Abstract
UL25 and UL17 are two essential minor capsid proteins of HSV-1, implicated in DNA packaging and capsid maturation. We used cryo-electron microscopy to examine their binding to capsids, whose architecture observes T=16 icosahedral geometry. C-capsids (mature DNA-filled capsids) have an elongated two-domain molecule present at a unique, vertex-adjacent, site that is not seen at other quasi-equivalent sites nor on unfilled capsids. Using SDS-PAGE and mass spectrometry to analyze wild-type capsids, UL25-null capsids, and denaturant-extracted capsids, we conclude that (i) the C-capsid-specific component is a heterodimer of UL25 and UL17; and (ii) capsids have additional populations of UL25 and UL17 that are invisible in reconstructions because of sparsity and/or disorder. We infer that binding of the ordered population reflects structural changes induced on the outer surface as pressure builds up inside the capsid during DNA packaging. Its binding may signal that the C-capsid is ready to exit the nucleus.
Introduction
Assembly and packaging of the nucleocapsid of herpes simplex virus is a complex, multistep, process involving the coordinated activity of some fifteen viral proteins (Baines and Duffy, 2006). Mechanistically, this pathway has much in common with capsid assembly of the tailed bacteriophages, suggesting common albeit distant evolutionary origins (Bamford et al., 2005; Steven et al., 2005; Baker et al., 2005). The capsid is first assembled as a precursor particle or procapsid into which DNA is then packaged. The procapsid’s surface shell is composed of hexamers and pentamers of the major capsid protein, UL19 (VP5), coordinated by triplexes of UL18 (VP23) and UL38 (VP19c) (Newcomb et al., 1993), with the UL6 portal at one vertex (Newcomb et al., 2001). Subsequently, the procapsid undergoes a major transition - maturation - involving profound structural and compositional changes (Newcomb et al., 1996; Trus et al., 1996) - Fig. 1. Its shape switches from spherical to polyhedral as the hexamers are remodeled (Heymann et al., 2003), such that a continuous inner “floor” layer is formed and sites are created around their outer tips to which the small protein UL35 (VP26) may bind (Booy et al., 1994; Zhou et al., 1995). Maturation is preceded by proteolytic cleavage of the internal scaffolding proteins. Because procapsids mature in vivo shortly after assembly, they are rarely captured in thin section electron micrographs of infected cells, but three capsids are observed in and may be isolated from the nuclei of infected cells: DNA-containing C-capsids; and two capsids that lack DNA - B-capsids, which retain the scaffolding protein, and A-capsids, which do not. All three have mature shells. The latter two capsids appear to be aberrant by-products, probably resulting from failure to complete (A-capsids) or to properly initiate (B-capsids) DNA packaging.
Figure 1. Alternative maturation pathways for the HSV-1 procapsid.
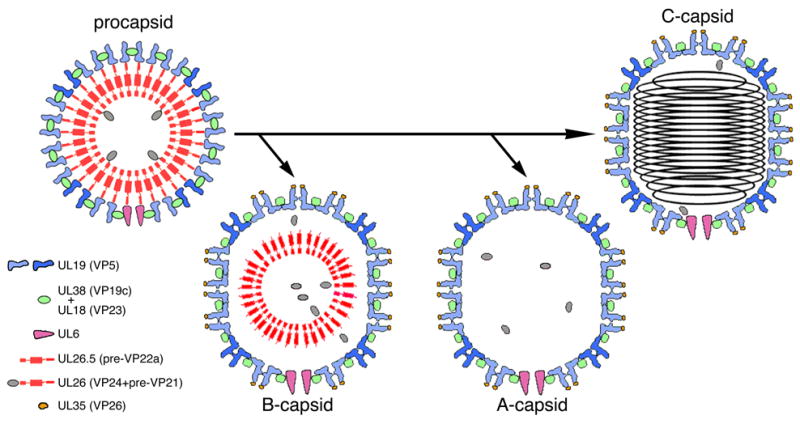
The procapsid surface shell is made up of hexamers (light blue subunits) and pentamers (mid-blue) of UL19, coordinated by triplexes, which are heterotrimers of UL18 and UL38. Its inner shell consists of the scaffolding proteins, UL26.5 and UL26, which is a fusion of the protease VP24 with VP21, an extended version of UL26.5. A single dodecamer of UL6 is present at the portal vertex. In maturation, the protease is released by autoproteolysis and processes the C-termini of the scaffolding proteins at the interface between the inner and outer shells. The surface shell then switches from round to polyhedral in shape, and UL35 binds to the outer tips of UL19 hexamers. The capsids are represented schematically in cross-sections that convey some but not all of their salient features.
In addition to the components listed above, several other viral proteins interact with the capsid during DNA packaging and/or in readying the nucleocapsid to exit from the nucleus. UL15, UL28, and UL33 are components of the terminase - the enzyme that recognizes and cleaves at specific sites on the replicating concatemer of viral DNA and translocates DNA into the capsid (Baines and Weller, 2005; Jacobson et al., 2006). UL17 and UL25 play roles in DNA maturation and packaging (Salmon and Baines, 1998). Some light has been cast by experiments with viruses deleted for these genes. In the absence of UL17, the replicating DNA concatemer is not cleaved to genome-sized pieces and DNA is not packaged (Thurlow et al., 2005). The absence of UL25 is less prohibitive (Stow, 2001): packaging takes place but this DNA does not appear to be stably encapsidated, fewer C-capsids being seen in the nuclei of infected cells (McNab et al., 1998). Observations on cells infected with a UL25-null mutant of pseudorabies virus (PRV, another alpha-herpesvirus) detected C-capsids inside but not outside the nucleus, implying that they are stabler than their HSV-1 counterparts but unable to embark on nuclear export (Klupp et al., 2006).
Previous studies have determined that UL17 and UL25 are incorporated into capsids in roughly equal amounts, estimated at 15–40 copies per capsid in B-capsids and A-capsids, and somewhat more in C-capsids (Suppl. Table 1). Absence of one protein (by mutation) impairs incorporation of the other, and when B-capsids are treated with increasing amounts of urea, the proteins are extracted in parallel (Thurlow et al., 2006). These observations suggest that they might form a complex. However, there is also evidence for non-coupled populations of these molecules (Thurlow et al., 2006).
Capsid-associated UL25 and UL17 are sensitive to trypsin (Newcomb et al., 2006; Wills et al., 2006), indicating that they are exposed on the outer surface, an inference corroborated by EM studies demonstrating that anti-UL25 and anti-UL17 antibodies bind to the capsid exterior (Newcomb et al., 2006; Thurlow et al., 2006).
Motivated by the prospect that identifying the capsid locations of UL25 and UL17 might yield insight as to their function(s), we have approached this problem by cryo-EM and image reconstruction, complemented with biochemical analysis. Initially, we focused on C-capsids, which have the highest content of UL25. For reference, we also analyzed unfilled capsids (A-capsids) from a UL25-null mutant and from the wild-type virus. These studies visualized an additional structural component present on C-capsids but not on any other capsid examined: we refer to it as the C-capsid-specific component (CCSC). Correlation of its presence with protein composition was not straightforward, as unfilled capsids also contain UL25 and UL17 but do not exhibit the CCSC. Nevertheless, combining our cryo-EM data with biochemical data and other observations led to the conclusions that the CCSC is a heterodimer of UL25 and UL17.
Results
Detection of an additional component, the CCSC, on the outer surface of C-capsids
C-capsids were isolated from the nuclear fraction of infected cells harvested at 20 h post-infection. Examination by cryo-EM confirmed that the capsids were highly purified (data not shown). The resulting micrographs were used to calculate two independent reconstructions (Table 1A). They consistently revealed an additional component - the CCSC - on the outer surface of the capsid (cf. Figs. 2B & 2A). This molecule is ~ 14 nm long by ~ 3 nm wide and appears to consist of two domains. It interacts with the two triplexes closest to the UL19 penton – triplexes Tc and Ta in the nomenclature of (Zhou et al., 1994); makes glancing contact with the adjacent UL19 hexamer (the P-hexon in the nomenclature of (Steven et al., 1986)), and extends towards the UL19 penton (Fig. 2C). The molecule is visibly present only at this site (apart from repetitions generated by icosahedral symmetry) and not at any of the other quasi-equivalent sites that are distributed over the capsid surface (Figs. 2B, 3A).
Table 1.
(A) Presence of CCSC in cryo-EM reconstructions of HSV-1 capsids, and (B) Mass spectrometric analysis of bands from SDS-PAGE of capsids.
| A: | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type of capsid | Resolution (Å) [1] | No. of particles | CCSC occupancy [2] | Copy number | ||||||
| C-capsids | 19.9 (0.5) 17.6 (0.3) | 643 pairs | 0.54, 0.55 (0.55) | 33 | ||||||
| C-capsids | 26.8 (0.5) 24.5 (0.3) | 538 | 0.45, 0.44 (0.45) | 27 | ||||||
| A-capsids (wildtype) [3] | 22.9 (0.5) | 2111 | 0.15, 0.07 (0.11) | 7 | ||||||
| A-capsids (wildtype) [3, 4] | 27.5 (0.5) 26.8 (0.3) | 1100 | 0.18, 0.08 (0.13) | 8 | ||||||
| A-capsids (UL25-null) | 22.7 (0.5) 21.9 (0.3) | 1088 | 0.00, 0.00 (0.00) | 0 | ||||||
| C-capsids (+ 0.5M GHCl) | 25.0 (0.5) 24.1 (0.3) | 595 | 0.00, 0.00 (0.00) | 0 | ||||||
| B : | ||||||||||
| Protein | M.W | peptides matched/submitted | coverage | AMD [5] | Score [6] | P value [7] | ||||
| UL6 | 74,046 | 10/20 | 23% | 0.009 | 127 | 1.99E-13 | ||||
| UL17 | 74,536 | 9/12 | 16% | 0.012 | 105 | 3.16E-11 | ||||
| UL25 | 62,631 | 19/28 | 45% | 0.017 | 211 | 7.93E-22 | ||||
| UL36 (VP1/3) | 335,655 | 33/59 | 15% | 0.017 | 155 | 3.16E-16 | ||||
Resolution is according to the Fourier Shell Correlation, thresholded at 0.5 or 0.3.
The two numbers correspond to measurements made in the vertex-distal and the vertex-proximal parts of the CCSC and (in bold), their mean.
reported by Cheng et al (2002)
In this reconstruction, grayscale sections show some faint density at the CCSC site, but it is at the level of background noise. If genuine, it would correspond to an occupancy of 3-fold to 4-fold lower than is observed on C-capsids.
Average mass deviation (absolute values) in Daltons for all matched peptides from theoretical peptides
Score is determined as −10*LOG10(P)
P is a probability-related measure that the identified protein is a random match
Figure 2. The CCSC is present on C-capsids but not on UL25-null A-capsids.
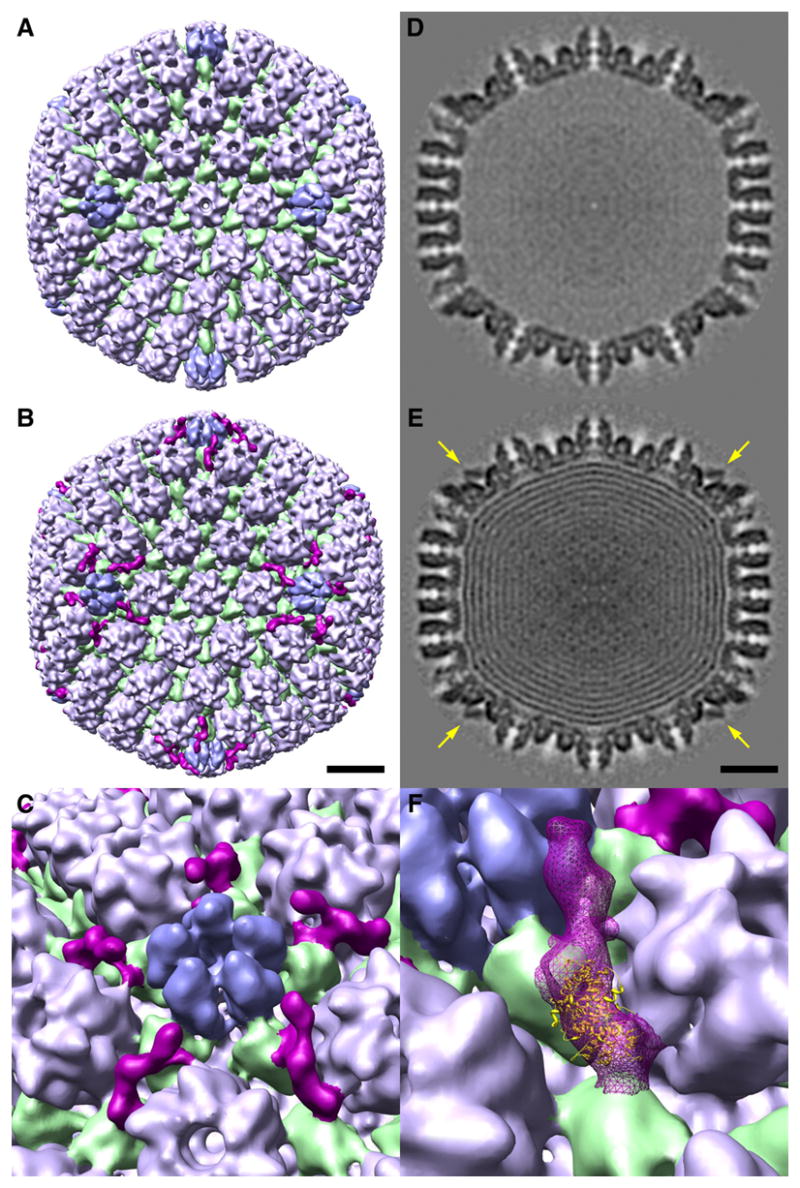
Surface renderings of cryo-EM reconstructions are shown in B (C-capsid) and A (UL25-null A-capsid). Triplexes are green; hexons (UL19 hexamers with six VP26 molecules around their outer tip) are light blue; pentons (UL19 pentamers) are mid-blue. The CCSC in B is purple. The capsids are viewed along a 2-fold axis of icosahedral symmetry. C shows a blow-up of the region around a penton, presenting five different views of the CCSC molecules clustered around this vertex. Bar (B, E) = 20 nm. D and E show central sections of the two capsids. High density is dark. The UL25-null A-capsid is empty; the C-capsid contains nested shells of DNA (Booy et al., 1991). Arrows in E point to the CCSC which is sampled in this plane. F shows the optimal fit of a UL25 fragment into the CCSC.
Figure 3. The CCSC is visible at only one of six quasi-equivalent sites on the HSV-1 C-capsid.
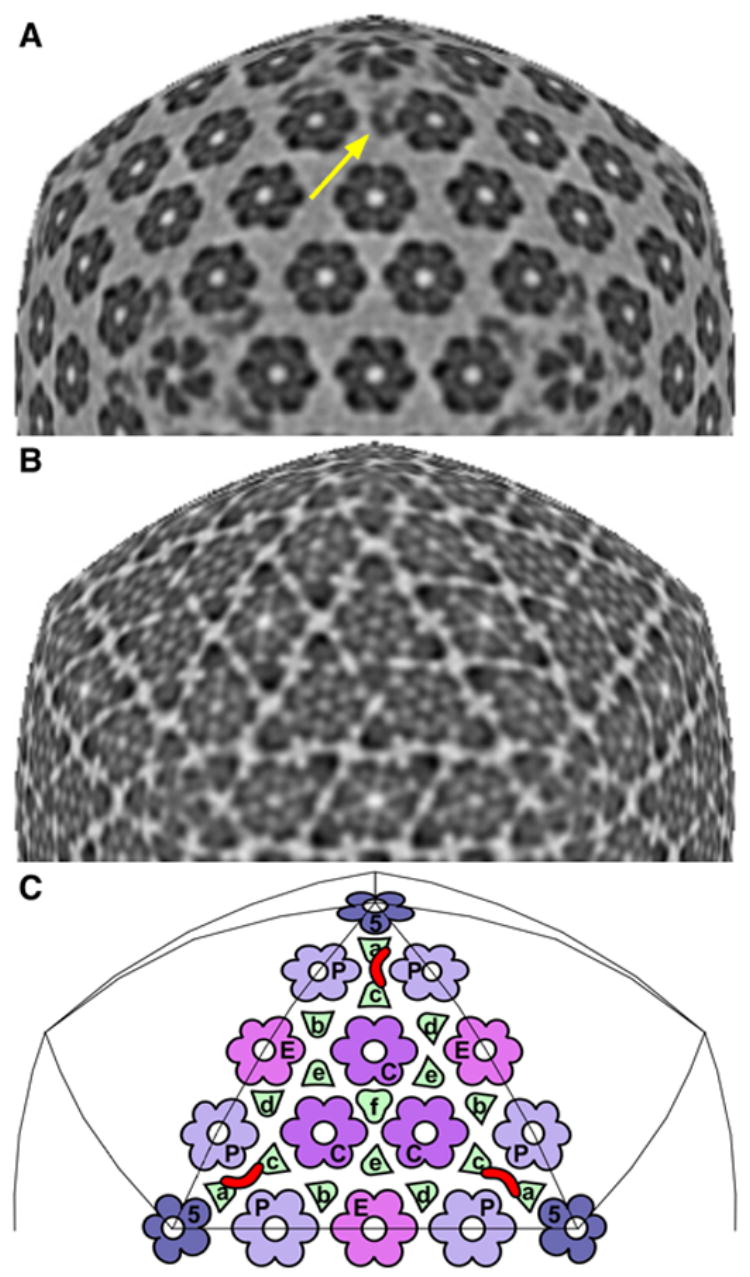
A & B show icosahedral sections through the C-capsid at radii of 61 nm and 53 nm, respectively, along the 3-fold axis. In them, the sampling surface maintains the same radial position relative to the capsid shell (as opposed to the capsid center). Section A is rescaled so that features in it line up with underlying features in section B. Section A samples the CCSC (yellow arrow), whereas section B samples the triplexes as well as the capsomer protrusions, just above the floor. In C are mapped the positions of the four kinds of capsomers and six kinds of triplexes on one triangular facet. Each CCSC (one is marked in red) overlies a pair of triplexes (Tc, Ta). Other quasi-equivalent sites may be found by substituting another kind of triplex for Tc. There are 320 triplexes, but maximum CCSC binding would correspond to 160 copies, because one CCSC occludes two triplexes. The CCSC is seen on C-capsids at only one of the six possible sites, at 50% occupancy. This occupancy may increase in virions, whose UL25 content is higher than that of C-capsids (Thurlow et al., 2006). Bar (B) = 15 nm.
The CCSC is also evident in grayscale sections (arrows, Figs. 2E, 3A, 4A). Its average occupancy may be estimated in terms of the value of its peak density relative to that in the capsid shell (assumed to be 100% occupancy) and the average density outside the capsid (0% occupancy). In this way, we measured the average occupancy of the CCSC to be 50 + 5%, corresponding to an average copy number of 30 per capsid (0.5 x 60) (Table 1A).
Figure 4. The CCSC is removed from C-capsids by 0.5 M guanidine.HCl and is absent from wild-type A-capsids.
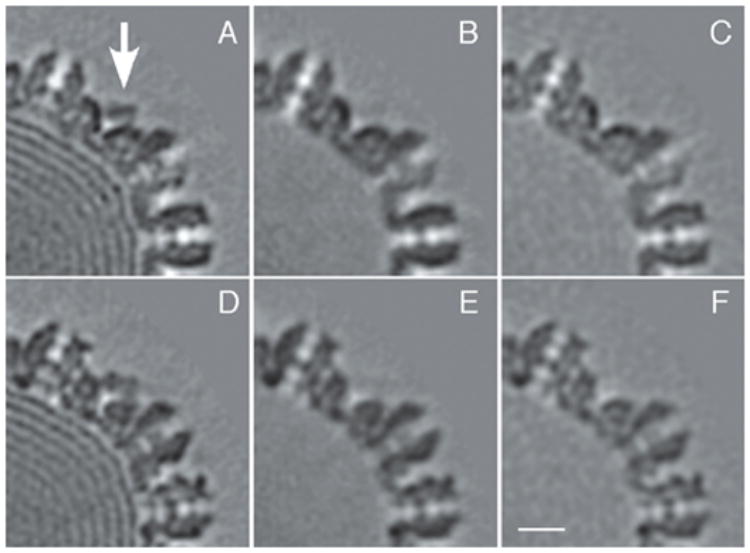
A, D: two sections from a C-capsid reconstruction in which the CCSC is sampled (arrow). B, E: corresponding sections from C-capsids, after treatment with 0.5 Guan.HCl. Note the absence of both the DNA and of the CCSC. C, F: corresponding sections from a reconstruction of wild-type A-capsids. Bar = 10 nm.
The CCSC is not seen on UL25-null A-capsids
Because our working hypothesis was that it might be possible to visualize UL25 on C-capsids (Introduction), we also analyzed capsids from a UL25-null deletion mutant as a negative control. However, we have not been able to recover C-capsids from this mutant. Accordingly, bearing in mind that wild-type A-capsids also contain UL25 (Newcomb et al., 2006), we isolated UL25-null A-capsids and analyzed them by cryo-EM. The capsid structure, otherwise normal, shows no trace of the CCSC (Figs. 2A, 2D).
The CCSC is not seen on wild-type A-capsids
The CCSC has not been reported in previous cryo-EM studies of A-capsids or B-capsids. Accordingly, to clarify the significance of its absence from UL25-null A-capsids, we scrutinized two reconstructions of wild-type A-capsids in the vicinity of the CCSC binding site but observed no significant density there (Fig. 4C; Table 1A).
Minor capsid proteins detected by biochemical analysis
In Figure 5, we compare the protein compositions of wild-type C-capsids, A-capsids, and virions, as resolved by SDS-PAGE with Coomassie Blue staining. Also compared are wild-type A-capsids and A-capsids from the UL25-null mutant. The gels confirm the high degree of purity of these preparations. The major proteins – UL19, UL38, and UL18 – contribute strong bands. The C-capsids show only three additional, much less intense, bands in the region between UL19 and UL38. We used mass spectrometry to identify them as UL17 (band c-1); UL6 (band c-2); and UL25 (band c-3) - Table 1B, Supp. Fig. 1. The UL25 and UL17 bands are also present in A-capsids: although even weaker than in C-capsids, they are in the same relative amounts, i.e. the UL25 band is somewhat stronger. The faint UL6 band is of the same intensity in both capsids - as expected, since each should contain a single dodecamer of this protein. The UL25 band is indeed missing from UL25-null A-capsids (Fig. 5, right panels).
Figure 5. Identification of minor protein components of HSV-1 capsids.
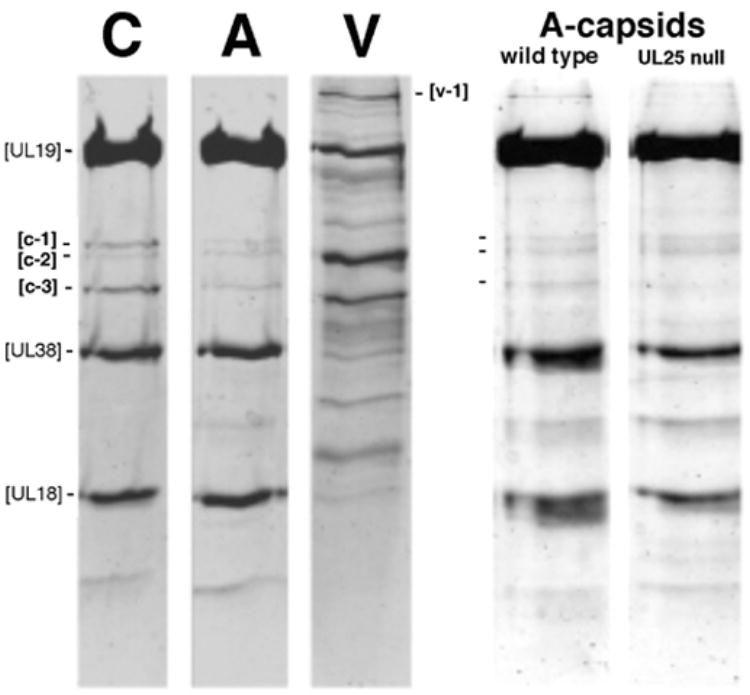
At left, C-capsids (C), wildtype A-capsids (A), and virions (V) are compared by SDS-PAGE with Coomassie Blue staining. Equal amount of protein were loaded in each lane. The major capsid proteins, UL19, UL38 and UL18 contribute strongly staining bands. The weak bands, c1, c-2, and c-3 were identified by mass spectrometry as UL17, UL6, and UL25, respectively. The v-1 band was identified as UL36. At right, wild-type A-capsids are compared with A-capsids from a UL25-null mutant. Again, c1, c-2, and c-3 are marked. The mutant capsids do not contain UL25, as confirmed by Western blotting (McNab et al., 1998). The very faint band running just below the UL25 position in the UL25-null mutant lane represents an unknown contaminant.
The intensities of these gel bands were used to estimate the copy numbers of UL25 and UL17. These quantitations were calibrated relative to both triplex proteins, yielding copy numbers of 82 + 19 (UL25) and 36 + 8 (UL17) for C-capsids (Supp. Table 1).
The protein composition of virions is considerably more complex than those of capsids -(Gibson and Roizman, 1972) and Fig 5. Because the UL36 (VP1-3) protein has been invoked as a prominent component attached to the capsid outer surface in virions (Zhou et al., 1999; see Discussion), we sought to identify the corresponding band by mass spectrometry. It is the most intense of the several high-MW bands in the virion lane (band v-1 in Fig. 5; see Supp. Fig. 1 and Table 1B). In contrast, there is essentially no staining at this position in the C-capsid or A-capsid lanes (Fig. 5). From these data, we put the VP1-3 content of C-capsids, if any, at less than 1 molecule per capsid.
Extraction of UL25 and UL17 from C-capsids
The copy numbers of UL25 and UL17 in C-capsids estimated by biochemical analysis are more than sufficient to match the number of CCSC molecules determined by cryo-EM. Conversely, UL25-null A-capsids have zero (UL25) or very small (UL17) contents of these proteins by SDS-PAGE (Fig. 5) and the CCSC is absent by cryo-EM (Fig. 2). However, the significance of the latter correlation is clouded by the absence of the CCSC from wild-type A-capsids which contain these proteins. It appeared, therefore, that the presence of the CCSC on C-capsids might depend on their having encapsidated DNA. To explore this hypothesis, we tested the effect of releasing their DNA by a relatively mild treatment that otherwise leaves the capsid shell minimally perturbed. Treatment of C-capsids with 0.5M guanidine hydrochloride has this effect (Newcomb and Brown, 1994).
After a 30-min incubation of C-capsids with 0.5 M Guan.HCl, they were separated from the denaturant and examined by cryo-EM for the CCSC, as well as biochemically, for the presence of UL25 and UL17. Almost all capsids were either empty or retained only vestigial amounts of DNA. Image reconstruction revealed that they retained no detectable amounts of the CCSC (Fig. 4B; Table 1A). A control reconstruction of C-capsids from the same preparation that were treated identically, apart from omitting the Guan.HCl, confirmed that they contained the CCSC (data not shown). That the capsid shells were otherwise minimally perturbed was attested by their retention of UL35, which is known to be relatively easily detached (Newcomb et al., 1993). Their UL35 was visible as characteristic “horns” around the tips of the UL19 hexons but not the pentons (data not shown, cf. Fig. 2C).
According to quantitative Western analysis, these C-capsids lost ~ 69% of their UL25 an a similar fraction (~ 59%) of their UL17 as a result of treatment with 0.5M guanidine-HCl.
Taken together, therefore, the cryo-EM observations and the biochemical data support the view that the CCSC is a heterodimer of UL25 and UL17.
Docking the crystal structure of a UL25 fragment into the CCSC
The C-terminal portion of UL25, residues 134 to 580, has been crystallized and its structure determined (Bowman et al., 2006). We tried docking this fragment (PDB: 2F5U), which is a monomer of ~ 5.5 x 3.5 x 4 nm, into the molecular envelope of the CCSC. Its volume, which we estimate as 122 nm3, corresponding to a mass of ~ 100 kDa, is too large for a monomer of either UL25 (63 kDa; the solved crystal structure has a mass of 44 kDa) or UL17 (75 kDa). The most plausible interpretation, in view of their copy numbers, is that the two lobes of the CCSC should accommodate, respectively, monomers of these two proteins. We tried docking UL25 into both lobes and in both orientations. The docking was performed both manually and with an automated docking program. In both cases, the most satisfactory fit was obtained with the UL25 fragment in the lobe that is distal to the adjacent 5-fold axis, oriented as shown in Fig. 2D. There, its center of mass is ~ 14 nm from the closest 5-fold axis and ~ 23 nm from the closest 3-fold axis of the capsid.
Discussion
We have visualized the CCSC - an elongated molecule, comprising at least two domains or subunits - on the outer surface of C-capsids (Figs. 2 & 3). Its primary points of contact are on the two triplexes closest to a vertex. There are no visible indications of its presence at other quasi-equivalent sites (Figs. 2B, 3A, 3C). Correlation of biochemical data with structural data affords persuasive evidence that the CCSC is a heterodimer of UL25 and UL17. However, these proteins are also present in lower but comparable amounts on unfilled capsids, i.e. A-capsids and B-capsids, on which the CCSC is not seen. To reconcile these observations, we infer that there are two populations of capsid-associated UL25 and UL17 molecules: the visible population seen on C-capsids; and populations that are invisible in averaged density maps but are nevertheless present on A- and B-capsids as well as C-capsids. In the following, we discuss this scenario and its implications.
The CCSC is not seen on unfilled capsids
In this context, it is noteworthy that, in immuno-gold EM labelling experiments of intracellular PRV capsids with anti-UL25 antibodies, Klupp et al. (2006) observed labelling of C-capsids but not of A-capsids nor B-capsids. Based on the present results, we suspect that their antibodies were conformationally sensitive, specifically recognizing UL25 in the PRV version of the CCSC.
The CCSC is present in virions and is not UL36 (VP1-3)
We reproducibly observed the CCSC in C-capsid reconstructions. In retrospect, it is also discernible in our earlier C-capsid reconstruction, despite its lower resolution (Booy et al., 1991). Like C-capsids, capsids inside virions are fully packaged with DNA. Consistent with this trend, the CCSC appears to be present in the nucleocapsid reconstructed from intact enveloped virions by Zhou et al. (1999). In this comparison - cf. Fig. 2 and Fig. 2 of (Zhou et al., 1999) - allowance should be made for the fact that the capsid in the latter reconstruction has a different hand. (The hand of the HSV-1 capsid was determined later in an EM tilting experiment (Cheng et al., 2002)). The inferred interaction of UL25 with the capsid inside virions is supported by the observation that it remains capsid-associated when virions are extracted with Triton X-100 (Wolfstein et al., 2006).
Zhou et al. (1999) assigned this feature of their reconstruction as part of a single protein, UL36 - a large tegument component of 335 kDa. However, as our C-capsids are devoid of UL36, that assignment is inapplicable. We posit that three proteins contribute to this feature: the two lower lobes are monomers of UL25 and UL17. The CCSC-like feature in virions extends further on to the penton crown than does its counterpart on C-capsids. (Penton-crowning density has also been detected on intraviral nucleocapsids by cryo-electron tomography - Grünewald et al., 2003). The latter density may represent a small portion of UL36 but this proposition remains unproven.
Evidence for two populations of UL25 and UL17
It is difficult to measure the copy numbers of minor components of such complex particles as HSV-1 virions and capsids with high precision by SDS-PAGE. Nevertheless, estimates have been made for UL25 and UL17 by several investigators, with fair consistency (Suppl. Table 1). Taken together, these data support the following view: (i) both proteins are present in wild-type A-capsids and B-capsids in copy numbers of ~ 15 to ~ 40 per capsid. (Different batches of A-capsids appear to be more variable in this respect than B-capsids – WWN, unpublished observations). (ii) Both proteins have higher contents in C-capsids, corresponding to ~ 50 – 80 copies of VP25 per capsid. (iii) All capsids have somewhat more UL25 than UL17.
From cryo-EM, we obtained an average of ~ 30 copies of the CCSC per C-capsid. The CCSC is too large for a monomer of either protein, and its shape does not subdivide into equal parts so as to suggest that it is a homodimer or a homotrimer. Accordingly, the most likely explanation is that it is a heterodimer of UL25 and UL17. This proposal is consistent with prior observations indicating that the bindings of some UL25 and UL17 molecules to capsids are coupled, which in turn suggests that they interact (Introduction). As the crystal structure for a major fragment of UL25 fits well into the vertex-distal portion of the CCSC (Fig. 2F), it follows that the vertex-proximal portion is likely to be UL17. This interpretation will be tested more conclusively if it proves possible to label the CCSC with anti-UL25 or anti-UL17 antibodies.
The average C-capsid should contain ~ 30 copies of both proteins, configured as CCSC molecules. According to the biochemical data (above), it should have additional populations of both proteins that are not seen in reconstructions. The latter resemble the populations that are present in A-capsids and B-capsids in size and (in)visibility. As noted above, the UL25 content appears to be somewhat greater than that of UL17 in all three capsids. The observed departure from equimolarity is not large and may simply reflect a difference in dye-binding affinity and/or experimental uncertainties. However, there is also evidence that capsids can bind some UL25 molecules in the absence of UL17 partners and vice versa (Newcomb et al., 2006; Thurlow et al., 2006).
Where are the invisible molecules located? And why are they invisible?
If they were at the same site as the CCSC and configured in the same way, UL25 and UL17 molecules are abundant enough in A-capsids and B-capsids to be seen in cryo-EM reconstructions (they would correspond to occupancies of 25% – 66%). The occupancy threshold for visibility in a cryo-EM density map depends on the residual noise level which, in turn depends on several factors including the number of particles included in the analysis. Acknowledging this variability, experience with the binding of Fab fragments to hepatitis B virus capsids (Conway et al., 2003) suggests that an occupancy of ~ 20% is a useful rule-of-thumb for a visibility threshold.
One explanation for the non-visibility of UL25 and UL17 molecules in reconstructions of A-and B-capsids is that they may be scattered around other quasi-equivalent sites. There are six variants of this site, if one counts by letting triplexes Ta, Tb, Td, Te or Tf substitute for Tc in its CCSC-binding role (Fig. 3C). Altogether, there are 320 potential binding sites per capsid. If heterodimers of UL25 and UL17 were randomly distributed around these sites, the average occupancy would be < 10% and thus, below the threshold of detectability. A second possibility invokes disorder instead of sparsity. If single UL25 molecules – which may be in excess over UL17 (see above) – were to bind to triplexes but remain free to pivot around their point of attachment, they would be seen poorly if at all in a reconstruction, even with relatively high occupancy. Third, it is not ruled out that some UL25 and/or UL17 molecules engage in an entirely different mode of binding.
Allosteric regulation of CCSC binding: a precedent in UL35 (VP26) binding
To account for the presence of the CCSC only on C-capsids, we suggest that, as DNA packaging progresses and pressure from encapsidated DNA builds up on the inner surface of the capsid wall, it eventually reaches a point where a structural transition is elicited on the outer surface that affects the CCSC binding site, resulting in an enhanced affinity. The envisaged temporal control of CCSC binding in the maturation pathway by allostery has a precedent in the binding of UL35. The latter protein does not bind to the procapsid but, as the procapsid matures, the six protrusion domains of each UL19 hexon, initially splayed apart (Heymann et al., 2003), cluster into a 6-fold symmetric ring on which bipartite binding sites for UL35 monomers are created in the crevices between neighboring protrusions (Wingfield et al., 1997). In UL19 pentons, on the other hand, the protrusion domains remain apart so that UL35 binding sites are not created. This exclusion of UL35 from pentons leaves their crowns open to engage in other interactions (Chen et al., 2001).
Nature of the allosteric switch that promotes CCSC binding
The binding site of a CCSC molecule on the underlying capsid is tripartite, with contact points on the Ta triplex, the Tc triplex, and the sidewall of the P-hexon. Our working hypothesis assigns the first and third of these contacts to UL25 and the second one to UL17, with the complex being further stabilized by the interface between UL25 and UL17. The inferred association of UL25 with a triplex is consistent with the report by Ogasawara et al. (2001) of an interaction with UL38 (VP19c). This arrangement - a network of interactions that appear, individually, to be rather weak but, collectively, consolidate the binding of the CCSC - is reminiscent of the situation pertaining to clathrin-coated vesicles (Wakeham et al., 2003).
The key transition in the maturation pathway is from the procapsid state to the C-capsid state, A- and B-capsids being abortive particles. On comparing the procapsid with the C-capsid (cf. Figs. 6A and 6B), two kinds of changes are seen to take place; the triplex structures are altered, as are the relative positions of the three CCSC contact points. Maturation of the procapsid is a progressive event, proceeding through multiple intermediate states (Heymann et al., 2004). The final maturation step, undergone only by the C-capsid and crucial for CCSC binding, involves similar but smaller structural changes that bring the three contact points into juxtaposition (Supplementary movie 1). The relative importance of the two kinds of structural changes for CCSC binding will be clarified if it proves possible to extend the resolution of the procapsid and C-capsid significantly.
Figure 6. Changes affecting the CCSC binding site on maturation of the HSV-1 procapsid.
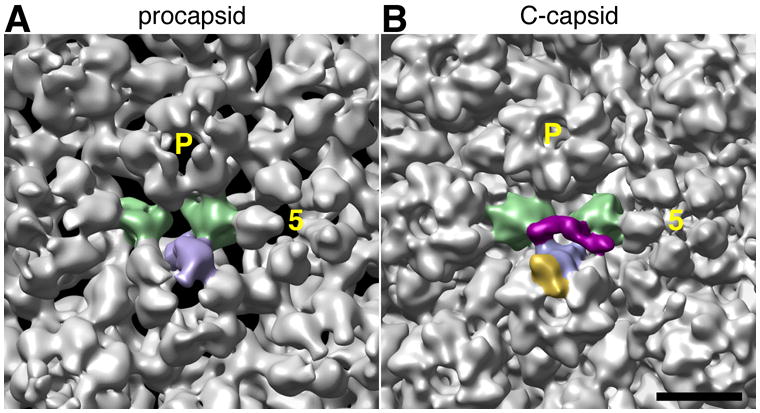
The regions of capsid surface surrounding the CCSC binding site are compared in cryo-EM reconstructions at ~ 2 nm resolution of the procapsid (A – from Cheng et al., 2002) and the C-capsid (B). In both cases, the two triplexes and one UL19 subunit that form the binding site for a single representative CCSC are shaded green and blue, respectively; this CCSC is purple in panel B and the UL35 subunit that tops the interacting UL19 subunit (but is not present on the procapsid) is gold. As reference points, the 5-fold axis of the adjacent UL19 penton is marked “5”, and a neighboring P-hexon with P. Bar = 10 nm.
Functional implications: signalling and nuclear exit
At least two functions have been considered for UL25: stabilization of the DNA-filled capsid (Newcomb et al., 2006) and a role in nuclear exit (Klupp et al., 2006). It is noteworthy that the two triplexes to which the CCSC binds, Tc and Ta, are also the two that are most susceptible to extraction by denaturants, at concentrations (6M urea or 2M Guan.HCl) that also remove the pentons but otherwise respect the capsid framework (Newcomb et al., 1993).Thus, these triplex sites might be considered as weak points that are in need of reinforcement. However, since the CCSC is already extracted under less rigorous conditions (0.5M Guan.HCl), it would not be still in place to provide reinforcement when denaturant concentrations rise to levels that threaten the Tc/Ta triplexes and the pentons. To the extent that resistance to denaturants is representative of the challenges against which the nucleocapsid needs to be stabilized - it is hard to see a role for the CCSC in this process. However, less extreme perturbations might also threaten the nucleocapsid, for instance, by causing it to lose its DNA and CCSC binding might offer some protection against them.
The allosteric switch in the capsid shell that we infer to precede CCSC binding is reminiscent of signalling across a cell membrane (Alberts et al., 1989). In the latter case, an event on one side of the membrane, such as binding of a ligand to a receptor, elicits a downstream event on the other side, such as a phosphorylation cascade. In HSV-1 packaging, the signal comes from pressure imposed on the capsid inner surface by packaged DNA and elicits a structural change on the outer surface that facilitates CCSC binding.
It is tempting to envisage CCSC binding as a signal for nuclear export. Deferring this signal until late in packaging would avoid premature export of unpackaged or incompletely packaged capsids. It would also enhance overall replication efficiency for A-capsids and B-capsids - which remain within the nucleus - not to be allowed to embark on the tegumentation/envelopment pathway, and therefore not committing further resources to the production of sterile particles. In this context, we note that UL36 is not needed for C-capsids to exit the nucleus but is required for them to achieve envelopment and exit the cytoplasm (Desai, 2000). The scenario outlined above relates to the observation by Thurlow et al. (2006) that virions contain twice as much UL25 as do nuclear C-capsids. This additional complement might represent an increase in average occupancy of the CCSC sites from ~ 50% to ~ 100%. Conversely, nuclear C-capsids may be still in the nucleus because they have not yet bound sufficient copies of the CCSC to enable their exit.
Noting that this scenario does not account for the phenotype of UL17-null mutants which fail to initiate DNA packaging (Salmon and Baines, 1998), we suggest that UL17 and possibly also UL25 may exercise additional functions, earlier in the nucleocapsid assembly pathway.
With respect to other regulatory mechanisms that may be used to signal that packaging of nucleic acid into virus particles is at or approaching completion, Lander et al. (2006) have proposed a different allosteric mechanism, involving interaction of DNA with the portal, as a signal for termination of packaging into bacteriophage P22.
Experimental procedures
Preparation of capsids
Wild-type C-capsids and A-capsids and UL25-null A-capsids were prepared as described (McNab et al., 1998; Newcomb et al., 2006).
Gel electrophoresis and Western blot analyses
These assays were performed as described (Newcomb et al., 2006). The antibodies used in the immunoblotting experiments were polyclonal antiserum raised against purified UL25 (McNab et al., 1998) and an anti-UL17 monoclonal, MAb 203 (Thurlow et al., 2006), a kind gift of Dr V. Preston.
Extraction of C-capsids with 0.5M guanidine hydrochloride
50 μl of 6M Guan.HCl in TNE buffer, pH 7.5, were mixed with 440 μl of TNE and 10 μl of 1M DTT followed by the addition of 100 μl of purified C-capsids at 0.5 mg/ml in TNE. After incubation on ice for 30 min, the solution was pelleted through a 20 % sucrose cushion, supernatant was discarded, and the pellet was resuspended in 50 μl of TNE.
The fractions of UL25 and UL17 extracted were measured from a Western blot. The extracted capsids were solubilized and run on an SDS-PAGE gel; then blotted on to PVDF and stained with 1% Ponceau S (Newcomb et al., 2006)). The blot was stained with monoclonal antibody 2D9 (1:2000 dilution) specific for UL25 as described (Sheaffer et al., 2001). The blot was then washed and restained with an anti-UL17 antibody (MAb 203).
Mass spectrometry
Designated bands were excised from gels and extracted with 0.2 ml 50% (w/v) acetonitrile in 25 mM Tris–HCl (pH 8.1), followed by 0.2 ml 100% acetonitrile, and dried in vacuum. The gel pieces were re-hydrated with 0.04 ml 20 μg/ml modified trypsin (Promega V5111) in 25 mM Tris–HCl (pH 8.1) and digested overnight at 37° C. Peptides were extracted with 0.1 ml 0.2% (w/v) trifluoroacetic acid, extracts were freed from salts with zip-tip pipette tips (Millipore) and eluted with 3 μl 50 mM alfa-cyano-4-hydroxycinammic acid in 50% (v/v) acetonitrile/0.1% (w/v) TFA directly on the mass-spectrometer sample plate.
Peptides were analyzed on an Applied Biosystems Voyager DE-Pro time-of-flight mass spectrometer in the reflectron mode. Internal mass calibration of raw spectra was performed with PeakErazor software (http://welcome.to/gpmaw/) on contaminant trypsin/keratin autolysis peaks. This resulted in mass accuracy < 50 ppm in the subsequent mass search. Protein identification was performed on a locally run Mascot Search program (http://www.matrixscience.com/search_form_select.html) against the SProt/TrEMBL database.
Cryo-electron microscopy was performed as described (Cheng et al., 1999). Microscopy was performed, using low-dose procedures, at defocus values described below, on a CM200-FEG microscope (Philips, Eindhoven, Netherlands). The same preparations of capsids were examined both by cryo-EM and by SDS-PAGE and mass spectrometry.
Three-dimensional image reconstruction
Micrographs were digitized on an SCAI scanner (Z/I Imaging, Huntsville, AL) giving a pixel size of 3.68Å. Particles were extracted and preprocessed with X3D (Conway et al., 1993), and contrast transfer function (CTF) parameters were estimated with Bsoft (Heymann and Belnap, 2007). CTF correction was performed by phase-flipping. Defocus settings were such that the first CTF zeros were at (20Å) −1 – (24Å) −1or (16Å) −1 – (20Å) −1 if a first exposure was taken. Initial estimates of the orientation angles were determined with the PFT algorithm (Baker and Cheng, 1996), starting from a pre-existing A-capsid reconstruction (Cheng et al., 2002) adjusted in scaling to match the current data. Refinement were performed with PFT2 (Bubeck et al., 2005). Density maps were calculated with EM3DR2 (http://people.chem.byu.edu/belnap/). For resolution assessment (Saxton and Baumeister, 1982), the radial zone between 42.7 nm and 67 nm was used.
Visualization and molecular modeling
Surface representations were produced, using Chimera (Goddard et al., 2005). Icosahedral sections (Böttcher et al., 1997) were calculated using a program of Dr J F Conway. The pentons, hexons, triplexes, and the CCSC were automatically segmented using a flooding algorithm implemented in bsegment/Bsoft (Heymann and Belnap, 2007). Interfaces were checked visually and refined manually. Thereconstructions were visualized at the threshold of 1 standard deviation. In the molecular modeling the threshold was adjusted for the CCSC to take into account its lower occupancy.
The crystal structure of UL25 (PDB 2F5U) was docked manually using Chimera, and the result checked by automated correlation-based fitting, with colores in the SITUS program (Chacon and Wriggers, 2002). All top-ranked solutions placed the UL25 fragment in the vertex-distal lobe of the CCSC.
Supplementary Material
Acknowledgments
We thank Dr J. Conway for providing his icosahedral section program, Drs B. Heymann and D. Belnap for reconstruction software, and Dr. V. Preston for a gift of M203 antibody. This work was supported in part by the Intramural Research Programs of NIAMS and CIT and the NIH Intramural Targeted Antiviral Program (ACS), and NIH grants to JCB (AI41644-10) and FLH (AI060836).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J. Molecular Biology of the Cell. 2. New York: Garland Publishing Inc; 1989. pp. 645–647. [Google Scholar]
- Baines JD, Duffy C. Nucleocapsid assemby and envelopment of Herpes Simplex Virus. In: Sandri-Goldin R, editor. Alpha Herpesviruses: Molecular and Cell Biology. Norwich, UK: Horizon Press; 2006. [Google Scholar]
- Baines JD, Weller SK. Cleavage and packaging of herpes simplex virus 1 DNA. In: Catalano C, editor. Viral Genome Packaging Machines. Kluwer Academic/Plenum Publishers; 2005. [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. Liquid-crystalline, phage-like, packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy FP, Trus BL, Newcomb WW, Brown JC, Conway JF, Steven AC. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Nat'l Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher B, Kiselev NA, Stel'Mashchuk VY, Perevozchikova NA, Borisov AV, Crowther RA. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J Virol. 1997;71:325–330. doi: 10.1128/jvi.71.1.325-330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman BR, Welschhans RL, Jayaram H, Stow ND, Preston VG, Quiocho FA. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J Virol. 2006;80:2309–2317. doi: 10.1128/JVI.80.5.2309-2317.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM. Structure of the poliovirus 135S cell-entry intermediate at 10Å resolution reveals the location of an externalized polypeptide that binds to membranes. J Virol. 2005;79:7745–7755. doi: 10.1128/JVI.79.12.7745-7755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- Chen DH, Jakana J, McNab D, Mitchell J, Zhou ZH, Dougherty M, Chiu W, Rixon FJ. The pattern of tegument-capsid interaction in the herpes simplex virus type 1 virion is not influenced by the small hexon-associated protein VP26. J Virol. 2001;75:11863–11867. doi: 10.1128/JVI.75.23.11863-11867.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N, Conway JF, Watts NR, Hainfeld JF, Joshi V, Powell RD, Stahl SJ, Wingfield PE, Steven AC. Tetrairidium, a four-atom cluster, is readily visible as a density label in three-dimensional cryo-EM maps of proteins at 10–25 Å resolution. J Struct Biol. 1999;127:169–176. doi: 10.1006/jsbi.1999.4120. [DOI] [PubMed] [Google Scholar]
- Cheng N, Trus BL, Belnap DM, Newcomb WW, Brown JC, Steven AC. Handedness of the herpes simplex virus capsid and procapsid. J Virol. 2002;76:7855–7859. doi: 10.1128/JVI.76.15.7855-7859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Trus BL, Booy FP, Newcomb WW, Brown JC, Steven AC. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J Struct Biol. 1993;111:222–233. doi: 10.1006/jsbi.1993.1052. [DOI] [PubMed] [Google Scholar]
- Conway JF, Watts NR, Belnap DM, Cheng N, Stahl SJ, Wingfield PT, Steven AC. Characterization of a conformational epitope on hepatitis B virus core antigen and quasiequivalent variations in antibody binding. J Virol. 2003;77:6466–73. doi: 10.1128/JVI.77.11.6466-6473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PJ. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J Virol. 2000;74:11608–11618. doi: 10.1128/jvi.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W, Roizman B. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Grünewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol. 2003;10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- Jacobson JG, Yang K, Baines JD, Homa FL. Linker insertion mutations in the herpes simplex virus type 1 UL28 gene: effects on UL28 interaction with UL15 and UL33 and identification of a second-site mutation in the UL15 gene that suppresses a lethal UL28 mutation. J Virol. 2006;80:12312–12323. doi: 10.1128/JVI.01766-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klupp BG, Granzow H, Keil GM, Mettenleiter TC. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J Virol. 2006;80:6235–6246. doi: 10.1128/JVI.02662-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- McNab AR, Desai P, Person S, Roof LL, Thomsen DR, Newcomb WW, Brown JC, Homa FL. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Brown JC. Induced extrusion of DNA from the capsid of herpes simplex virus type 1. J Virol. 1994;68:433–440. doi: 10.1128/jvi.68.1.433-440.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Homa FL, Brown JC. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J Virol. 2006;80:6286–6294. doi: 10.1128/JVI.02648-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Homa FL, Thomsen DR, Booy FP, Trus BL, Steven AC, Spencer JV, Brown JC. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75:10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Trus BL, Booy FP, Steven AC, Wall JS, Brown JC. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- Ogasawara M, Suzutani T, Yoshida I, Azuma M. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J Virol. 2001;75:1427–1436. doi: 10.1128/JVI.75.3.1427-1436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon B, Baines JD. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of U(L)15-encoded proteins with B capsids requires at least the U(L)6, U(L)17, and U(L)28 genes. J Virol. 1998;72:3045–3050. doi: 10.1128/jvi.72.4.3045-3050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J Microsc . 1982;127(Pt 2):127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Sheaffer AK, Newcomb WW, Gao M, Yu D, Weller SK, Brown JC, Tenney DJ. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J Virol. 2001;75:687–698. doi: 10.1128/JVI.75.2.687-698.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr Opin Struct Biol. 2005;15:227–236. doi: 10.1016/j.sbi.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven AC, Roberts CR, Hay J, Bisher ME, Pun T, Trus BL. Hexavalent capsomers of herpes simplex virus type 2: symmetry, shape, dimensions, and oligomeric status. J Virol. 1986;57:578–584. doi: 10.1128/jvi.57.2.578-584.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow ND. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J Virol. 2001;75:10755–10765. doi: 10.1128/JVI.75.22.10755-10765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow JK, Murphy M, Stow ND, Preston VG. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J Virol. 2006;80:2118–2126. doi: 10.1128/JVI.80.5.2118-2126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow JK, Rixon FJ, Murphy M, Targett-Adams P, Hughes M, Preston VG. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J Virol. 2005;79:150–158. doi: 10.1128/JVI.79.1.150-158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC. The herpes simplex virus procapsid: Structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- Wakeham DE, Chen CY, Greene B, Hwang PK, Brodsky FM. Clathrin self-assembly involves coordinated weak interactions favorable for cellular regulation. EMBO J. 2003;22:4980–4990. doi: 10.1093/emboj/cdg511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills E, Scholtes L, Baines JD. Herpes simplex virus 1 DNA packaging proteins encoded by UL6, UL15, UL17, UL28, and UL33 are located on the external surface of the viral capsid. J Virol. 2006;80:10894–10899. doi: 10.1128/JVI.01364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Thomsen DR, Homa FL, Booy FP, Trus BL, Steven AC. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J Virol. 1997;71:8955–8961. doi: 10.1128/jvi.71.12.8955-8961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfstein A, Nagel CH, Radtke K, Dohner K, Allan VJ, Sodeik B. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic. 2006;7:227–237. doi: 10.1111/j.1600-0854.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, He J, Jakana J, Tatman JD, Rixon FJ, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nature Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Prasad BV, Jakana J, Rixon FJ, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


