Abstract
Routine typing of a potential bone marrow donor by sequence-specific oligonucleotide probes and sequence based typing produced inconclusive subtyping results, suggesting a new allele. A magnetic bead-based method, haplotype specific extraction (HSE), was used to separate the diploid sample into its haploid components. The sample was then re-typed using standard sequence-based typing (SBT), revealing a new human leukocyte antigen (HLA) allele, since named B*1576. HSE used in conjunction with standard SBT is a convenient and simple tool for resolving ambiguous and novel allele combinations without the need for amplification or subcloning.
Keywords: Haplotype Specific Extraction, HSE, molecular haplotyping, new HLA-B allele
INTRODUCTION
We have identified a new human leukocyte antigen (HLA)-B allele, B*1576, in a potential bone marrow donor using a technique for isolating haploid DNA, called haplotype specific extraction, (HSE). Preliminary typing of the subject by sequence-specific oligonucleotide probes and sequence based typing indicated a possible HLA-B*15, 56 allele combination but provided inconclusive results, suggesting a novel allele. Haplotype specific extraction was then used to resolve the ambiguity. HSE probes were designed to target two of the heterozygous differences that were identified between the likely B*1501 allele and the suspected new allele during the initial typing. The probes were used to separate the diploid DNA into its haploid fractions through an automated process of capture by magnetic beads and extraction of the HLA-B region, including exons 2 and 3. The extracted DNA was then sequenced, resulting in the conclusive identification of the new allele by its haploid sequence.
HSE is a simple, rapid and automated means to determine new alleles without cloning by converting the diploid state of the original sample into its haploid fractions. The method provides the distinct advantage in that it can produce true (non-statistically derived) haplotype data without familial information.
MATERIALS AND METHODS
The sample discussed here, ID #111542, is identified as an anonymous caucasoid bone marrow donor participant typed by the Labor für Immungenetik of the University Munich. DNA was obtained from blood using automated DNA isolation (BioRobot M48 workstation, Qiagen) and was initially typed for the HLA-B locus by sequence-specific oligonucleotide probes (SSOP-Elpha, Biotest AG, Dreieich, Germany) and direct sequence-based typing (SBT) [1]. All conventional typing assays failed to give conclusive results and suggested the likely presence of a new allele. When considering Helmberg-SCORE analysis [2] of exons 2 and 3 and allowing for up to three mismatches, the closest typing of the sample appeared to be of a low-resolution type B*15 and B*56.
Haplotype specific extraction was performed on the sample using standard HLA-B HaploPrep™ separation kits and protocols available through Qiagen. Haplotype specific extraction utilizes three factors to generate haploid genomic DNA: Genetic variations in the form of polymorphic markers (i.e. heterozygous positions such as single nucleotide polymorphisms, SNPs), copy number variations (insertions/deletions) or other unique sequence elements (such as transposons) are used for the design of HSE probes that distinguish between the targeted genomic region and any other sequence. The sample DNA and the HSE probes are then combined with a polymerase, buffer and a mix of regular and biotinylated nucleotides. The DNA is denatured and then annealed in order for the probes to specifically bind to a genomic region or polymorphic allele. The conditions of the reaction favor hybridization and subsequent elongation of the probe to a perfectly matched sequence over any other, unmatched sequence. An example is one of the two alleles of a heterozygous single nucleotide polymorphism. In addition, the polymerase’s fidelity ensures a high level of specificity for extension of only the matched, hybridized probe. The extended, biotinylated probe and the bound genomic target are then captured by magnetic beads through an automated mixing process that removes non-targeted material. The resulting DNA is then directly usable in down-stream genotyping assays.
In this example, a nucleotide alignment of a polymorphic section of HLA-B exon 2, in which all nucleotide differences between allele B*1501 and the suspected new allele (now named B*1576) were located, was performed to identify suitable sites for probe selection (Fig. 1). B*070201 is shown as a reference sequence. All positions corresponding to exons 2 and 3 of HLA-B were controlled for heterozygosity using the MT Navigator PCC software (Applied Biosystems). There were eight heterozygous positions available to separate the two alleles, located at nt 259, 261, 272, 277, 280, 282, 283 and 292 (red box in Figure 1).
Figure 1.

Sequence alignment of HLA-B exons 2 and 3 of B*1501 and B*1576 (see [3]). The reference sequence shown on top is B*070201. All nucleotide differences between the B*1501 and B*1576 alleles are located in the region 259–292 of exon 2 (red box). The changes in this region lead to a difference in the expression of six amino acids (bottom line in red box). The pattern is consistent with an epitope seen in several other HLA-B allele groups (i.e. 07,42,54,55,56,67,81,82,83) from which the B*1576 allele may have arisen by recombination.
The polymorphisms at positions 261 and 277 were selected for separating the two observed variants by four probes from the HLA-B ‘HaploPrep™’ kit (Qiagen): HSE probes B261G and B261C, respectively, target the G allele of the known allele B*1501 and the C allele of the suspected new allele (B*1576). Probes B277A and B277G2, respectively, target the A allele of B*1501 and the G of the suspected new allele (B*1576). Each probe was used individually to isolate its respective targeted allele in a separate HSE reaction.
The purpose of a redundant probe selection targeting multiple polymorphic positions is to avoid a potential failure of separation that, in principle, could occur if a unique combination of two novel alleles were present that would not be complementary to at least one HSE probei. However even if this were to occur, sequencing after HSE would not produce an incorrect typing, but only result in a failed separation (i.e. diploid sequence). By selecting more than one separation point, it becomes very unlikely that multiple separations would fail due to such unexpected target sequences. As can be seen from the alignment in Figure 1, it is possible that the new allele seen here arose through a paired recombination event with breakpoints located at nucleotide positions < 258 and > 292 that substituted this segment of the B*1501 allele with a segment of a second allele identical with the B*070201 reference sequence.
For each HSE separation, about 250 ng of genomic DNA (8.5 μl of approx. 30 ng/μl DNA) was incubated in 200 μl tubes with 1.5 μl probe (Qiagen), 15 μl hybridization buffer (H-Buffer, Qiagen) and 5 μl water on a T3 thermalcycler (Biometra, Göttingen, Germany). The sample was then denatured at 95C for 15 minutes, followed by incubation at 64C for 10 minutes on a heat block with heated lid (Hybex™ Microsample Incubator, Scigene; formerly Trutemp™, Robbins Scientific). The sample was then combined with 50 μl HaploPrep beads (Qiagen), magnetically mixed, washed three times and eluted in 50 μl water on a BioRobot M48 (Qiagen). 10 μl of the haplo-separated DNA bound to the magnetic beads were then used as template for the generic amplification and sequencing of HLA-B exons 2 and 3, as described earlier [1].
RESULTS
The presence of a new allele was confirmed by consistent sequencing results for the HSE-extracted DNA targeted with HSE probes B261C and B277G2. The sequencing peaks at multiple polymorphic positions after haploseparation were hemizygous (haploid), with the second allele occasionally visible as small, underlying traces in the background of the electropherogram (Figure 2). The data shows that all four HSE probes independently produced allelic separation. The new allele resulted in a final sample typing of B*1501, 1576.
Figure 2.
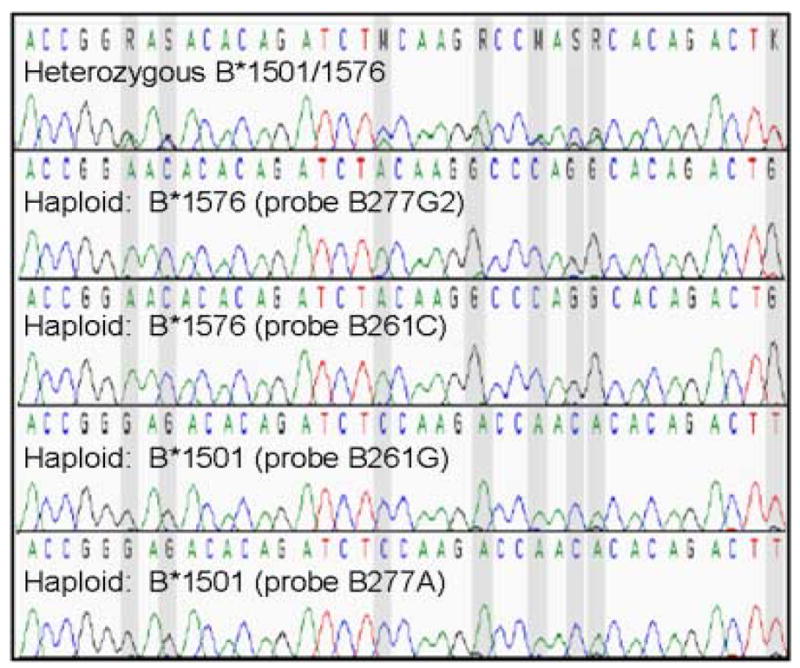
A diploid sample unable to be typed by conventional means was suspected to contain a new HLA allele. Multiple HSE extractions using HSE probes B277G2, B277A, B261G and B261C (Qiagen) produced consistent haploid sequencing results that resolved all heterozygous allele combinations and identified a new HLA-B allele, B*1576. The gray vertical bars identify heterozygous single nucleotide polymorphisms in the diploid sample (top) that were resolved to their haploid genotype in each haploseparated DNA.
DISCUSSION
A convenient method for determining ambiguous typing results for HLA alleles is critical in light of the rapid increase in allele variants being detected [3]. Haplotype specific extraction provides a rapid, automated method for determining true (i.e. non-statistically derived) and previously unknown haplotypes, directly from genomic DNA, without requiring any familial information [4–10]. The method does not rely on amplification-based phasing methods which can be affected by polymorphic variation and eliminates the need for cloning or subcloning for the characterization of new alleles. The method’s utility is emphasized by its ability to resolve all known HLA-A, -B, -C, -DR and –DQ alleles as offered through Qiagen’s HaploPrep reagent kit. Further, the flexibility and specificity of the biochemistry allows for new HSE probes to be easily developed based upon genomic variation as small as single nucleotide changes (SNPs).
HSE provides a number of benefits to determining ambiguous or novel alleles in that it can be applied to most forms of genomic DNA preparations. Only a single probe and reagent is required for separation of a given allele type, and the automation of HSE provides an efficient means to process samples for haplotype analysis. The DNA resulting from the method contains both the local genetic content (the area being targeted by the probe) and information distal to the site of capture, allowing for typing to be performed across an entire gene region with one HSE reaction. The size of a region that can be isolated in one haploextraction depends largely on the size of the (diploid) genomic DNA used as starting material for HSE and the method that was used for its preparation. Linkage distances of 40–50kb in one direction (upstream as well as downstream from the capture point) can reliably be achieved for DNA that was isolated from blood by magnetic beads. Finally, the extracted HSE product can be directly applied to most down-stream genotyping assays such as sequence based typing, SSO, SSP, PCR/real-time PCR, pyrosequencing and array based comparative genomic hybridization without further modification. This, in conjunction with the method’s ability to unambiguously determine haplotype information, provides a versatile tool for resolving tissue typing ambiguities and identifying new alleles.
Acknowledgments
This work was funded by the NIAID (AT SBIR grant R44-AI51036 to Generation Biotech) and by Qiagen. We would like to thank Mark Kunkel and Dimitrios Monos for help in preparing this manuscript.
Footnotes
The nucleotide sequence data reported in this paper was given the accession number AJ535668 by the EMBL nucleotide sequence database. The name B*1576 was officially assigned to this new allele by the World Health Organization (WHO) Nomenclature Committee in January 2003. This follows the agreed policy that, subject to the conditions stated in the most recent Nomenclature Report, names will be assigned to the new sequences as they are identified. Lists of such new names will be published in the following WHO Nomenclature Report [3].
For example, a hypothetical recombination event between nucleotide positions 259 and 261 could have been present if two new alleles were present in this sample: New Allele 1: A-G, New Allele 2: G-C; instead of G-G (B*1501) and A-C (B*1576) (see Figure 1). In this highly unlikely case, HSE probes targeting this region would not have properly annealed to either one of the alleles.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Witter K, Wolpl A, Zahn R, Klein HG, Albert ED. Sequence-based typing confirmed a novel B*40 allele, B*4046, which was identified through sequence-specific oligonucleotide hybridization routine typing. Tissue Antigens. 2004 Apr;63(4):378–81. doi: 10.1111/j.0001-2815.2004.00156.x. [DOI] [PubMed] [Google Scholar]
- 2.Helmberg W, Lanzer G, Zahn R, Weinmayr B, Wagner T, Albert E. Virtual DNA analysis--a new tool for combination and standardised evaluation of SSO, SSP and sequencing-based typing results. Tissue Antigens. 1998 Jun;51(6):587–92. doi: 10.1111/j.1399-0039.1998.tb03000.x. [DOI] [PubMed] [Google Scholar]
- 3.Marsh SG, Albert ED, Bodmer WF, et al. Nomenclature for Factors of the HLA System 2004. In: Marsh Steven GE., editor. Hum Immunol. 5. Vol. 66. Anthony Nolan Research Institute, Royal Free Hospital; Pond Street, Hampstead, London NW3 2QG, UK: 2005. May, pp. 571–636. Epub 2005 Mar 3; and: IMGT/HLA database, www.ebi.ac.uk/imgt/hla/ambig.html. [DOI] [PubMed] [Google Scholar]
- 4.Mrazek F, Fae I, Ambruzova Z, Raida L, Indrak K, Petrek M, Fischer GF. A novel HLA-B*420502 allele identified by PCR-SSO/SSP routine typing and confirmed by Sequencing-based typing. Tissue Antigens. 2005 Mar;65(3):275–7. doi: 10.1111/j.1399-0039.2005.00376.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang CY, Pan Q, Xue M, Miao KR, Fei XM, Zhou XY, Zhao X, Kukuruga D, Osowski L, Poore B, Beattie R, Shi WX, Zhang H. Identification of an HLA-B*07 allele variant (B*0740) in the Chinese Han population. Tissue Antigens. 2005 Aug;66(2):148–50. doi: 10.1111/j.1399-0039.2005.00438.x. [DOI] [PubMed] [Google Scholar]
- 6.Lebedeva TV, Huang A, Ohashi M, Sibilia P, Alosco SM, Kempenich J, Yu N. The recombinant HLA-B*5518 allele supports the evidence of conserved haplotype association of rare alleles. Tissue Antigens. 2005 Aug;66(2):156–9. doi: 10.1111/j.1399-0039.2005.00442.x. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y, Aubrey MT, Zhang G, Ji Y, Freed BM. HLA-B*1586 is a novel, hybrid HLA-B15/B22 allele with unique serology and haplotypic association. Tissue Antigens. 2006 Feb;67(2):176–7. doi: 10.1111/j.1399-0039.2006.00531.x. [DOI] [PubMed] [Google Scholar]
- 8.Dapprich J, Cleary MA, Gabel HW, Akkapeddi A, Iglehart B, Turino B, Beaudet L, Lian J, Murphy NB. A rapid, automatable method for molecular haplotyping. In: Hansen JA, editor. Immunobiology of the Human MHC; Proceedings of the 13th International Histocompatibility Workshop and Congress, IHWC 2001; Seattle, WA, USA, IHWG Press. 2006. pp. 93–96. [Google Scholar]
- 9.Nagy M, Entz P, Otremba P, Schoenemann C, Murphy N, Dapprich J. Tissue Antigens. 2006. Haplotype specific extraction: a universal method to resolve ambiguous genotypes and detect new alleles – demonstrated on HLA-B. accepted for publication. [DOI] [PubMed] [Google Scholar]
- 10.Dapprich J, Magira E, Samonte MA, Lind C, Rossman MD, Rosenman K, Hsu S, Monos D. Tissue Antigens. 2006. Identification of a Novel HLA-DPB1 Allele (DPB1*1902) by Haplotype Specific Extraction and Nucleotide Sequencing. accepted for publication. [DOI] [PubMed] [Google Scholar]


