as template for GrailEXP [11] analysis. The analysis predicted 11 exons including a putative promoter region for the human (Homo sapiens, Hs) XRRA1 gene (Fig. 2, Fig. 3A). These 11 exons spliced perfectly to create the mRNA for Hs XRRA1. The exon-intron junctions, comprising the 5' splice donor and 3' splice acceptor, follow Kozak consensus (Table 3) [12]. Cloning of the 5' region of XRRA1 gene that comprised the first four exons from HCT116Clone2_XRR and HCT116Clone10 revealed that Hs XRRA1 had those exons. However, in addition to the one from HCT116Clone2_XRR, we obtained one splice variant that lacked exon three and ended on exon four (GenBank accession no. AY163836) (Fig. 3A).
Figure 2.
Promoter region of human XRRA1 gene as predicted by GrailExp [11]. Two putative sites of TATA box were found at -506 and -400 from the start codon. Predicted transcription factor binding sites revealed several members of bZIP family such as C/EBPa, c-Jun, c-Fos, AP-1 and CREB.
Figure 3.
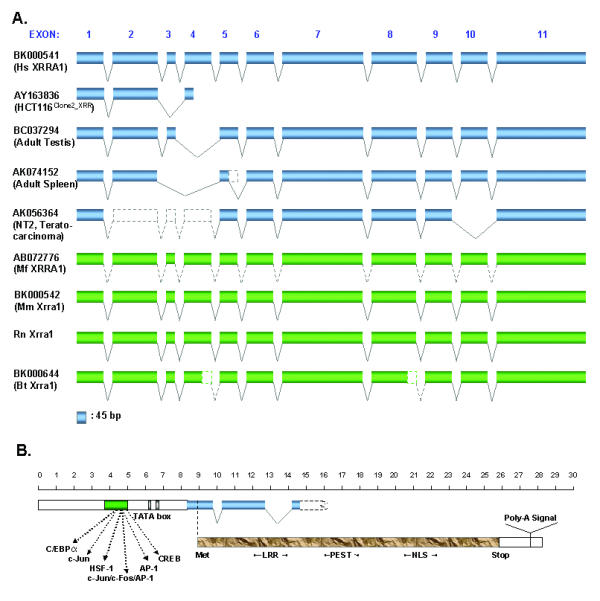
Representation of the XRRA1 genomic structure. (A) The XRRA1 gene spans 64 kb and comprises 11 exons. HCT116Clone2_XRR cells contain 2 splice variants, one of which lacks exon 3 and is truncated. Other cDNA clones such as BC032794 lack exon 4, AK074152 lacks exon 3–4 and possible partial deletion of exon 5, and AK056364 lacks exon 2–4 and 10. The gene is located between genomic markers D11S916-D11S911 (80.1–84.2 cM) or at position 625.9 cR of NCBI RH Map. (B) The assembled Hs XRRA1 comprises a putative promoter region, start/stop codons and poly-A signal, and predicted a protein with motifs such as LRR, PEST, and NLS. Scale bar is multiplied by 100.
Table 3.
Exon-intron junctions of human (Hs) XRRA1, murine (Mm) Xrra1 and rat (Rn) Xrra1
| Exon No. | Exon Size (bp) | 5' Splice Donor | Intron No. | Intron Size (bp)a | 3' Splice Acceptor | |
| 1 | Hs XRRA1 | 128 | GGCTCAGGAGgtaaagccataa | ttgcactcacagGGAGGCATCT | ||
| Mm Xrra1 | 133 | GGCTCAGGAGgtattgtcacag | ctgtgcccacagGGAGGCATCC | |||
| Rn Xrra1 | 133 | GGCTCAGGAGgtattatcacag | ctgtgcccacagGGAGGCATCT | |||
| 2 | Hs XRRA1 | 219 | GACCGGACAGgtgagtgacacc | 1 | 687 | ccttaattgaagACTGAAGAAG |
| Mm Xrra1 | 206 | GACCGCACAGgtgagctatggc | 685 | ccttaattgaagACTGAAGAAG | ||
| Rn Xrra1 | 206 | GACCGCACAGgtgagcccgccc | - | tcttaattgaagACTGAAGAAG | ||
| 3 | Hs XRRA1 | 43 | AACCTCAGAGgtaggaactgac | 2 | 43215 | cttctgtttcagAGGTTGTGTT |
| Mm Xrra1 | 44 | AACCTCAGAGgtaggaggacct | 4118 | tttctgtttcagAGGTTGTGTT | ||
| Rn Xrra1 | 44 | AACCTCAGAGgtaggaggaacc | - | tttatgtttcagAGGTTGTGTT | ||
| 4 | Hs XRRA1 | 127 | CTACAACAAGgtgactttctgc | 3 | 3696 | cttctttctaagACAACCAAGA |
| Mm Xrra1 | 126 | CTACAACAAGgtaactgctttg | 3706 | tttcctctgaagACAGCCAAGG | ||
| Rn Xrra1 | 126 | CTACAACAAGgtaaactgcttt | - | ttccctcctaagACAACCAAGG | ||
| 5 | Hs XRRA1 | 96 | CATACACGAGgtatagtgcccg | 4 | 7073 | cattcattagagATCGCAAAAG |
| Mm Xrra1 | 94 | CACACACGAGgtatggtggcca | 4393 | cattcattagagATCGCAAAAG | ||
| Rn Xrra1 | 94 | CACACACGAGgtatggtggcca | - | tgcactttccagATCGCAAAGG | ||
| 6 | Hs XRRA1 | 123 | ATCATGGAAGgtagggcttcca | 5 | 779 | tcatggcaccagGGGTCCCTCC |
| Mm Xrra1 | 116 | GTCACGGAAGgtaagccttcca | 736 | ccttcataccagGGATCCACAC | ||
| Rn Xrra1 | 116 | ATCGCGGAAGgtaagccttcca | - | ccctcacaccagGGATCCCACC | ||
| 7 | Hs XRRA1 | 351 | CCTGACCCAGgtacctgtatcc | 6 | 2622 | tctccatgacagGTGAAAAGCG |
| Mm Xrra1 | 324 | CCTGACCCAGgtgacagccccc | 2256 | tctctcatccagGTGAAGACCT | ||
| Rn Xrra1 | 324 | CCTGACCCAGgtgacagccccc | - | tctctcatacagGTGAAAACCT | ||
| 8 | Hs XRRA1 | 202 | GAGCCAAAGGgtatgtgagggc | 7 | 2839 | tcctgggcacagGTGAGTGAAC |
| Mm Xrra1 | 196 | GAGCCAAAGGgtatgggattaa | 1421 | ttcctggcacagGTGAATGAGC | ||
| Rn Xrra1 | 199 | GAGCCAAAGGgtacgggattaa | - | ttcctggcacagGTGAATGAGC | ||
| 9 | Hs XRRA1 | 117 | AGAGAAACGGgtaaacatccag | 8 | 788 | ttcttgccctagGGAATCCAGA |
| Mm Xrra1 | 116 | GGAGAAGAGGgtaagcacttca | 594 | tgcatcctctagGCATCCAGAA | ||
| Rn Xrra1 | 116 | AGAGAAGAGGgtaagcacttca | - | tgcatcctccagGCATCCAGAA | ||
| 10 | Hs XRRA1 | 108 | GCTCCACTAGgtacggctctgc | 9 | 207 | accttctgccagGCCCAGAGAA |
| Mm Xrra1 | 106 | GCTCCACTGGgtatggtgccag | 169 | tgcatcctctagGCCGGGAGGA | ||
| Rn Xrra1 | 106 | GCTCCACTGGgtatggtgccag | - | cgtttgtgccagGTTGCGAGGA | ||
| 11 | Hs XRRA1 | 484 | 10 | 397 | cttttcctgcagGTGCTGTCCT | |
| Mm Xrra1 | 445 | 294 | tcattcctgtagGCACTGTGTT | |||
| Rn Xrra1 | 445 | - | tcatccctgtagGCGCTGTGTG |
aThe intron sizes of Rn Xrra1 gene could not be determined since the sequencing of rat genomic clone CH230-188L13 (GenBank AC127923) containing the gene was in progress. Nevertheless, their splicing sites can be identified, thus the exon sizes are also known.
To test whether these exon-intron borders were actually unique to XRRA1, we searched for possible XRRA1 homologues from other organisms. Thus far, we could only retrieve Xrra1 containing genomic clones from mouse (Mus musculus, Mm) (GenBank accession no. NW_000328) and rat (Rattus norvegicus, Rn) (GenBank accession no. AC127923). Mm Xrra1 gene is located on chromosome 7. GrailEXP analyses showed that Mm Xrra1 gene also contained 11 exons. Alignment of mouse and rat genomic sequences resulted in information for rat exons, which also consist of 11 exons. Interestingly, all the exon-intron borders of Hs XRRA1, Mm and Rn Xrra1 genes are well conserved (Table 3). We again employed EBOA to look for all possible ESTs of mouse and other organisms (Table 2). We used the longest human EST containing XRRA1 cDNA sequence as template for the analysis. From the mouse dbEST, we obtained 13 ESTs as primary sequences, which assembled Mm Xrra1 (GenBank accession no. BK000542). However, unlike the GrailEXP analysis, Mm Xrra1 lacks exon one and part of exon two. Nevertheless, these two approaches verified the cDNA sequence for Mm Xrra1. We could not find any publicly available ESTs for rat. Other ESTs found to be homologues to XRRA1 were from pig (Sus scrofa, Sc), horse (Equus caballus, Eq) and bovine (Bos taurus, Bt). We assembled Bovine (Bt) Xrra1 (GenBank accession no. BK000644) from 5 ESTs. The Bt Xrra1 gene is likely to have 11 exons with truncated exons 4 and 8 (Table 2, Fig. 3A).
Table 2.
ESTs of Mm, Rn, Bt, Sc, and Eq Xrra1 with corresponding nucleotides and exons to Hs XRRA1
| Organism | EST | Corresponding Nucleotides of Hs XRRA1 | Corresponding Exons of Hs XRRA1 | Genomic Clone | Exons |
| Mus musculus Xrra1 (1903 nt) | BI990168.1 | 312 – 776 | 2 – 7 | NW_000328 | 1 – 11 |
| AV046823.1 | 660 – 1011 | 6 – 7 | |||
| BE650319.1 | 1017 – 1093, 1215–1414 | 7 – 8, 8 – 10 | |||
| BU610007.1 | 1215 – 1383, 1400 – 1491 | 8 – 9, 10 | |||
| AA144723.1 | 1215 – 1414 | 8 – 10 | |||
| AI647383.1 | 1215 – 1383, 1400 – 1491 | 8 – 9, 10 | |||
| BE956870.1 (+/-) | 1437 – 1491, 1529 – 1599, 1668 – 1737, 1933–1966 | 10, 11 | |||
| BQ840215.1 | 1436 – 1599 | 10 – 11 | |||
| BE631695.1 | 1529 – 1599 | 11 | |||
| BQ175761.1 (+/-) | 1529 – 1599, 1668 – 1737, 1933 – 1966 | 11 | |||
| BE956083.1 (+/-) | 1529 – 1599, 1668 – 1737, 1933 – 1966 | 11 | |||
| AI449753.1 (+/-) | 1668 – 1737, 1933 – 1966 | 11 | |||
| AW125246.1 (+/-) | 1933 – 1966 | 11 | |||
| Rattus norvegicus Xrra1 (1909 nt) |
AC127923.2 | 1 – 11 | |||
|
Bos taurus Xrra1 (1759 nt) |
BM106446.1 | 1 – 422 | 1 – 4 | ||
| BI540915.1 | 542 – 703 | 5 – 6 | |||
| BE722755.1 | 797 – 1248 | 7 – 8 | |||
| BM254248.1 | 1308 – 1760 | 9 – 11 | |||
| BE751905.1 | 1366 – 1761 | 9 – 11 | |||
| Sus scrofa Xrra1 (970 nt) | BF442510.1 | 407 – 708 | 4 – 6 | ||
| BF442490.1 | 407 – 703 | 4 – 6 | |||
| BF444411.1 | 407 – 514, 1208 – 1269 | 4, 8 – 9 | |||
| BI341438.1 | 581 – 810, 877 – 978, 1006 – 1079 | 5 – 7, 7, 7 – 8 | |||
| Equus caballus Xrra1 (621 nt) | BM735121.1 | 1336 – 1575, 1658 – 1761, 1902 – 1970 | 9 – 11, 11, 11 |
We used cDNA libraries from macaque (Macaca fascicularis, Mf) brain and testis to obtain Mf XRRA1. Since we have created and sequenced these libraries, we were able to look for the Mf XRRA1. We found only three testis clones (QtsA-12093, 10089 and 20433) from 10,400 testis and 53,000 brain cDNAs that were homologous to Hs XRRA1. One of the clones, QtsA-20433 (GenBank accession no. AB072776) had the full-coding sequence and showed strong identity to Hs XRRA1, sharing 96 and 95% identities in cDNA and protein sequences, respectively. SIM4 [13] and BLAST analysis showed that Mf XRRA1 sequence dispersed properly into 11 exons on human chromosomal band 11q13 (Fig. 3A).
XRRA1 is conserved among human, macaque, mouse, rat, pig and bovine
Multiple alignments of both XRRA1 cDNA and protein sequence from human, macaque, mouse, rat, pig, and bovine Xrra1s showed a conservative molecule. Percent identities at both the nucleotide and protein level among these mammals resulting from two by two alignments are listed in Table 4. As expected, phylogenetically, human XRRA1 is closer to macaque XRRA1 and both of them are closer to bovine and pig Xrra1s than murine and rat Xrra1s. We failed to find possible XRRA1 homologues in other organisms ranging from microorganisms, plants, and animals. The latter included organisms whose genomes have been sequenced completely such as yeast, plasmodium, fruit fly, zebra fish and fugu.
Table 4.
Percent identities of XRRA1 cDNA and protein among several mammals
| Percent Identity (cDNA)* | |||||||
| Percent Identity (Protein)* | ORGANISM | Hs XRRA1 | Mf XRRA1 | Mm Xrra1 | Rn Xrra1 | Sc Xrra1 | Bt Xrra1 |
| HsXRRA1 | - | 96.0 | 75.9 | 75.9 | 83.4 | 76.8 | |
| Mf XRRA1 | 95.0 | - | 76.2 | 76.3 | 83.6 | 77.2 | |
| MmXrra1 | 71.8 | 74.1 | - | 93.6 | 74.6 | 70.9 | |
| Rn Xrra1 | 69.6 | 71.7 | 90.2 | - | 74.7 | 70.1 | |
| Sc Xrra1 | 77.7 | 79.4 | 67.7 | 62.6 | - | 86.4 | |
| Bt Xrra1 | 68.3 | 69.5 | 62.0 | 61.6 | 70.1 | - | |
*Hs, Homo sapiens; Mf, Macaque fascicularis; Mm, Mus musculus; Rn, Rattus norvegicus; Sc, Sus scrofa; Bt, Bos taurus.
The Hs XRRA1 gene produces a novel 559 aa protein (Fig. 3B). The XRRA1 protein contains two possible sites of leucine-rich repeats (aa 20–41 and aa 138–164), a PEST sequence at aa 247–264, two tyrosine phosphorylation sites at aa 81 and aa 142, and bipartite nuclear targeting sequences are found at aa 357–373 and aa 485–501. Besides human, Mf XRRA1, Mm, Rn, and Bt Xrra1s retain the above-mentioned motifs.
XRRA1 responded differentially to XR in clones of varying radiation responses
The RT-PCR results confirmed earlier observation from our cDNA microarray analysis that unirradiated HCT116Clone2_XRR cells had lower basal expression of XRRA1 than unirradiated HCT116Clone10 cells (Fig. 4, Table 5). However, XRRA1 expression in HCT116Clone2_XRR cells increased almost immediately (~10 minutes) after XR and decreased thereafter. By contrast, HCT116Clone10 cells down regulated XRRA1 expression approximately ten minutes following XR. Interestingly, XRRA1 expression in HCT116CloneK_XRS cells was initially similar to HCT116Clone10 cells but regained the basal levels by 24 hours following XR.
Figure 4.
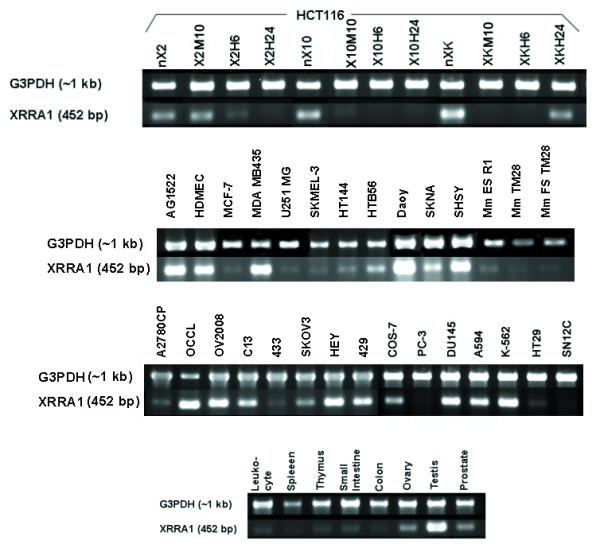
RT-PCR of XRRA1 and a house-keeping gene G3PDH from HCT116 clones treated with X-radiation, various cancer and normal cells, and normal tissues/organs. The same set of primers was used to amplify corresponding mRNA from mouse embryonic stem cells and fibrosarcoma cells/tumor.
Table 5.
Differential expression of human XRRA1 in XR-treated HCT116 clones, in normal tissues/organs and cells, as well as in various cancer cell types.
| Cells/Tissues/Organs | Relative Expression Level (Normalized against G3PDH)* | Cell/Tissue/Organ type; Cancer Type |
| HCT116Clone2_XRR | +++ | Colorectal, more resistant to X-radiation than HCT116 |
| HCT116Clone10 | ++++ | Colorectal, similar radiation response to HCT116 |
| HCT116CloneK_XRS | ++++++ | Colorectal, more sensitive to X-radiation than HCT116 |
| HCT116Clone2_XRR | +++ | Without X-radiation |
| HCT116Clone2_XRR M10 | ++++ | 10 Minutes after X-radiation |
| HCT116Clone2_XRR H6 | ++ | 6 Hours after X-radiation |
| HCT116Clone2_XRR H24 | + | 24 Hours after X-radiation |
| HCT116Clone10 | ++++ | Without X-radiation |
| HCT116Clone10 M10 | + | 10 Minutes after X-radiation |
| HCT116Clone10 H6 | + | 6 Hours after X-radiation |
| HCT116Clone10 H24 | + | 24 Hours after X-radiation |
| HCT116CloneK_XRS | ++++++ | Without X-radiation |
| HCT116CloneK_XRS M10 | + | 10 Minutes after X-radiation |
| HCT116CloneK_XRS H6 | + | 6 Hours after X-radiation |
| HCT116CloneK_XRS H24 | ++++ | 24 Hours after X-radiation |
| Testis | ++++++++ | Normal |
| Prostate | +++++ | Normal |
| Ovary | +++++ | Normal |
| Leukocyte | ++ | Normal |
| Spleen | + | Normal |
| Thymus | ++ | Normal |
| Small Intestine | ++ | Normal |
| Colon | + | Normal |
| AG1522 | ++++ | Normal fibroblast (immortalized) |
| COS-7 | +++ | Normal fibroblast (immortalized) |
| HDMEC | ++++ | Normal microvascular endothelial (primary) |
| MCF7 | + | Breast cancer |
| MDA MB435 | +++++ | Breast cancer |
| U251MG | + | Glioma |
| SKMEL-3 | + | Melanoma |
| HT144 | + | Melanoma |
| HTB56 | +++ | Lung cancer |
| A594 | ++++ | Lung cancer |
| Daoy | ++++ | Neuroblastoma |
| SKNA | +++ | Neuroblastoma |
| SHSY | ++++ | Neuroblastoma |
| A2780CP | + | Ovarian cancer, resistant to cisplatin |
| OCCL | ++++ | Ovarian cancer |
| OV2008 | ++++ | Ovarian cancer |
| C13 | +++ | Ovarian cancer |
| 433 | + | Ovarian cancer |
| SKOV3 | ++ | Ovarian cancer |
| HEY | ++++ | Ovarian cancer |
| 429 | +++ | Ovarian cancer |
| PC-3 | + | Prostate cancer |
| DU145 | +++++ | Prostate cancer |
| SN12C | + | Renal cancer |
| HT29 | + | Colorectal cancer |
| K-562 | +++++ | Leukemia |
| Mm TM28 | +++ | (Murine) Fibrosarcoma |
| Mm FS-TM28 | + | (Murine) Fibrosarcoma biopsy |
| Mm ES-R1 | ++ | (Murine) Embryonic Stem Cell |
*+, very low; ++, low; +++ moderate; >++++, high.
XRRA1 was expressed predominantly in the testis in human and macaque
We carried out DNA microarray where PCR-amplified QtsA-20433 (Mf XRRA1) was included. Because we found that Mf XRRA1 was more commonly present in testis than in brain cDNA libraries, we further investigated whether Mf XRRA1 was predominantly expressed in the testis. The ratio of Mf XRRA1 expression in testis to that in mixed tissues was 3.02. We also compared the expression of XRRA1 between human and macaque testis. The ratio of XRRA1 expression in human testis to macaque testis was 1.04. In addition, we examined Hs XRRA1 expression from various normal human tissues/organs. We found that Hs XRRA1 was expressed predominantly in testis followed by prostate and ovary (Fig. 4, Table 5). Other tissues/organs such as peripheral blood leukocyte, spleen, thymus, small intestine and colon demonstrated low expression of Hs XRRA1.
XRRA1 was expressed ubiquitously in various types of cancer cells
We evaluated the expression of Hs XRRA1 in various cancer cell lines (neuroblastoma, glioma, breast, lung, leukemia, renal, ovarian, prostate, another colorectal). XRRA1 was present in all of those cells, although the expression level was variable (Fig. 4, Table 5). We also detected XRRA1 expression in immortalized normal fibroblast AG1522 and COS-7 cells, as well as in a dermal microvascular endothelial cell line (HDMEC). Surprisingly, besides mouse fibrosarcoma cells and tumor biopsy from a mouse model of fibrosarcoma, we were also able to detect Mm Xrra1 expression in pluripotent cells such as murine embryonic stem cells R1 (Fig. 4, Table 5).
Over-expression of GFP-XRRA1 fusion protein was only achieved transiently and showed nucleo-cytoplasmic protein distribution
To understand further the possible function of the XRRA1 gene, we fused the XRRA1 cDNA immediately downstream of a green fluorescent protein (GFP) cDNA. We over-expressed the GFP-XRRA1 in HCT116 clones, but after 48 hours post-transfection the fluorescence clearly decreased both in the number of cells and intensity of fluorescence. We obtained similar results when we over-expressed GFP-XRRA1 in COS-7 cells. After three weeks selection in 500 μg/ml G418, very few COS-7 cells remained fluorescent (Fig. 5). Only transient expression of the GFP-XRRA1 was observed in both types of cells. The fluorescence of the GFP-XRRA1 did not show an exclusive nuclear localization (although the predicted XRRA1 protein contains NLS motif). Instead, the fluorescence appeared in most cells to be nucleo-cytoplasmic (Fig. 5).
Figure 5.
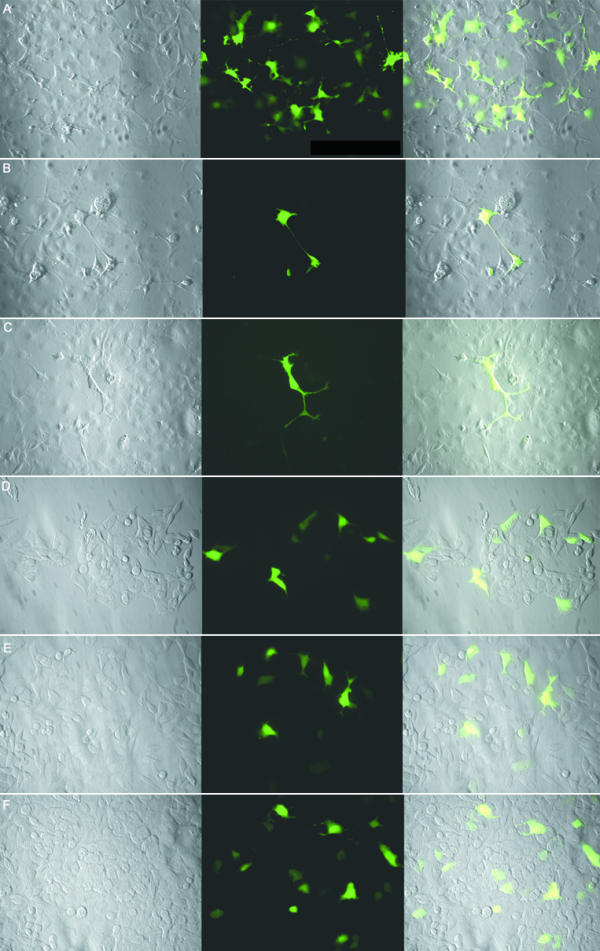
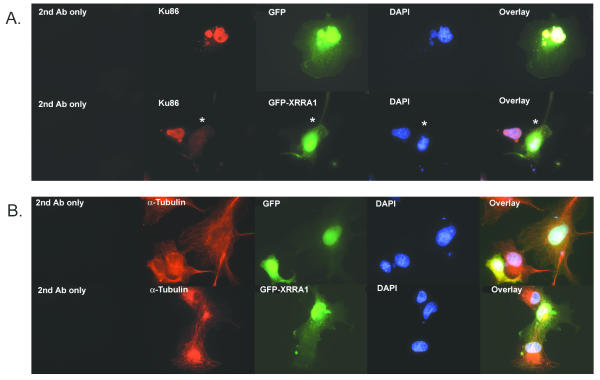
GFP-XRRA1 fusion protein in COS-7 and HCT116 clones. A, B, and C are transfected COS-7 cells. D, E, and F are transfected HCT116Clone2_XRR cells. Control with GFP cassette alone is on panel A and D. Expression of GFP-truncated XRRA1 is shown in panels B and E. Expression of GFP-full length XRRA1 is shown in panels C and F. Transfected COS-7 cells were examined by fluorescence microscopy after 3 weeks of G418-selection. Expression of GFP-XRRA1 fusion protein was detected only transiently in HCT116 clones. The leftmost panel is a phase-contrast picture of the cells. The middle panel is green fluorescence shown with FITC filter. The rightmost panel is an overlay of the two previous panels. Magnification used was 400×.
Over-expressed COS-7 cells with GFP-XRRA1 might modulate Ku86 expression
Ku86 was exclusively localized in nuclei with or without XR (Fig 6). The over-expression of GFP-XRRA1 abolished nuclear immunostaining of Ku86 in COS-7 (Fig 6A). Immunostaining of α-Tubulin, as additional control for the experiment, did not change in the presence of GFP-XRRA1 (Fig 6B). The secondary antibody conjugated with Cyanine-3 (Cy3) was used for both Ku86 and α-Tubulin immunostaining.
Figure 6.
Immunocytochemistry of COS-7 cells over-expressing GFP-XRRA1 with Ku86 (A) and α-Tubulin (B). (A) Ku86 (red fluorescence) was localized in nuclei. Nuclear Ku86 immunostaining was eliminated (asterisk) in the presence of GFP-XRRA1 (green fluorescence), but not with the GFP alone. (B) Neither GFP nor GFP-XRRA1 changed the immunostaining of α-Tubulin (red fluorescence). DAPI-stained nuclei are shown in blue fluorescence. Magnification used was 1000×.
Discussion
EBOA and in silico analysis helped us to identify a novel gene XRRA1 from a previously unknown EST R40588 not only in human, but also in mouse, bovine, and rat. Recently, several clones that contained Hs XRRA1 spliced variants have been deposited to GenBank database by others. Hs XRRA1 clone from testis (GenBank accession no. BC037294) lacked exon 4; the one from spleen (GenBank accession no. AK074152) lacked exons 3, 4 and a possible partial deletion of exon 5, and the one from NT2 cells (GenBank accession no. AK056364) lacked exons 2, 3, 4, and 10 (Fig. 3A). The testis clone with GenBank no. BC037294 did not appear to have a proper open reading frame. EBOA and GrailExp methods failed to produce a start site further upstream of Hs XRRA1 (GenBank no. BK000541).
Surprisingly, when we searched from approximately 63,400 macaque brain and testis cDNAs, we found only three testis clones that were homologous to Hs XRRA1, suggesting that the Mf XRRA1 gene is expressed only at a very low level in brain. The Mf XRRA1 is highly homologous with Hs XRRA1. Because the Hs XRRA1 gene consists of 11 exons, it is likely that the Mf XRRA1 gene also contains 11 exons (see Fig. 3A). In general, the XRRA1 gene seems to be highly conserved among mammals (i.e. human, macaque, mouse, rat, pig, and bovine), suggesting that it has similar function(s). Moreover, we were unable to find any convincing evidence of XRRA1 gene from fully sequenced genomic DNA of fish, fly, worm, yeast and microbes. This suggests that XRRA1 might be specific for, or evolutionary-enriched in, mammals. A specific role(s) for XRRA1 in mammals was further supported by the observation that the predicted protein sequences for motifs such as LRR, PEST, and NLS, as well as Tyr phosphorylation sites, are preserved among the above-mentioned mammals. Additionally, these motifs suggested that the XRRA1 protein may interact dynamically with other proteins, or might be subjected to rapid degradation via the proteasomal pathway, or might suggest a possible role for XRRA1 in cellular signaling.
Although the protein contains a NLS motif, it appears that XRRA1 is not exclusively located in the nucleus, as shown by the over-expression of the GFP-XRRA1 fusion protein. HCT116 clones and COS-7 cells containing the fusion protein (either with full length or truncated XRRA1 protein) had lost the auto-fluorescence. This suggested that the protein concentration above the basal expression levels of XRRA1 might be lethal to the cells.
It is noteworthy that the expression of XRRA1 in HCT116 clones was variably affected by 4 Gy XR. The results suggested that the XRRA1 gene might be involved in the immediate response following XR and might further be linked to XR resistance and/or sensitivity. Thus the return of XRRA1 to almost pre-irradiated basal levels by 24 hours post-irradiation in radiosensitive HCT116CloneK_XRS cells (but not in the other two clones) might be necessary for the subsequent manifestation of enhanced sensitivity to XR by this clone. One possibility is that the XRRA1 gene may play a role either in the expression of cell death and/or signaling of DNA damage following XR stress. Therefore, genetic modulation of the expression of XRRA1 in these clones may, specifically lead to either enhanced or decreased XR-induced lethality, and, generally, may be relevant for determining cellular response to XR in both tumor and normal cells.
In mammalian cells, DNA double strand breaks caused by XR are preferentially repaired by non-homologous end joining (NHEJ) mechanism. The major molecules involved in the NHEJ pathway are heterodimeric DNA end-binding complexes Ku86/Ku70 [14]. Therefore, we used Ku86 as the first candidate to test whether XRRA1 is involved in the modulation of the NHEJ pathway. The immunocytochemistry study on both HCT116Clone2_XRR and COS-7 cells showed that there was no significant modulation of Ku86 with XR. However, COS-7 cells over-expressing GFP-XRRA1 were devoid of nuclear Ku86. This phenomenon suggested that over-expression of XRRA1 might have resulted in the down-regulation of Ku-86. Further studies are required to determine whether there is a negative correlation between XRRA1 and Ku86 expressions during cellular response to XR.
The varying expression of XRRA1 following XR described above could be related to the role of transcription factors. In fact, we were able to detect a possible region between nt -506 and -400 from the start codon for transcription factor binding sites that regulate the expression of XRRA1. These factors were C/EBPα, HSF-1, c-Jun, c-Fos, AP-1, and CREB. Interestingly, most of the previously mentioned transcription factors are members of the bZIP superfamily. The latter is associated with various functions, including mediating G1 arrests [15,16], regulating responses against environmental stresses including IR and UV [17], acting as an immediate-early response factor [18], controlling wide range plasticity processes [19], and serving as a promoter element that mediates transcriptional activation in response to increased levels of intracellular essential secondary messenger cAMP [20]. Thus, putative binding sites for these transcription factors further strengthen the suggestion that the expression of XRRA1 was rapidly affected after XR treatment.
XRRA1 was expressed predominantly in the testis in both human and macaque. It may be relevant to note that the testis is generally regarded as one of the most sensitive organs to XR [21]. Thus upregulation of XRRA1 in an XR-sensitive organ would appear to be generally consistent with our observation that it is downregulated in the XR-resistant HCT116Clone2_XRR cells. Because there was comparable expression level of XRRA1 in both human and macaque, it suggests that there is a functional necessity for XRRA1 in mammalian testis (i.e. XRRA1 might serve as a testis-specific molecule). By contrast, the low expression of XRRA1 in healthy colon and relatively much stronger expression of XRRA1 in HCT116 cells, may suggest an upregulation of this gene in cancer development. The ubiquitous expression of XRRA1 in numerous cancer cells and in immortalized normal cells might be indicative of a role of this gene in tumor development.
Interestingly, from the SAGEmap, we identified three tags that specifically represent XRRA1 anchored with restriction enzyme NlaIII. The tags were TATTCAGGGG, ACCTGGTGCC, and GAATCAAGTG. SAGE libraries where XRRA1 was less than 20 tags per million were excluded. We found that the XRRA1 was present in mammary gland epithelium ductal in situ carcinoma, normal cerebellum, normal gastric body epithelium, well differentiated oligodendroglioma, pancreatic epithelium ductal adenocarcinoma, metastasis mammary gland carcinoma, ovarian clear cell poorly differentiated carcinoma, brain glioblastoma multiform cell line, ovarian serous adenocarcinoma, mammary gland ductal in situ high grade carcinoma, prostate carcinoma, brain juvenile ependymoma, and medulloblastoma cerebellum. The foregoing data further suggested that XRRA1 might be correlated with carcinomas in breast, brain, prostate, pancreas and ovary. Finally, the expression of Mm Xrra1 expression in mouse embryonic stem (ES) cells (R1) as well as in mouse fibrosarcoma cells and in fibrosarcoma biopsy suggest that XRRA1 might function as an early expressed gene.
Conclusions
XRRA1 is a novel molecule that is expressed selectively in normal healthy tissues/organs. It is expressed at relatively higher levels in primary sex organs such as testis (of both human and macaque), prostate and ovary. The expression of XRRA1 in normal proliferative cells, embryonic stem cells, and various tumor cells is also generally high and comparable to testicular expression of XRRA1. These findings suggest that XRRA1 expression may be important for cell proliferation, development, and differentiation as well as carcinogenesis. In regard to XR, we found that XRRA1 expression was rapidly modulated after treatment, suggesting the potential involvement of this gene/protein in the manifestation/expression of radioresistance/sensitivity by tumor cells such as human colorectal cancer cells. Over-expression of GFP-XRRA1 fusion protein was located in both nucleus and cytoplasm of HCT116 clones or COS-7 cells. A possible correlation with NHEJ pathway for repairing DNA double strand break after XR is shown by the lack of Ku86 immunostaining in COS-7 cells that over-expressed the GFP-XRRA1. Further experiments to determine the function of XRRA1 in DNA damage sensor, repair and apoptosis following radio- and or chemotherapy are ongoing. Also the function of XRRA1 in carcinogenesis as well as embryogenesis and reproduction biology needs to be elucidated.
Methods
Normal/Cancer cell cultures
Clones of colorectal cancer cells (HCT116Clone2_XRR, HCT116Clone10, and HCT116CloneK_XRS), normal fibroblast cells (COS-7 and AG1522), breast cancer cells (MCF-7 and MDA MB435), glioma cells (U251 MG), melanoma cells (SKMEL-3 and HT144), lung cancer cells (HTB56 and A549), ovarian cancer cells (A2780CP and A2780S), prostate cancer cells (PC-3 and DU145), leukemia cells (K-562), colorectal cancer cells (HT29), renal cancer cells (SN12C), and mouse fibrosarcoma cells (TM28) were grown in DMEM/Ham' s F12 1:1 mix (Wisent Inc., St. Bruno, QC, Canada) supplemented with 10% fetal bovine serum (FBS) (Wisent Inc.) and 15 mM HEPES in a humidified atmosphere of 95% air, 5% CO2 at 37°C. Various individuals from the ORCC, Ottawa, Canada, kindly provided us with the cells. They were: J.C. Bell (COS-7, PC-3, DU145, K-562, HT29, and SN12C cells), C.L. Addison (MCF-7 and MDA MB435 cells, neuroblastoma cells (Daoy, SKNA, and SHSY), normal human dermal microvascular cells (HDMEC)), HC Birnboim (TM28 cells), B. Vanderhyden (ovarian cancer cells OCCL, OV2008, C13, SKOV3, 433, HEY, and 429), and M. McBurney (mouse embryonic stem cells R1). H.C. Birnboim also donated a C57BL/6 mouse with a solid fibrosarcoma (TM28) growing in the flank. A. Gatignon (Lady Davis Insitute, Montreal, Canada) kindly provided the U251 MG cells. Clones of HCT116, AG1522, SKMEL-3, HT144, A2780CP, and A2780S cells were from our own laboratory collection (C.E. Ng, ORCC, Ottawa, Canada).
Cloning and sequencing of Hs XRRA1 splice variant
A specific primer pair was created to amplify the 5' region of the XRRA1 cDNA. They were 5'-GCGCTGGAGACACTGATGCTGG ATGACAAC-3' and 5'-GGCCAGGCTAAGGTATCTCAGCTCTGGG-3'. These primers generated a product of 452-bp that comprised the first 4 exons. Total RNA from HCT116Clone2_XRR and HCT116Clone10 cells were used as template for RT-PCR. PCR fragments were gel purified using Gel Extraction Kit (Qiagen Inc., Mississauga, ON, Canada) and cloned into pTZ57R plasmid using InsT/Aclone PCR Product Cloning Kit (MBI Fermentas, Burlington, ON, Canada). Sequencing of the clones was performed by DNA Sequencer model 4000L according to the manufacturer' s protocol (LI-COR Biosciences, Lincoln, NE, USA) using M13 primer.
Construction of macaque cDNA libraries and DNA sequencing of their cDNA inserts
Oligo-capped cDNA libraries were made from macaque brain and testis. cDNAs were isolated according to the method described previously [22,23]. The 5'-end sequences of the clones were sequenced using ABI 3700 sequencer (Applied Biosystems, Tokyo, Japan) and categorized using DYNACLUST (Dynacom Co., Chiba, Japan) based on a BLAST search against the GenBank database. The entire sequences of clones were determined by the primer walking method with an ABI PRISM BigDye Terminator Sequencing kit (Applied Biosystems) according to the manufacturer' s instructions.
Construction of plasmid expressing Mf XRRA1 fused to GFP
Macaque cDNA clone of QtsA-20344 was found from the cDNA libraries to contain Mf XRRA1 transcript. A 1709-bp XhoI fragment of Mf XRRA1 was removed from pME18S-FL3 and sub-cloned into XhoI sites of pEGFP-C1 (BD Biosciences, Mississauga, ON, Canada), immediately downstream of the GFP sequence (pFM1709). To create a truncated Mf GFP-XRRA1 (pFM584), 1.1-kb EcoRI fragment was removed from pFM1709 leaving a 584-bp of the 5' XRRA1 cDNA fused with GFP.
Transfection of GFP-XRRA1 into HCT116Clone2_XRR, HCT116Clone10, HCT116CloneK_XRS, and COS-7 cells
HCT116 clones and COS-7 cell were transfected with a mixture of pFM584, pFM1709 or pEGFP-C1 and lipid complexes FuGENE 6 (Roche Diagnostics, Laval, QC, Canada) transiently or stably. Briefly, HCT116 clones and COS-7 were grown on cover slips in a 6-well plate and reached 60–70% confluences before transfection. The ratio of FuGENE 6 and pure plasmid was 6:1 in serum free medium. G418 (Sigma-Aldrich, Oakville Ltd., ON, Canada) at a concentration of 400 μg/ml was used for selection. Cells were fixed with 3.7% formaldehyde in PBS for 10 min, and then washed three times with 1x PBS. Coverslips were mounted upside down onto slides with Dako Fluorescent Mounting Medium (DAKO, Carpinteria, CA, USA). Visualization was performed using a fluorescence microscope Axioskop 2 MOT (Carl Zeiss MicroImaging Inc., Thornwood, NY, USA). Images of cells were captured with a CCD camera fitted with a FITC filter.
Immunocytochemistry
Transfected COS-7 and HCT116Clone2_XRR cells with pFM1709 or pEGFP-C1 were grown on cover slips until they reached 70–80% confluency before being treated with 4 or 10 Gy XR. Cells were fixed in 2% formaldehyde in 1x PBS for 15 min 24 h after XR treatment. Cells were then permeabilized with 0.2% Triton X-100 for 10 min. Ku86 antibody was obtained from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA. The secondary antibody was conjugated with Cy3 (Amersham Biosciences Corp, Baie d' Urfe, QC, Canada). Controls for the immunostaining were the secondary antibody alone, and control for the experiment was α-Tubulin antibody that was obtained from Calbiochem, San Diego, CA, USA.
RT-PCR
Total RNAs of normal/cancer cells were extracted using RNAeasy kit (Qiagen Inc.) according to the manufacturer' s instructions. TRIZOL Reagent (Invitrogen Inc., Burlington, ON, Canada) was used to isolate RNA of mouse fibrosarcoma tumor biopsy. RNAs of human peripheral blood leukocyte, spleen, thymus, small intestine, colon, ovary, testis and prostate were purchased from BD Biosciences. First Strand cDNA Synthesis kit (MBI Fermentas) was used to create first strand cDNA from RNA samples. A specific primer pair that amplified ~1 kb house keeping gene glyceraldehyde-3-phosphate dehydrogenase (G3PDH) was used as the control. The primers were 5'-TGAAGGTCGGTGTGAACGGATTTGGC-3' and 5'-CATGTAGGCCA TGAGGTCCACCAC-3'. Conditions of PCR were as follows: 94°C for 30 s, 61°C for 15 s, and 72°C for 40–60 s for 35 cycles. The PCR products were then separated on 1.5% agarose gels, stained with ethidium bromide, and visualized with the gel documentation imager Epi Chemi II (UVP Inc., Upland, CA, USA). Densitometry of bands were analyzed using Labworks™ software (UVP). Relative expression of XRRA1 was calculated as percentages of the mean raw densities of grayscale level of XRRA1 to G3PDH.
X-radiation of HCT116Clone2_XRR, HCT116Clone10, and HCT116CloneK_XRS cells
Clones of HCT116 cells were cultured at 1 × 105 cells/ml. Cultures reached 70–80% confluence before being treated with XR. Each clone was treated with a single dose of 4 or 10 Gy using a 250 kVp X-ray unit (Pantak, CT, USA) at a dose rate of 150 cGy/min. Total RNAs were collected 5 minutes, 6 hours and 24 hours following the XR. Specific primers for Hs XRRA1 as mentioned above were used. G3PDH was used as control. Relative expression of XRRA1 to G3PDH was evaluated
cDNA microarray of Mf XRRA1 from testis versus mixed tissues or human testis
512 fully sequenced testis cDNA clones were amplified using 5'-CTTCTGCTCTAAAAGCTGCG-'3 as a forward primer and 5'-CGACCTGCAGCTCGAGCACA-'3 as a reverse primer. Successful amplification was confirmed by agarose gel electrophoresis. Approximately 300 μg /ml DNA in 2 × Solution-T reagent (Takara Bio Inc., Shiga, Japan) were printed on duplicate glass-slides with a GMS 417 arrayer (Genetic MicroSystems, Woburn, MA, USA). RNA was isolated with TRIZOL Reagent (Invitrogen K.K., Tokyo, Japan) and purified with Oligo-Tex (Takara Bio Inc.). Macaque testis RNA was labeled with Cy3-dUTP (Pharmacia K.K., Tokyo, Japan). A mixture of macaque RNA was also obtained from 10 tissues: brain, heart, skin, liver, spleen, renal, pancreas, stomach, small intestine, large intestine, and labeled with Cy5-dUTP (Pharmacia). We mixed equal amount of RNA of these ten tissues. In a separate experiment, RNA from human testis (BD Biosciences) was labeled with Cy5-dUTP. Both labeled RNAs were co-hybridized to DNA spots. After the hybridization and washing procedure, slides were scanned with ScanArray (PerkinElmer Life Sciences Co., Tokyo, Japan). Beta actin and/or total signal intensity was used as the control for hybridization. The methods and results of DNA microarray have been deposited in the Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo with the accession numbers GPL206, GSM2387, and GSM2388.
ESTs-based ORF assembling (EBOA) and other in silico analysis
In order to elucidate what gene R40588 might represent, we employed EBOA to obtain the full-length cDNA containing start-, stop-codon, and poly-A signal or a complete open reading frame (ORF). The sequence of R40588 was used as a template to do BLAST search for all possible overlapping ESTs from human dbEST http://www.ncbi.nlm.nih.gov/dbEST. These retrieved ESTs, either sense-antisense or 5'- and 3'-direction, were selected and were again used as template for searching other overlapping ESTs. To confirm the ORF candidates, (i) we carried out a BLAST search against full-length cDNAs that were available on public databases, and (ii) genomic sequences comprising the ORF candidates were analyzed with GrailEXP version 3.3 http://compbio.ornl.gov/grailexp[11]. Multiple alignments were done according to ClustalW method [24]. Transcription factors and their binding sites were predicted by employing the Alibaba2 version 2.1 program http://www.gene-regulation.com/pub/programs.html that used the TRANSFAC database http://transfac.gbf.de/TRANSFAC[25]. Possible tissues/organs expression of XRRA1 was explored from the SAGEmap http://www.ncbi.nlm.nih.gov/SAGE[26]. Protein motifs were searched using software available on these web sites: http://www.expasy.ch/prosite[27], http://www.at.embnet.org/embnet/tools/bio/PESTfind[28], and http://www.cbs.dtu.dk/services/NetPhos[29].
Data deposition
The sequences reported in this paper have been deposited in the DDBJ/EMBL/GenBank databases under the accession numbers BK000541, AY163836, AB072776, BK000542, and BK000644, respectively. HUGO Gene Nomenclature Committee and MGD Nomenclature Committee have approved the names and symbols of the XRRA1 and Xrra1 genes.
List of abbreviations
Bt, Bos taurus (bovine); EBOA, ESTs-based ORF assembling; EST, expressed sequence tag; Hs, Homo sapiens (human), Gy, Gray; Macaca fascicularis (macaque); Mm, Mus musculus (mouse); ORF, open reading frame; Rn, Rattus norvegicus (rat); SAGE, serial analysis gene expression; Sc, Sus scrofa (pig); XRRA1, X-ray radiation resistance associated 1, XR, X-radiation.
Authors' contributions
FMM carried out the in silico analysis including the multiple sequence alignments, cloning of the splice variant, XR assay, cells cultures, RT-PCR of XR treated, various human/mouse cancer and normal cells/tissues/organs, construction and expression of the fusion protein, immunocytochemistry study, and drafted the manuscript. NO carried out the construction and sequencing of macaque testis and brain cDNA libraries, and cloning and cDNA microarray of Mf XRRA1. KH was funded to carry out the construction of cDNA libraries and cDNA microarray of macaque brain and testis, conceived of the Mf XRRA1 study and the in silico analysis. QYL contributed technical expertise to the cDNA microarray analysis of HCT116 clones. CEN is the Principal Investigator and was funded to carry out the cDNA microarray of HCT116 clones, supervised this work, provided the technical radiobiological expertise and contributed to the writing of this manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgments
We thank Dr. A. Lagarde for assistance with the DNA sequencing. This work was supported in part by grants from the National Cancer Institute of Canada with funds from the Canadian Cancer Society (to CEN) and from the Ministry of Health, Labor, and Welfare of Japan (to KH).
Contributor Information
Felix M Mesak, Email: Felix.Mesak@orcc.on.ca.
Naoki Osada, Email: nosada@uchicago.edu.
Katsuyuki Hashimoto, Email: khashi@nih.go.jp.
Qing Y Liu, Email: Qing_Yan.Liu@nrc.ca.
Cheng E Ng, Email: Cheng.Ng@orcc.on.ca.
References
- Zhivotovsky B, Joseph B, Orrenius S. Tumor radiosensitivity and apoptosis. Exp Cell Res. 1999;248:10–17. doi: 10.1006/excr.1999.4452. [DOI] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- Pandita TK. ATM function and telomere stability. Oncogene. 2002;21:611–618. doi: 10.1038/sj.onc.1205060. [DOI] [PubMed] [Google Scholar]
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Jeggo PA. DNA breakage and repair. Adv Genet. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- Marcou Y, D'Andrea A, Jeggo PA, Plowman PN. Normal cellular radiosensitivity in an adult Fanconi anaemia patient with marked clinical radiosensitivity. Radiother Oncol. 2001;60:75–79. doi: 10.1016/S0167-8140(01)00370-X. [DOI] [PubMed] [Google Scholar]
- Mirzayans R, Aubin RA, Bosnich W, Blattner WA, Paterson MC. Abnormal pattern of post-gamma-ray DNA replication in radioresistant fibroblast strains from affected members of a cancer-prone family with Li-Fraumeni syndrome. Br J Cancer. 1995;71:1221–1230. doi: 10.1038/bjc.1995.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. Genes required for ionizing radiation resistance in yeast. Nat Genet. 2001;29:426–434. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Xu Y, Uberbacher EC. Automated gene identification in large-scale genomic sequences. J Comput Biol. 1997;4:325–338. doi: 10.1089/cmb.1997.4.325. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Nelsen C, Hendrickson EA. Ku86 is essential in human somatic cells. Proc Natl Acad Sci USA. 2002;99:832–837. doi: 10.1073/pnas.022649699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AD. Runx1, c-Myb, and C/EBPalpha couple differentiation to proliferation or growth arrest during hematopoiesis. J Cell Biochem. 2002;86:624–629. doi: 10.1002/jcb.10271. [DOI] [PubMed] [Google Scholar]
- Wang H, Goode T, Iakova P, Albrecht JH, Timchenko NA. C/EBPalpha triggers proteasome-dependent degradation of cdk4 during growth arrest. EMBO J. 2002;21:930–941. doi: 10.1093/emboj/21.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Chen C, Stevenson MA, Auron PE, Calderwood SK. Heat shock factor 1 represses transcription of the IL-1beta gene through physical interaction with the nuclear factor of interleukin 6. J Biol Chem. 2002;277:11802–11810. doi: 10.1074/jbc.M109296200. [DOI] [PubMed] [Google Scholar]
- Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Sandstrom DJ, Hoeffer CA, Ramaswami M. AP-1 functions upstream of CREB to control synaptic plasticity in Drosophila. Nature. 2002;416:870–874. doi: 10.1038/416870a. [DOI] [PubMed] [Google Scholar]
- Servillo G, Della Fazia MA, Sassone-Corsi P. Coupling cAMP signaling to transcription in the liver: pivotal role of CREB and CREM. Exp Cell Res. 2002;275:143–154. doi: 10.1006/excr.2002.5491. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, van de Kant HJ, Dol R, Wagemaker G, van Buul PP, van Duijn-Goedhart A, de Jong FH, Broerse JJ. Long-term effects of irradiation before adulthood on reproductive function in the male rhesus monkey. Biol Reprod. 2002;66:486–494. doi: 10.1095/biolreprod66.2.486. [DOI] [PubMed] [Google Scholar]
- Osada N, Hida M, Kusuda J, Tanuma R, Iseki K, Hirata M, Suto Y, Hirai M, Terao K, Suzuki Y, Sugano S, Hashimoto K, Kusuda J. Assignment of 118 novel cDNAs of cynomolgus monkey brain to human chromosomes. Gene. 2001;275:31–37. doi: 10.1016/S0378-1119(01)00665-5. [DOI] [PubMed] [Google Scholar]
- Osada N, Hida M, Kusuda J, Tanuma R, Hirata M, Hirai M, Terao K, Suzuki Y, Sugano S, Hashimoto K. Prediction of unidentified human genes on the basis of sequence similarity to novel cDNAs from cynomolgus monkey brain. Genome Biol. 2002;3:1–006. doi: 10.1186/gb-2001-3-1-research0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, Krull M, Matys V, Michael H, Ohnhauser R, Pruss M, Schacherer F, Thiele S, Urbach S. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash AE, Tolstoshev CM, Wagner L, Schuler GD, Strausberg RL, Riggins GJ, Altschul SF. SAGEmap: a public gene expression resource. Genome Res. 2000;10:1051–1060. doi: 10.1101/gr.10.7.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. doi: 10.1016/0968-0004(96)10031-1. [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]