Abstract
CFTR is a cyclic AMP (cAMP)-activated chloride (Cl−) channel and a regulator of outwardly rectifying Cl− channels (ORCCs) in airway epithelia. CFTR regulates ORCCs by facilitating the release of ATP out of cells. Once released from cells, ATP stimulates ORCCs by means of a purinergic receptor. To define the domains of CFTR important for Cl− channel function and/or ORCC regulator function, mutant CFTRs with N- and C-terminal truncations and selected individual amino acid substitutions were created and studied by transfection into a line of human airway epithelial cells from a cystic fibrosis patient (IB3–1) or by injection of in vitro transcribed complementary RNAs (cRNAs) into Xenopus oocytes. Two-electrode voltage clamp recordings, 36Cl− efflux assays, and whole cell patch-clamp recordings were used to assay for the Cl− channel function of CFTR and for its ability to regulate ORCCs. The data showed that the first transmembrane domain (TMD-1) of CFTR, especially predicted α-helices 5 and 6, forms an essential part of the Cl− channel pore, whereas the first nucleotide-binding and regulatory domains (NBD1/R domain) are essential for its ability to regulate ORCCs. Finally, the data show that the ability of CFTR to function as a Cl− channel and a conductance regulator are not mutually exclusive; one function could be eliminated while the other was preserved.
CFTR is a transmembrane protein involved in the regulation of several processes, including the activation of outwardly rectifying Cl− channels (1, 2) and the inhibition of Na+ channels by cAMP-dependent protein kinase A (PKA) (3–5). Mutations in CFTR cause cystic fibrosis (CF). Both channels lose this pattern of PKA sensitivity when CFTR is absent or its function is severely compromised in mutant forms. Other members of the ATP-binding cassette (ABC) transporter superfamily also regulate other processes. For example, the multidrug transporter, MDR, may regulate volume-activated chloride channels (6–9). The sulfonylurea receptor (SUR) binds sulfonylurea compounds such as glybenclamide and confers sulfonylurea inhibition upon a separate ATP-gated K+ channel protein in pancreatic β cells (10–15). More recent results suggest that CFTR can act as a SUR for ATP-gated K+ channels in kidney (16).
We have shown previously that CFTR regulates outwardly rectifying Cl− channels (ORCCs) by an autocrine mechanism involving ATP release that is CFTR dependent. The ATP released binds to purinergic receptors to stimulate ORCCs (17, 18). The mechanism of how ATP is released, either through CFTR itself or by a separate mechanism, remains highly controversial (19–21). Two possibilities are that CFTR either transports ATP directly or activates an alternate ATP-release pathway.
A key question in CF research is: How does CFTR allow protein kinase A to activate separate populations of ORCCs and inhibit a distinct family of Na+-conductive channels? In this study, we tested the hypothesis that the complex, multidomain structure of CFTR supports its multifunctional behavior and that separate domains within the CFTR protein perform Cl− channel function independent of its regulatory functions. We show that the ability of CFTR to regulate ORCCs is not dependent upon CFTR’s Cl− channel function and that conductance regulation is separate from CFTR’s ability to conduct Cl−.
MATERIALS AND METHODS
Site-Directed Mutagenesis and CFTR cDNA Truncation.
Single-stranded DNA for CFTR (CFTR cDNA clone pBQ4.7) (22) was propagated in a dut− ung− strain of CJ236 Escherichia coli cells with M13 helper phage and was extracted from the bacteria. A single batch of single-stranded DNA was used for mutagenesis of all CFTR constructs used in this study by standard methods. Briefly, a mutagenic oligonucleotide that introduces the restriction site necessary for truncating the cDNA or the desired point mutation and a silent restriction site for selection of mutated cDNA colonies was annealed to single-stranded DNA. Using the oligonucleotide primer to synthesize the complementary strand, we re-created double-stranded DNA and used it to transform competent JM109 cells. Candidate colonies were screened for the truncating or silent restriction site introduced by the mutagenesis primer for selection, and these candidate clones were confirmed for correct mutagenesis by dideoxynucleotide-termination DNA sequencing (Sequenase; United States Biochemical).
For N-terminal truncation mutations, the M265V missense mutation and a silent mutation to create a unique SpeI site were introduced into the CFTR cDNA with a mutagenic oligonucleotide, 5′-GAC TAG TGA TTA CCT CAG AAG TGA TTG-3′. A truncation at the 5′ end of the cDNA, Δ259, was performed by digesting with SpeI, which cut in the pBluescript multiple cloning site and at the new site introduced by the oligonucleotide to remove that segment of the cDNA, and religating the plasmid. This created the Δ259-M265V construct. For Δ259-M265, the identical procedure was performed with introduction of an SpeI site without a missense mutation at methionine-265 with a mutagenic oligonucleotide, 5′-GTG AAA GAC TAG TGA TTA CC-3′. Digestion with SpeI created the Δ259 truncation, creating the Δ259-M265 construct.
For the “dual arginine” construct, R334W/R347P, the two appropriate point mutations and a silent mutation creating a unique NcoI site were introduced into the cDNA with a single mutagenic oligonucleotide, 5′-GGA ATC ATC CTC TGG AAA ATA TTC ACC ACC ATC TCA TTC TGC ATT GTT CTG CCC ATG GCG GTC ACT CGG CAA TTT CCA TGG GC-3′. NcoI was used to screen for mutagenized cDNAs, and the positive cDNA preparations were sequenced to determine whether both point mutations were introduced. This dual arginine mutation was shuttled into the pRSV-CFTR plasmid as described below. For null the construct, R334W/R347P in the transmembrane domain (TMD)-1 background (R334W/R347P-TMD-1), the identical mutations were introduced along with a silent NcoI site (as above) as well as a stop codon and an EcoRV site slightly downstream with a longer mutagenic oligonucleotide, 5′-GGA ATC ATC CTC TGG AAA ATA TTC ACC ACC ATC TCA TTC TGC ATT GTT CTG CCC ATG GCG GTC ACT CGG CAA TTT CCA TGG GCT GTA CAA ACA TGG TAT GAC TCT CTT GGA GCA ATA AAC TAA ATA CAG GAT ATC TTA C-3′. Digestion with EcoRV, which cut the cDNA at the new site introduced and at a site 3′ to the end of the cDNA and 5′ to the simian virus 40 polyadenylation signal, deleted the majority of the cDNA, leaving only that cDNA that encodes TMD-1. The plasmid was religated to form the truncated cDNA.
For the C-terminally truncated constructs, TMD-1 CFTR or K370XEcoRV was created by introducing a stop codon followed by an EcoRV restriction site by using the mutagenic oligonucleotide 5′-GCA ATA AAC TAA ATA CAG GAT ATC TTA C-3′. Digestion with EcoRV truncated the cDNA severely as for the TMD-1 construct described above. The plasmid was religated to form the truncated cDNA. T-N-R CFTR or D835XEcoRV, the artificial half-molecule construct of CFTR, was created by introducing a stop codon followed by an EcoRV restriction site by using the mutagenic oligonucleotide 5′-GTG CCT TTT TTA AGA TAT CGA GAG CAT ACC A-3′. Elimination of the 3′ half of the cDNA was performed with an EcoRV digest followed by a religation of the plasmid.
For all truncation mutants, these digestions and religations were performed in the pBQ4.7 vector used to make in vitro transcribed cRNA for oocyte injection and in the pRSV vector for mammalian cell transient lipofection.
Shuttling of Mutated CFTR cDNA Fragments.
As for the dual arginine mutant and all other point mutations or introduced restriction sites, mutations were subcloned from pBQ4.7 into the mammalian expression vector pRSV-CFTR. Mutations within nucleotides 1–645 of the CFTR cDNA were “shuttled” to pRSV-CFTR by digesting each plasmid with XbaI and ligating the appropriate cDNA fragment into pRSV-CFTR. Mutations within nucleotides 488-1130 of the CFTR cDNA were shuttled with a double digestion with NruI and Kpn2I. Mutations within nucleotides 1130–2461 of the CFTR cDNA were shuttled with a double digestion of Kpn2I and HpaI. Mutations within nucleotides 2461–4173 of the CFTR cDNA were shuttled with a double digestion of HpaI and NcoI. All mutations and subcloning were verified by DNA sequencing after shuttling. Subcloning of full-length CFTR after mutagenesis of the pBQ4.7 vector was extremely difficult and inefficient.
Two-Electrode Voltage Clamp Recording.
These methods have been described in detail previously (23).
In Vitro Transcription of CFTR Construct cRNA.
These methods have been described in detail previously (23). EcoRV was used to linearize all cDNA construct templates for in vitro transcription with a T7 Megascript kit (Ambion, Austin, TX). RNA cap analog (United States Biochemical) was used to cap and protect the 5′ end of the cRNA.
Transient Lipofection of IB3–1 CF Airway Epithelial Cells with CFTR cDNA.
Lipofection of IB3–1 CF airway epithelial cells with a Rous sarcoma virus (RSV) promoter-driven mammalian expression vector containing wild-type, mutant, or truncated CFTR was performed according to the manufacturer’s protocol (GIBCO/BRL) with some modifications. Optimization of transfection efficiency was performed with a similar pRSV construct containing the lacZ reporter gene. Transfection of IB3–1 CF cells at 50–75% confluence yielded 20–30% positively transfected cells as determined by staining with X-Gal (Promega β-galactosidase enzyme assay kit) and was performed for 36Cl− efflux and [γ-32P]ATP release/trapping assays. Transfection of IB3–1 cells at approximately 30% confluence resulted in higher efficiency and was performed for cells to be used in patch-clamp recordings. Two to 3 μg of wild-type, mutant, or truncated cDNA in the pRSV mammalian expression vector was mixed with OptiMEM-I reduced-serum medium (GIBCO/BRL) in one tube and 12–15 μl of Lipofectin Reagent (GIBCO/BRL) was mixed with OptiMEM-I in a second tube. The mixtures were mixed gently and incubated for 30 min at room temperature. After this incubation, the contents of each tube were mixed together and incubated for an additional 30 min at room temperature. IB3–1 cells were incubated with cDNA and Lipofectin for 6–8 hr at 37°C. A deviation from the manufacturer’s protocol was that the transfecting solution was left in the well after this 37°C incubation and fresh medium containing serum was added to the wells and mixed with the transfecting solution. After the overnight incubation, the solution was aspirated and fresh medium was added. The cells were studied at 72 hr; this was the peak of expression observed with lacZ reporter gene expression.
Chloride-36 Efflux Assay.
Cells were seeded at 50–75% confluence and transfected as above. Cells were washed three times with Ca- and Mg-free PBS (GIBCO/BRL) to remove serum. Thirty microliters of 36Cl− solution (sodium salt from NEN/DuPont; 1 μCi/μl; 1 μCi = 37 kBq) was diluted in 9 ml of Ringer’s solution, and 1.5 ml of this loading solution was added to each well of a six-well plate. The plate was incubated for 2–3 hr in a 37°C warm room. The Ringer’s solution for these experiments was a standard HCO3−-free, Hepes- and phosphate-buffered 140 mM NaCl Ringer’s solution supplemented with 5 mM glucose and titrated to pH 7.45 with 1 M NaOH. All efflux runs were performed in a 37°C warm room. Each well served as its own control. At time 0, Ringer’s solution without cAMP agonists was added and removed immediately. A fresh aliquot of Ringer’s solution was added immediately after that and the efflux run was started. This process was repeated every 15 sec until the time point at 1 min, when Ringer’s solution with forskolin (2.5 μM), 8-bromo-cAMP (250 μM), and 8-(4-chlorophenylthio)-cAMP (CPT-cAMP; 250 μM) was added; these compounds were not added for the remaining 4 min of the efflux run. At the end of the run, 0.5 M NaOH was added in two aliquots to lyse the cells, and all of the cell lysate was recovered to determine how much 36Cl− had remained in the cells to standardize the data. Each sample was diluted in scintillation cocktail, and its radioactivity was measured in a scintillation counter and normalized on a Microsoft Excel spreadsheet as the amount of 36Cl− lost from the cells per min.
Whole Cell Patch-Clamp Recording.
Cells were seeded at 30% confluence and transfected as above with wild-type, mutant, or truncated CFTR cDNA. The night before recording and approximately 48 hr after transfection, transfected cells were trypsinized from six-well plates, seeded, and concentrated onto Vitrogen (human collagen, Celtrix Laboratories, Palo Alto, CA)-coated glass coverslips (Bellco Glass, Vineland, NJ) for patch-clamp recording. Symmetrical 145 mM Tris⋅HCl solutions were used to study Cl− currents exclusively. Whole cell recording has been described in detail previously (18).
Single Channel Patch-Clamp Recording.
These methods have been described in detail previously (18).
RESULTS
Expression of Wild-Type and Mutant CFTR cRNA in Xenopus Oocytes.
Xenopus oocytes lack ORCCs that interact with CFTR. This makes Xenopus oocytes an ideal model system to study both wild-type and mutant forms of CFTR, both of which generate cAMP-activated Cl− currents that are specific for CFTR (see refs. 22 and 23).
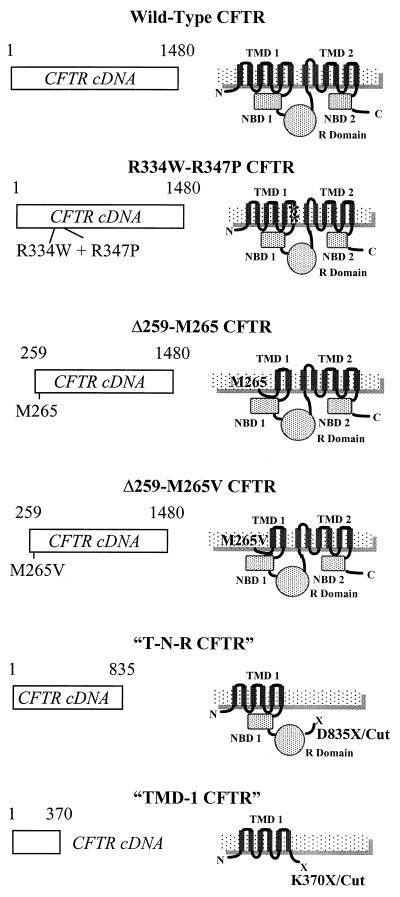
Fig. 1 illustrates the truncations and mutations created and characterized in this study. Three types of mutants were created: N-terminal truncations, C-terminal truncations, and conduction mutants. N-terminal truncated mutants were engineered to test the role of TMD-1. Δ259-M265 was truncated to eliminate the N terminus and the first four predicted transmembrane α-helices of CFTR (predicted amino acids 1–259) with methionine-265 intact. Δ259-M265V is identical to Δ259-M265 but with methionine-265 changed to a valine, shifting the translation initiation codon downstream within the coding sequence. Δ259-M265 generates currents that are typical of CFTR, suggesting that this mutant functions as a Cl− channel (Table 1). In sharp contrast, Δ259-M265V does not produce any currents (Table 1.). Similar results were obtained in our previous studies of the single channel properties of these mutants expressed in Xenopus oocytes (23). In that study, Δ259-M265 CFTR formed functional CFTR Cl− channels with the same halide permeability ratio of Cl− > I− as wild-type but with a smaller single channel conductance of 6.7 pS vs. 9.3 pS for wild type. No single channel events were observed from oocytes injected with the Δ259-M265V construct, suggesting that this mutant either does not conduct Cl− or is not processed normally in oocytes.
Figure 1.
Truncated and mutated cDNA constructs. Schematic illustrations of the mutations and truncations made to the CFTR cDNA and the putative protein product derived from that construct. NBD, nucleotide-binding domain; R domain, regulatory domain.
Table 1.
Cl− currents in CFTR cRNA-injected Xenopus oocytes
| cRNA injected | Current, nA
|
n | P value | |
|---|---|---|---|---|
| Basal | cAMP-stimulated | |||
| None | −89.3 ± 13.7 | −82.7 ± 13.5 | 9 | NS |
| Wild-type CFTR | −117.2 ± 27.7 | −828.1 ± 295.7 | 16 | <0.001 |
| Δ259-M265 | −133.4 ± 27.6 | −509.9 ± 159.9 | 8 | <0.01 |
| Δ259-M265V | −106.2 ± 32.1 | −103.7 ± 29. | 9 | NS |
| TMD-1 | −321.5 ± 75.6* | −587.7 ± 145.5 | 10 | <0.05 |
| T-N-R | −110.7 ± 27.6 | −316.3 ± 35.7 | 8 | <0.05 |
| R334W-R347P | 107.4 ± 16.1 | −104.7 ± 17.3 | 6 | NS |
| R334W-R347P-TMD-1 | −75.9 ± 20.1 | −117.3 ± 22.4 | 6 | NS |
Current values are shown for all mutants immediately before and 5 min after stimulation with cAMP agonists [forskolin, 10 μM; 3-isobutyl-1-methylxanthine (IBMX), 1 mM] at the −90-mV clamped voltage. Oocytes were used that had resting membrane potentials more negative than −30 mV. Of the panel of mutants, only TMD-1 (K370EcoRV) displayed basal or constitutively active currents before cAMP agonist addition (P < 0.05, ANOVA and Bonferroni ad hoc test). These currents were insensitive to 4,4′-diisothiocyanatostilbene-2,2′-disulfonic acid (DIDS) and differed in phenotype from endogenous Ca2+-activated Cl− currents (external Ca2+ concentration was 0.5 mM) (25). NS, not significant.
Significant constitutively active current for the construct compared to all other conditions and CFTR constructs listed.
C-terminal truncations were engineered to test the role of portions of CFTR beyond TMD-1. A “half molecule” of CFTR was created that contains TMD-1, NBD-1, and the R domain of CFTR (T-N-R CFTR or D835XEcoRV) but with the second half (C-terminal half; predicted amino acids 836-1480) of the molecule removed. This half molecule of CFTR is similar to the naturally occurring splice variant of CFTR found in kidney (23). When injected into Xenopus oocytes the T-N-R CFTR also generates Cl− currents with properties typical of CFTR (Table 1). Again this result is similar to what was shown previously for the T-N-R CFTR expressed in Xenopus oocytes (23). Our previous study showed that T-N-R CFTR (23) has a single channel conductance and halide selectivity identical to those of CFTR.
In the most severe C-terminal truncation, all domains of CFTR (predicted amino acids 372-1480) were removed except the first transmembrane domain of CFTR (TMD-1 CFTR or K370XEcoRV CFTR). This C-terminal truncation, TMD-1 CFTR, was created with the six membrane-spanning segments of TMD-1 and without the cytoplasmic regulatory domains of CFTR (NBDs, R domain). Expressed in oocytes (Table 1), TMD-1 CFTR had a relatively large basal (cAMP-independent) activity that was further enhanced by cAMP agonists. In single channel patch clamp studies of TMD-1 CFTR in Xenopus oocytes, this mutant has a single Cl− channel conductance of 8.3 ± 0.4 pS (n = 7), which is near to that of wild-type CFTR. TMD-1 CFTR Cl− channels were observed in seven of eight patches from injected oocytes (halide permeability was assessed by using whole cell currents; see the following section).
Finally, dual Cl− conduction mutations, R334W and R347P (referred to as dual arginine CFTR) were made. Each of these residues was shown to be important in Cl− conductance (24, 25). We tested the hypothesis that changing the two amino acids at these positions to arginine would “knock out” the Cl− conductance within CFTR. As expected the dual arginine mutants, R334W/R347P, either in full-length CFTR or within the TMD-1 construct, fail to produce Cl− currents when injected into Xenopus oocytes (Table 1). Again this is because the dual arginine mutation fails either to conduct Cl− or to be processed normally. Taken together, these results suggest that the transmembrane segments 5 and 6 are essential for the Cl− conduction and ion selectivity of CFTR.
Chloride Channel and ORCC Regulator Functions.
To assess the Cl− channel and ORCC regulator functions of CFTR, paired 36Cl− efflux assays without and with a cAMP agonist mixture (2.5 μM forskolin, 250 μM 8-bromo-cAMP, and 250 μMCPT-cAMP) were performed in IB3–1 cells. IB3–1 cells are a CF bronchial epithelial cell line used extensively to study CFTR function (26). The experiments were performed as an initial screen to assess whether an individual CFTR mutant could restore cAMP-stimulated 36Cl− efflux (Table 2). Mock-transfected IB3–1 cells failed to respond to cAMP agonists (Table 2), as is typical of CF cells. Wild-type, Δ259-M265-, TMD-1-, and T-N-R-transfected cells, however, all responded to cAMP agonists as predicted from oocyte recordings (Table 2).
Table 2.
cAMP-stimulated Cl− efflux in CFTR cDNA-transfected IB3-1 CF cells
| cDNA transfected | n | Cl− efflux, % lost per min
|
Paired P value | |
|---|---|---|---|---|
| Before agonists | After agonists | |||
| Mock | 42 | 33.01 ± 3.12 | 29.53 ± 2.22 | NS |
| Wild-type | 37 | 22.99 ± 1.47 | 46.51 ± 6.53* | <0.005 |
| Δ259-M265 | 30 | 21.85 ± 1.43 | 47.67 ± 5.95* | <0.005 |
| Δ259-M265V | 18 | 24.55 ± 1.17 | 29.25 ± 2.23** | <0.05 |
| TMD-1 (K370X) | 24 | 16.63 ± 1.80 | 53.51 ± 9.50* | <0.005 |
| TMD-1 (K370EcoRV) | 24 | 19.54 ± 1.67 | 41.27 ± 5.22* | <0.005 |
| T-N-R | 18 | 19.21 ± 1.89 | 28.05 ± 3.35** | <0.05 |
| R334W-R347P | 18 | 19.85 ± 3.20 | 31.16 ± 6.79** | <0.05 |
| R334W-R347P-TMD-1 | 18 | 23.12 ± 2.60 | 26.26 ± 3.42 | NS |
The Before agonists value is the rate of 36Cl− efflux immediately prior to stimulation with cAMP agonists (2.5 μM forskolin, 250 μM CPT-cAMP, and 250 μM 8-bromo-cAMP). The After agonists value shown is the peak stimulated rate after addition of cAMP agonists. For mutants Δ259-M265V, T-N-R, and R334W-R347P, the magnitude of cAMP stimulation is significantly less (P < 0.05, versus paired control value as denoted by two asterisks) than that for the wild type and other responding mutants [Δ259-M265, TMD-1 (K370X), TMD-1 (K370EcoRV), P < 0.005 as denoted by one asterisk], as determined by ANOVA followed by the Bonferroni ad hoc test. NS, not significant.
Importantly, Δ259-M265V-transfected IB3–1 cell cultures and R334W/R347P-transfected cultures also responded to cAMP in 36Cl− efflux assays, despite the lack of intrinsic Cl− channel function in oocyte recordings (Table 2). In contrast, the mutant R334W/R347P/TMD-1 failed to respond (Table 2). These results suggest that Cl− channel function of CFTR is not strictly required to restore cAMP-regulated Cl− transport in IB3–1 CF cells. It also demonstrates that these mutant forms of CFTR, which themselves cannot conduct Cl− in Xenopus oocytes, can still mediate Cl− efflux from the IB3–1 cells. As will be shown in more detail in the following section, this CFTR-induced stimulation of Cl− efflux occurs by stimulating a separate population of Cl− channels.
This point is shown more convincing in the whole cell current recordings, in which the activity of both CFTR and ORCC channel populations were measured simultaneously (27) in IB3–1 cells. This CF bronchial epithelial cell line possesses ORCCs, but they cannot be regulated by CFTR. The failure of ORCCs to be regulated by CFTR in this CF epithelial cell line, despite the presence of purinergic receptors, is caused by an inability of the cells to release ATP after cAMP stimulation. When recombinant wild-type CFTR cDNA is transfected into IB3–1 cells, Cl− currents generated by CFTR can be detected as expected. In addition, protein kinase A regulation of ORCCs is restored (see ref. 1). From our previous work, this restoration of ORCC regulation involves the cAMP-dependent release of ATP that occurs only after CFTR transfection into the IB3–1 cells (18).
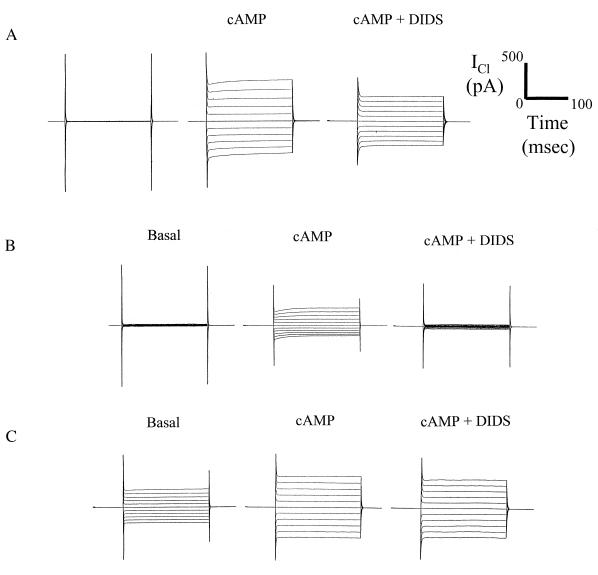
Expression of wild-type CFTR (Figs. 2 and 3) and Δ259-M265 CFTR resulted in a mixture of CFTR and ORCC whole cell Cl− currents in transfected IB3–1 cells prestimulated with cAMP. [No currents are expressed in either parental or mock-transfected cells (data not shown; also see refs. 18 and 27).] Extracellular addition of DIDS (500 μM) inhibited the outwardly rectifying currents while having no effect on the linear CFTR currents (Fig. 2). CFTR channels have been shown previously to be insensitive to DIDS (27), when DIDS is added to the extracellular compartment, whereas ORCCs are blocked by this agent (27). Thus, DIDS is ideal for dissecting out the contribution of CFTR and ORCCs to macroscopic Cl− currents. The presence of both linear Cl− currents and outwardly rectifying currents in transfected cells stimulated by cAMP indicates that both the wild-type and the Δ259-M265 CFTR are able not only to conduct Cl− but also to restore defective regulation of the ORCCs. A summary of DIDS-sensitive and DIDS-insensitive Cl− currents in whole IB3–1 cell recordings for each construct is shown in Fig. 3.
Figure 2.
Recordings of whole cell currents of wild-type and various mutant CFTRs expressed in cultured bronchial CF epithelial cells (IB3–1). (A) Wild-type CFTR: representative whole cell patch-clamp recordings of basal, cAMP-stimulated, and DIDS-inhibited cAMP-stimulated Cl− currents in wild-type CFTR-transfected IB3–1 cells. cDNA constructs were transfected transiently into IB3–1 CF cells and studied in physiological assays 72 hr after transfection in comparison to wild-type CFTR, mock-transfected cells, and parental IB3–1 cells. DIDS (500 μM) inhibited the outwardly rectifying current, whereas the remaining, underlying linear current (CFTR current) was unaffected (−100 mV to +100 mV in 20-mV increments from holding voltage of 0 mV). (B) Δ259-M265V CFTR: Typical whole cell patch-clamp recordings of basal, cAMP-stimulated, and DIDS-inhibited cAMP-stimulated Cl− currents from a Δ259-M265V-transfected cell. DIDS (500 μM) inhibited all of the current that was significantly outward rectified; no underlying linear current (CFTR current) was observed. (C) TMD-1 CFTR: Representative whole cell patch-clamp recordings of basal, cAMP-stimulated, and DIDS-uninhibited cAMP-stimulated Cl− currents from a TMD-1-transfected cell. DIDS (500 μM) failed to inhibit the current observed, indicating that the linear current was due to CFTR channel activity exclusively (see Fig. 3 for summarized data for all mutants).
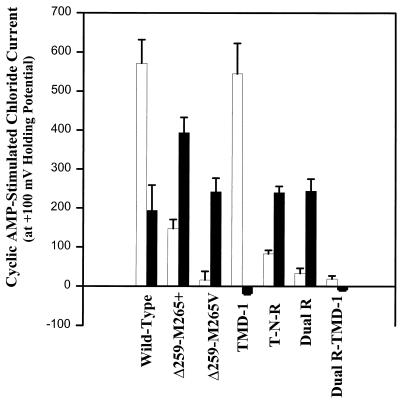
Figure 3.
Summary of whole cell currents of wild-type and various mutant CFTRs expressed in cultured bronchial CF epithelial cells (IB3–1). Summary of linear, DIDS (500 μM)-insensitive Cl− currents (open bars) and outwardly rectified, DIDS-sensitive Cl− currents (filled bars) measured in cells transfected with different CFTR constructs. The background Cl− current (56.2 ± 6.2 pA) was subtracted from these data. Summarized total whole cell currents (in nA) are presented as ICl- at −100 mV/ICl- at −100 mV with n in parentheses: parental IB3–1, −101.9 ± 12.1/66.3 ± 24.1 (8); nonresponders, −82.4 ± 15.9/57.2 ± 14.7 (71); wild-type, −676.2 ± 75.8/878.9 ± 76.0 (7); Δ259-M265, −316.8 ± 111.4/653.7 ± 63.3 (11); Δ259-M265V, −206.9 ± 52.3/371.1 ± 54.8 (8); TMD-1, −587.1 ± 83.0/582.5 ± 84.8 (8); T-N-R, −289.3 ± 27.3/435.4 ± 28.6 (6); dual arginine (Dual R), −177.5 ± 39.8/389.6 ± 57.7 (8); and Dual R-TMD-1, −150.3 ± 18.1/147.3 ± 15.3 (10).
In contrast, in IB3–1 cells transfected with Δ259-M265V, currents were more strongly outwardly rectified and were completely inhibited by DIDS, with no underlying linear CFTR currents (Fig. 3). Consistent with oocyte expression and the Cl− efflux studies, these results showed that elimination of the first four α-helices of CFTR and mutation of methionine-265 to a valine eliminated CFTR’s ability to generate Cl− currents in both oocytes and IB3–1 cells. The presence of strongly rectifying currents that are completely DIDS-sensitive indicates that this mutant can support cAMP regulation of ORCCs. Likewise, cAMP-stimulated whole cell Cl− currents generated by R334W-R347P (dual arginine) CFTR in transfected IB3–1 cells were strongly outwardly rectified and blocked fully by DIDS. Again, linear currents were absent, indicative of stimulation of ORCCs exclusively, again with no stimulation of Cl− conduction through CFTR (Fig. 3). Thus both of these mutants are processed sufficiently and contain the functional domains necessary to behave as conductance regulators and to support cAMP regulation of ORCCs.
In sharp contrast, however, TMD-1 CFTR-transfected cells had only linear currents, which were in part independent of cAMP stimulation. Although these IB3–1 cells do possess ORCCs, defective regulation is not restored after transfection of TMD-1 CFTR. The selectivity of TMD-1 CFTR was assessed in IB3–1 cells by using the difference in whole cell currents between solutions containing either Cl− or I−. The results showed that cAMP-activated CFTR currents at +100 mV were 1193.7 ± 199.3 pA in Cl−-containing solutions vs. 564.7 ± 49.6 pA in I−-containing solutions, suggesting that the Cl−:I− selectivity is 2:1 (n = 3) for TMD-1 CFTR. These results highlight two important points: first, that the selectivity filter of TMD-1 CFTR mutant is intact; second, that ORCC currents that have a halide selectivity of I− > Cl− (see ref. 27 for a discussion of this point) are not induced after transfection of the TMD-1 construct. The results show that a region distal to TMD-1 was essential for regulation of ORCCs.
Insertion of both R334W and R347P mutations into a TMD-1 background eliminated its ability to generate Cl− currents, as shown in Table 1, and its ability to activate ORCCs, as demonstrated by the complete lack of any currents when this construct was expressed in IB3–1 cells (Fig. 3).
Currents generated by T-N-R CFTR expressed in IB3–1 cells were also outwardly rectified; however, a small but significant linear current remained after DIDS inhibition (Fig. 3), illustrating, as in oocyte recordings, that this mutant maintains some Cl− channel and regulatory functions. More importantly, results with T-N-R CFTR suggest that the region of CFTR important for regulatory interaction with ORCCs lies distal to TMD-1 and proximal to TMD-2 within CFTR (i.e., the NBD-1/R domain portion of CFTR).
DISCUSSION
TMD-1 Is Essential for Proper Chloride Conduction and Selectivity of CFTR.
Several disease-causing mutations in CFTR located in TMD-1 result in mild or more severe reductions in single Cl− channel conductance (22, 24, 25). Those that cluster in transmembrane segment 6 of TMD-1 are those that have the greatest effects on Cl− channel conductance of CFTR (24, 25, 28). In this study, we show that a CFTR construct containing only TMD-1 produces constitutively active Cl− channels with single channel conductance and ion selectivity identical to wild type. Our previous work showed that removing transmembrane segments 1–4 of TMD-1 does not affect ion selectivity (22). Together these data show that transmembrane segments 5 and 6 of TMD-1 form an essential part of the Cl− conduction pore within CFTR. Surprisingly, despite the lack of the cytoplasmic regulatory domains, the currents produced by the TMD-1 CFTR variant were enhanced by cAMP stimulation. The most likely explanation for this surprising result is that cAMP is probably stimulating the recruitment of more channels in the plasma membrane of the oocyte, but more experiments will have to be performed to eliminate the possibility that some other more direct mechanism is involved.
NBD-1 and the R Domain Are Key Domains for Regulation of ORCCs.
A key component of this study was the results generated from two mutants, one where TMD-1 is truncated beyond methionine-265 and another where arginines are inserted into full-length CFTR at positions 334 and 347. Both are either not processed sufficiently or do not contain the necessary domains for them to conduct Cl− across the plasma membrane. On the other hand, they are processed sufficiently and do possess the domains necessary to regulate ORCCs. These findings show that the two functions of CFTR, Cl− conduction and ion channel regulation, are not mutually exclusive. Mutations and truncations can be made that eliminate CFTR’s ability to conduct Cl− but leave its ability to regulate ORCCs intact.
Which domains are critical for CFTR to interact with ORCCs? Our results show that a construct containing only TMD-1 cannot interact with ORCCs, whereas regulation by CFTR is intact with the expression of the first half of CFTR (T-N-R CFTR). Thus a domain independent of TMD-1 is important for regulation of ORCCs by CFTR within an area of CFTR that includes the NBD-1 and the R domain. Given that CFTR regulates ORCC activity by means of the release of ATP to the external medium, it is likely that this same region of CFTR plays an important role in facilitating ATP release either directly through CFTR or indirectly by controlling an alternate ATP release mechanism.
Interestingly, a similar portion of CFTR has been shown by the yeast two-hybrid technique to interact directly with epithelial Na+ channels (28). Thus this portion of CFTR may be important in a direct interaction with epithelial Na+ channels and in the indirect interaction that occurs between CFTR and ORCCs that operates by means of the release of extracellular ATP. It is important to note that mechanisms other than ATP release may also contribute to CFTR–ORCC regulatory interaction, including direct linkage in the intracellular membrane or indirectly by means of crosstalk between CFTR and ORCC via intracellular signal transduction pathways.
Conclusion.
It is well known that CFTR has a complex multidomain structure (29). To function normally all of the domains are important. However, specific domains play critical roles either in selective Cl− transport through CFTR or in the regulation of other channels such as ORCCs. This concept has implications for our understanding of CF. Those mutations that are expected to cause the most severe disease are those, such as G551D, that both drastically affect the ability of CFTR to move Cl− effectively itself and also eliminate its ability to regulate other channels such as the ORCC. Other mutations that retain at least one function or only partially reduce both functions may result in less severe pulmonary disease (30).
Acknowledgments
This study was funded by a CF Research Development Award (R-020 9C-1) and National Institutes of Health Grants (HL-47122 and DK-48977) to W.B.G. E.M.S. was funded by a National Research Service Award (HL-08832).
ABBREVIATIONS
- CFTR
cystic fibrosis transmembrane conductance regulator
- CF
cystic fibrosis
- SUR
sulfonylurea receptor
- ORCC
outwardly rectifying Cl− channel
- TMD
transmembrane domain
- NBD
nucleotide-binding domain
- R domain
regulatory domain
- CPT-cAMP
8-(4-chlorophenylthio)-cAMP
- DIDS
4,4′-diisothiocyanatostilbene-2,2′-disulfonic acid
References
- 1.Egan M E, Flotte T, Afione S, Solow R, Zeitlin P L, Carter B J, Guggino W B. Nature (London) 1992;358:581–584. doi: 10.1038/358581a0. [DOI] [PubMed] [Google Scholar]
- 2.Gabriel S E, Clarke L L, Boucher R C, Stutts M J. Nature (London) 1993;363:263–268. doi: 10.1038/363263a0. [DOI] [PubMed] [Google Scholar]
- 3.Boucher R C, Stutts M J, Knowles M R, Cantley L, Gatzy J T. J Clin Invest. 1986;78:1245–1252. doi: 10.1172/JCI112708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stutts M J, Canessa C M, Olsen J C, Hamrick M, Cohn J A, Rossier B C, Boucher R C. Science. 1995;269:847–850. doi: 10.1126/science.7543698. [DOI] [PubMed] [Google Scholar]
- 5.Ismailov I I, Awayda M S, Jovov B, Berdiev B K, Fuller C M, Dedman J R, Kaetzel M, Benos D J. J Biol Chem. 1996;271:4725–4732. doi: 10.1074/jbc.271.9.4725. [DOI] [PubMed] [Google Scholar]
- 6.Higgins C F. Cell. 1995;82:693–696. doi: 10.1016/0092-8674(95)90465-4. [DOI] [PubMed] [Google Scholar]
- 7.Higgins C F. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 8.Valverde M A, Diaz M, Sepulveda F V, Gill D R, Hyde S C, Higgins C F. Nature (London) 1992;355:830–833. doi: 10.1038/355830a0. [DOI] [PubMed] [Google Scholar]
- 9.Valverde M A, Bond T D, Hardy S P, Taylor J C, Higgins C F, Altamirano J, Alvarez-Leefmans F J. EMBO J. 1996;15:4460–4468. [PMC free article] [PubMed] [Google Scholar]
- 10.Philipson L H, Steiner D F. Science. 1995;268:372–373. doi: 10.1126/science.7716539. [DOI] [PubMed] [Google Scholar]
- 11.Inagaki N, Gonoi T, Clement J P, Namba N, Inazawa J, Gonzalez G, Aguilar-Bryan L, Seino S, Bryan J. Science. 1995;270:1166–1170. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- 12.Aguilar-Bryan L, Nichols C G, Wechsler S W, Clement J P, Boyd A E, Gonzalez G, Herrera H, Nguy K, Bryan J, Nelson D A. Science. 1995;268:423–426. doi: 10.1126/science.7716547. [DOI] [PubMed] [Google Scholar]
- 13.Inagaki N, Gonoi T, Clement J P, Wang C Z, Aguilar-Bryan L, Bryan J, Seino S. Neuron. 1996;16:1011–1017. doi: 10.1016/s0896-6273(00)80124-5. [DOI] [PubMed] [Google Scholar]
- 14.Isomoto S, Kondo C, Yamamada M, Matsumoto S, Higashiguchi O, Horio Y, Matsuzawa Y, Karachi Y. J Biol Chem. 1996;271:24321–24324. doi: 10.1074/jbc.271.40.24321. [DOI] [PubMed] [Google Scholar]
- 15.Ammala C, Moorhouse A, Gribble F, Ashfield R, Proks P, Smith P A, Sakura H, Coles B, Ashcroft S J H, Ashcroft F M. Nature (London) 1996;379:545–548. doi: 10.1038/379545a0. [DOI] [PubMed] [Google Scholar]
- 16.McNicholas C M, Guggino W B, Schwiebert E M, Hebert S C, Giebisch G H, Egan M E. Proc Natl Acad Sci USA. 1996;93:8083–8088. doi: 10.1073/pnas.93.15.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jovov B, Ismailov I I, Berdiev B K, Fuller C M, Sorscher E J, Dedman J R, Kaetzel M A, Benos D J. J Biol Chem. 1995;270:29194–29200. doi: 10.1074/jbc.270.49.29194. [DOI] [PubMed] [Google Scholar]
- 18.Schwiebert E M, Egan M E, Hwang T-H, Fulmer S B, Allen S S, Cutting G R, Guggino W B. Cell. 1995;81:1063–1073. doi: 10.1016/s0092-8674(05)80011-x. [DOI] [PubMed] [Google Scholar]
- 19.Reisin I L, Prat A G, Abraham E H, Amara J F, Gregory R J, Ausiello D A, Cantiello H F. J Biol Chem. 1994;269:20584–20591. [PubMed] [Google Scholar]
- 20.Reddy M M, Quinton P M, Haws C, Wine J J, Grygorczyk R, Tabcharani J A, Hanrahan J W, Gunderson K L, Kopito R R. Science. 1996;271:1876–1879. doi: 10.1126/science.271.5257.1876. [DOI] [PubMed] [Google Scholar]
- 21.Abraham E H, Okunieff P, Scala S, Vos P, Oosterveld M J S, Chen A Y, Shrivastav B. Science. 1997;275:1324–1325. doi: 10.1126/science.275.5304.1324. [DOI] [PubMed] [Google Scholar]
- 22.Piazza Carroll T, Morales M M, Fulmer S B, Allen S S, Flotte T R, Cutting G R, Guggino W B. J Biol Chem. 1995;270:11941–11946. doi: 10.1074/jbc.270.20.11941. [DOI] [PubMed] [Google Scholar]
- 23.Morales M M, Carroll T P, Morita T, Schwiebert E M, Devuyst O, Wilson P D, Lopes A G, Stanton B A, Dietz H C, Cutting G R, Guggino W B. Am J Physiol. 1996;270:F1038–F1048. doi: 10.1152/ajprenal.1996.270.6.F1038. [DOI] [PubMed] [Google Scholar]
- 24.Sheppard D N, Rich D P, Ostedgaard L S, Gregory R J, Smith A E, Welsh M J. Nature (London) 1993;362:160–164. doi: 10.1038/362160a0. [DOI] [PubMed] [Google Scholar]
- 25.Tabcharani J A, Rommens J M, Hou Y-X, Chang X-B, Tsui L-C, Riordan J R, Hanrahan J W. Nature (London) 1993;366:79–82. doi: 10.1038/366079a0. [DOI] [PubMed] [Google Scholar]
- 26.Zeitlin P L, Lu L, Hwang T-C, Rhim J, Cutting G R, Kieffer K A, Craig R, Guggino W B. Am J Respir Cell Mol Biol. 1991;4:313–319. doi: 10.1165/ajrcmb/4.4.313. [DOI] [PubMed] [Google Scholar]
- 27.Schwiebert E M, Flotte T, Cutting G R, Guggino W B. Am J Physiol. 1994;266:C1464–C1477. doi: 10.1152/ajpcell.1994.266.5.C1464. [DOI] [PubMed] [Google Scholar]
- 28.Kunzelmann K, Kiser G L, Schreiber R, Riordan J R. FEBS Lett. 1997;400:341–344. doi: 10.1016/s0014-5793(96)01414-7. [DOI] [PubMed] [Google Scholar]
- 29.Riordan J R, Rommens J M, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J-L, Drumm M L, Iannuzzi M C, Collins F S, Tsui L-C. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 30.Fulmer S B, Schwiebert E M, Morales M M, Guggino W B, Cutting G R. Proc Natl Acad Sci USA. 1995;92:6832–6836. doi: 10.1073/pnas.92.15.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]