Abstract
The mouse metallothionein-I (mMT-I) promoter is activated by the metal response element-binding transcription factor (MTF), which binds metal response elements (MREs) when stimulated with heavy metals. We analyzed eight K562 erythroleukemia cell clones, each carrying a single integrated copy of an mMT-I/β-geo construct, using a system that can independently assess the level of β-geo expression and the rate at which it is silenced. In these clones, basal expression and rate of silencing vary widely and independently with integration site. This implies that the rates of transcription and of silencing are separate properties determined by interaction of the regulatory elements of the transgene with the site of integration. Induction of the mMT-I promoter with zinc both increases expression level and strongly retards silencing of β-geo expression. At a given integration site, expression level and silencing are affected coordinately by induction. Taken together with earlier studies of distant metal-responsive elements, these results suggest that distance from the promoter may determine whether a factor can increase transcription rate. Stimulation of an MRE can both increase transcription and overcome repressive effects of chromatin; we suggest that these functions are linked.
Activating factors bound in upstream promoter regions and in more distant enhancers are thought to regulate the rate of transcription through direct interaction with the initiation complex (1). In this view of transcriptional control, activating factors bound close to the promoter or at more distant sites (enhancers) perform essentially the same function. Most of the evidence for the effect of enhancers on transcription rate is derived from systems in which populations of cells are analyzed (2, 3). In those systems, an increase in transcription products will result from either an increase in the rate of transcription of all of the templates or an increase in the proportion of templates that are actively transcribed. However, assay of expression in single cells suggests that enhancers act primarily to establish and maintain transcription, and that their direct effect on transcription rate is minimal (4, 5, 6, 7, 8). Similarly, evidence that factors binding to upstream promoter regions regulate the rate of transcription is derived primarily from experimental systems, such as transient transfection and in vitro transcription assays, that analyze the total transcribed product or reporter expression from a very large number of templates (1), and so the effect of promoter activation on transcription is in most cases unclear.
The mouse metallothionein-I (mMT-I) promoter is a well-characterized example of a simple inducible promoter. Heavy metals induce binding of the activating factor MTF to a series of metal response elements (MREs) in the mMT-I promoter; this increases expression of MT and of reporter genes driven by the mMT-I promoter (9, 10, 11, 12). Although this has been regarded as an indicator of an increase in transcription rate, the reporter systems used have not assayed the number of transfected cells expressing the reporter.
Genes translocated or exogenously introduced into the genome are subject to position effects (13, 14). These have been classified into two types: variegating effects, in which the gene is inactivated in a subpopulation of cells in a stochastic and clonally heritable fashion, and stable effects, in which the gene is active in all cells, but its expression level is influenced by the integration site. In an earlier study, we examined the effect of MREs (derived from the mMT-I promoter) on expression of the β-geo reporter gene, when the MREs were placed downstream of β-geo, 4 kb distant from the promoter (7). There was a strong tendency for expression of β-geo to be silenced, at a rate characteristic of the individual integration site; we have compared this to position-effect variegation and other silencing phenomena (13). We established that stimulation of the MREs had no effect on the level of β-geo expression. However, stimulation of the distant MREs drastically retarded silencing; a parallel study found that a globin enhancer also retarded silencing without significantly affecting expression level. Since other studies of expression in individual cells have also shown that enhancers have little effect on the level of transcription (4, 5, 6, 7, 8), we concluded that distant activating elements act primarily by creating and maintaining domains in which promoter activity is permitted. This left open the question of whether the same elements are able to regulate transcriptional level when placed near the promoter, and if so whether expression level is linked to silencing.
We now report a study of expression driven by the mMT-I promoter when stably integrated into K562 erythroleukemia cells. The basal level of expression is strongly affected by the integration site; metal stimulation does increase transcription from this promoter, although not to the degree predicted by earlier experiments with transient assays. In the absence of metal stimulation, expression of β-geo is silenced, and the rate of silencing is dependent on the integration site; there is no relationship between the rate of silencing and the level of β-geo expression when different integration sites are compared. Metal stimulation of the promoter counteracts silencing. At a given integration site, increasing concentrations of zinc have a parallel affect on expression level and rate of silencing. Taken together with the earlier study of a MT enhancer (7), this experiment suggests that distance from the promoter determines the ability of an activating factor to increase transcription, but that in either location the factor prevents formation of higher-order chromatin structures that silence transcription.
MATERIALS AND METHODS
Cell Culture and Transfection.
Conditions for growth and electroporation of K562 cells were as described (6). For induction of MT elements, zinc sulfate was added to the medium to a concentration of 80 μM, which we have established is not toxic to K562 cells; growth rates of induced and uninduced cells are equivalent. Following electroporation of K562 cells with the MT/β-geo plasmid (Fig. 1), G418-resistant clones were screened for single-copy integrants. Genomic DNA was digested with EcoRI, which cuts at a single site in the β-geo gene in MT/β-geo. Single integration events were confirmed by using hybridization probes located either upstream (900-bp ClaI/BamHI lacZ fragment) or downstream (1.2-kb EcoRI/XhoI β-geo fragment) of the EcoRI site in β-geo. Only those clones that generated a single hybridization signal greater than 4 kb in size (with a different size for each probe) were selected for subsequent analyses.
Figure 1.
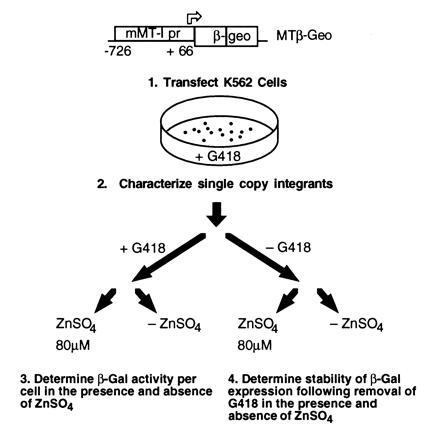
Strategy for study of zinc induction of the mMT-I promoter. The construct (top) contains the mMT-I promoter (from −727 to +66) driving expression of β-geo and is derived from pSAβ-geo (18). The mMT-I promoter contains a series of six MREs starting directly upstream of the TATA element (12). MT/β-geo was electroporated into K562 cells and eight single-copy integrants were characterized. Basal expression and inducibility were studied by splitting cells maintained in G418 into two aliquots, and adding zinc sulfate (to 80 μM) to one aliquot. Twenty-four hours later β-galactosidase (β-gal) activity in each aliquot was determined by 4-methylumbelliferyl β-d-galactoside (MUG) conversion in cell lysates. Stability of β-geo expression was determined by removing cells from G418 and dividing them into two aliquots. One aliquot was maintained in standard medium, the other in medium with 80 μM zinc sulfate. At intervals, the fluorescence-activated cell sorter–β-gal (FACS-gal) assay was performed to assay the proportion of expressing and silenced cells.
β-gal Assays.
The MUG assay was performed on bulk cellular lysates in 96-well plates on a Dynatech fluorimeter as previously described (16). The protein content of lysates was determined by the Bradford method, and this content was confirmed to lie in the linear portion of this assay before proceeding with the MUG assay. Fluorescence of each sample was measured in triplicate, and mean β-gal activity was determined following correction for protein content of the lysates. MUG assays were repeated at least three times, and mean activity and SEM were calculated. FACS-gal analysis was performed as previously described (15, 17).
RESULTS
Experimental Strategy.
To determine the mechanism by which an upstream activator increases reporter gene expression, we have studied the effect of stimulating an inducible promoter in a system that can independently assess the level and stability of expression (7). The level of expression is reflected in the level of the reporter gene product in a population of cells all of which are actively expressing. The stability of expression is reflected in the rate at which expression of the reporter is silenced, in cells within a population that, at a starting point, contains only expressing cells. β-geo is a fusion of β-gal and aminoglycoside phosphotransferase (neoR) and has activities of both proteins (18). We have used β-geo in conjunction with FACS-gal, an extremely sensitive flow cytometric method which detects β-gal expression in single cells (7, 15, 16, 17). Clones that stably express β-geo can be selected with G418; when maintained in G418 all cells express β-geo, and the level of β-geo expression in these clones can then be quantitated by conversion of MUG (6, 16). When cells are removed from G418, expression is no longer required, and silencing may occur; the percentage of expressing cells can be measured with FACS-gal.
We derived K562 erythroleukemia cell lines carrying a construct (kindly provided by R. Palmiter) in which expression of β-geo is driven by the mMT-I promoter, and we screened clones to identify those carrying single integrated copies of MT/β-geo (Fig. 1). These clones were analyzed in the same way as those in our earlier study of a metal-inducible enhancer derived from the mMT-I promoter (7). The effect of zinc stimulation on transcription rate was observed while cells were maintained in G418, to ensure that only expressing cells were analyzed. The effect of zinc on the silencing of transcription was studied by removing clones from G418 and splitting them into two aliquots, one maintained in standard medium and the other in medium supplemented with zinc. At intervals up to a year the proportion of expressing cells was measured with FACS-gal. Eight clones were analyzed in this way.
Basal and Inducible Expression of MT/β-geo.
The basal level of β-geo expression varied as much as 6-fold among clones carrying single copies (Fig. 2A). It is unlikely that this effect is due entirely to integration near endogenous enhancers, as even a powerful erythroid enhancer has at most a 2-fold effect on expression of a closely linked reporter (7). Zinc stimulation of the mMT-I promoter increased β-gal activity from 2- to 5.5-fold depending on the clone (Fig. 2A). This amount of induction is less than that reported with MT promoters using assays that did not separate effects on expression from effects on the number of expressing cells (12). In this system G418 selection ensures that all cells are expressing the reporter.
Figure 2.
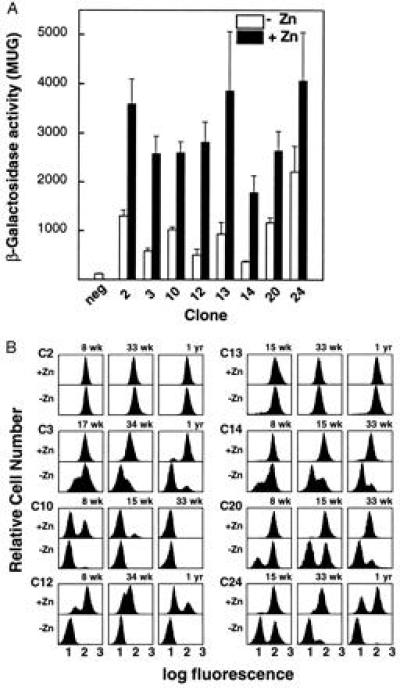
Effects of zinc stimulation on expression level and silencing of β-geo in K562 cell clones. (A) The basal and induced expression of β-geo in eight clones carrying single integrated copies of MT/β-geo; bars denote SEM. Results shown are means of at least three experiments, and are normalized to the negative control. There is a 5-fold variation of basal expression level between clones (open bars). Induced levels (solid bars) are 24 h after addition of 80 μM zinc sulfate. The basal and induced levels of β-gal in cells maintained free of G418 do not differ significantly from those shown, except in cases in which silenced cells exist in the assayed population; in those cases the levels are proportional to the percentage of expressing cells. (B) Silencing of β-geo expression in MT/β-geo clones is retarded by zinc stimulation. Cells were continuously expanded with (+Zn) or without (−Zn) 80 μM zinc sulfate in the culture medium. The x axis in these histograms represents a three-decade log scale of fluorescence, and the y axis represents the relative cell number. All clones were maintained in G418 until time 0 and then split into induced and uninduced aliquots, so that at time 0 all were expressing and the two populations were identical. There is a steady accumulation of silenced cells over time. Weeks and years are denoted as wk and yr, respectively. The histograms show progressive silencing over a period of up to 1 yr. Two clones (C2 and C13) show no silencing. The other clones show significant, but varying, proportions of silent cells, but in each case zinc stimulation slows or completely prevents silencing. In the case of clone 10, silencing in both aliquots was complete by 33 wk so that later time points are not shown. The uninduced (−Zn) aliquots of clones 14 and 20 were lost after 33 wk and so there is no 1-yr time point.
The influences of the integration site on expression level in these clones are comparable to stable position effects in Drosophila, in which all cells may express but expression level is influenced by the integration site (14). Expression from the mMT-I promoter is affected to approximately the same extent by the integration site and by metal induction of the promoter elements (Fig. 2A); the greatest difference in basal expression between clones was 6-fold, and the greatest induction (in clone 12) was nearly 6-fold.
Silencing of β-geo.
Expression of β-geo was silenced in six of the eight clones; at successive points after removal of G418 selection, increasing numbers of nonexpressing cells were found in the population (Fig. 2B). The silencing we observe appears similar to effects noted in a variety of systems, including classical position-effect variegation, telomeric silencing in yeast and trypanosomes, mating-type silencing in yeast, and silencing of transgenes in mice and cell lines (7, 13, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28). Silencing in our K562 clones occurs at a highly variable rate that is related to the site of integration; in some (clones 10 and 12) nearly all unstimulated cells were silenced at the first assay point, while in two (clones 2 and 13) expression was stable (without zinc stimulation) for a year following removal of G418 selection.
Significantly, stimulation with zinc strongly retarded silencing in all of the six clones in which silencing occurred (Fig. 2B). Clones that silenced more rapidly in the absence of zinc also silenced more rapidly in the presence of zinc. Once silenced, expression of β-geo was generally not responsive to stimulation with zinc. Clone 3 is an exception; expression in this clone is reactivated by zinc stimulation, although the mMT-I promoter in the silenced cells is methylated (not shown).
There is no relationship between the basal level of expression and the rate of silencing. For example, clone 10 is the most rapidly silenced, while clone 13, with a similar basal expression level, is not silenced at all; this demonstrates that transcription level alone does not determine stability of expression. However, although apparently independent of each other, rates of both transcription and silencing are very much affected by the integration site.
Linkage Between Silencing and Expression Level.
No relationship between β-geo expression level and rate of silencing is evident when clones are compared (Fig. 2). This suggests that transcription rate and silencing of transcription are independent manifestations of the interaction between the regulatory elements in the transgene and the site of integration. However, both are affected when the mMT-I promoter is stimulated. We thus asked if transcription and silencing rates are linked at a given integration site. The effect of increasing levels of zinc on transcription and silencing was analyzed in the two clones (clones 10 and 12) in which silencing was most rapid (Fig. 3). The effect of increasing levels of zinc on the level and stability of expression was analyzed in the two clones (clones 10 and 12) in which silencing was most rapid (Fig. 3). In both clones, the induction of increased β-geo expression (Fig. 3A) corresponds to a decrease in the rate of silencing (Fig. 3B) at all concentrations of zinc. For example, zinc had slight effects on level of expression and silencing at 20 μM, but greater effects on both at 60 μM (Fig. 3). Thus retardation of silencing and an increased level of transcription appear to be linked when the MREs are situated within a promoter.
Figure 3.
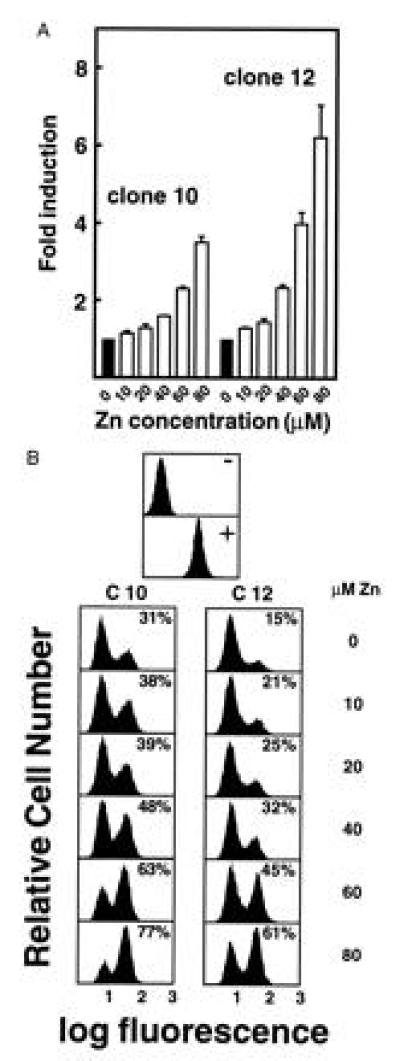
Titration of zinc shows parallel effects on level of expression and silencing within clones. (A) Clones 10 and 12 were induced for 24 h with increasing concentrations of zinc sulfate up to 80 μM. β-gal activity in each of the aliquots was then assayed by MUG conversion as in Fig. 2. (B) Clones 10 and 12 were removed from G418 selection and maintained in medium containing varying concentrations of zinc sulfate, up to 80 μM, as in A. FACS-gal was performed at weekly intervals; the histograms shown are derived from the 2-wk time point. Concentration of zinc is shown at the right. The percentage figures within the histogram boxes represent the proportion of cells in the positive peak. The effect of zinc on silencing parallels the effect on expression level in both clones.
DISCUSSION
We find that the action of MTF on the MREs of the mMT-I promoter has two measurable effects: retardation of silencing and an increase in the level of reporter gene expression. A comparison of individual clones containing single copies of the mMT-I/β-geo construct integrated at different sites demonstrates two types of position effect: a stable effect that influences the level of transcription and a silencing (variegating) effect that is independent of the transcription level. The lack of any apparent relationship between expression level and silencing rate when clones are compared suggests that transcription level and stability are independently influenced by the integration site. Each of the two position effects is modified by zinc stimulation of the promoter, and at a given integration site this modification is coordinate. Taken together with evidence that MTF does not increase transcription level from a distance, this leads us to the conclusion that MTF affects both transcription level and the assembly of repressive chromatin structures through a single mechanism which affects transcription rate only over short distances.
Other instances in which transcriptional activators may directly counteract the repressive effects of chromatin packaging have been noted, although these have not correlated promoter activity and silencing. In Drosophila, the establishment of clonally heritable states of repression in homeotic genes by members of the Polycomb group (Pc-G) is antagonized by transcriptional activators of the trithorax group (29, 30). Silencing of URA3 expression by the yeast telomere is counteracted by overexpression of Ppr1, a transcriptional activator of URA3 (31). We have also used the system described in this paper to show that deletion of an enhancer from an integrated construct has only a slight effect on the level of reporter expression, but drastically destabilizes expression, and that distant MREs powerfully retard silencing without any effect on transcription (7). Taken together, these findings suggest that an important function of a transcriptional activator, independent of the location of its binding site relative to the promoter, is to prevent the formation of repressive higher-order chromatin structures.
Much evidence has accumulated that transcriptional activators displace or reorder nucleosomes, and this has been interpreted as resulting in increased transcription. On naked DNA templates activator effects are rapidly attenuated with distance, suggesting that chromatin structure mediates long-range activation (32, 33, 34, 35, 36). Early work with the simian virus 40 enhancer also suggested that its effect on transcription rate is rapidly attenuated if it is moved away from the promoter (37). The findings presented here, taken together with work on distant enhancer elements (7), suggest that an activator may affect chromatin structure in such a way as to keep a transcription unit in an active state, without increasing the rate of transcription; this may be the primary function of activating elements. When MTF binds in the promoter, effects on both expression level and maintenance of the active state are seen; when bound at a distant site, only maintenance of the active state is affected. Proximity to the site of transcriptional initiation thus appears to determine whether MTF can regulate the level of transcription, and this may apply generally to transcriptional activators.
Genes in their endogenous contexts are flanked by elements that may ensure expression in appropriate lineages. The work discussed above suggests that the primary function of nonpromoter control elements is to ensure expression rather than regulate transcription rate; this view predicts that deletion of critical enhancer elements from an endogenous locus would result in variegated expression (although such elements might also affect the chromatin milieu that creates stable position effects on transcription rate). Loss of an upstream promoter element in the context of an intact locus may instead result in decreased transcription, as is known to be the case for several β-globin promoter mutations (38). Little information on the effects of endogenous enhancer deletions is currently available. Deletion of individual enhancer elements in the β-globin locus control region results in decreased β-globin expression (39, 40), but the assay used in these studies did not examine individual cells, so that variegation could not be assessed. Furthermore, erythroid cells undergo nuclear condensation during their terminal differentiation, and we have speculated that deletion of individual enhancers might result in premature cessation of transcription during terminal differentiation rather than the complete failure of gene expression that is typical of variegation (41). A more detailed understanding of these issues will require study of the effects of mutating transcriptional control elements in their native context, as well as the ability to distinguish expression in individual cells.
Retardation of silencing is not directly correlated with higher transcription level, as revealed by comparison of different integration sites, and of distant binding sites with proximal ones. The activity of MTF appears to affect both coordinately, but only when close to the promoter. How does increased binding of MTF to the upstream activator elements mediate both an increase in transcription level and an increase in stability? While effects on transcription and on the active state could be separable functions of this factor, we suggest that a single function related to modulation of chromatin structure could produce effects that differ with proximity to the initiation site. In each case the formation of repressive higher-order structures is antagonized, but in addition the change in structure near the initiation site may increase the efficiency of initiation or elongation of transcription.
Acknowledgments
We thank Richard Palmiter for providing the MTβ-geo construct, Ron Reeder, Mark Walters, and Emma Whitelaw for helpful discussions and support, and Jeff Eidemiller for help with the FACS assays. This work was supported by National Institutes of Health Grants 5RO1HL48356 and 5RO1DK44746 (to M.G.), 1F32HL08732 (to S.F.), and 5RO1HL48790 (to D.I.K.M.). D.I.K.M. was supported by the James S. McDonnell Foundation and is a Scholar of the Leukemia Society of America.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
Abbreviations: β-gal, β-galactosidase; FACS-gal, fluorescence-activated cell sorter–β-gal assay; MT, metallothionein; MRE, metal response element; MTF, MRE-binding transcription factor; MUG, 4-methylumbelliferyl β-d-galactoside; β-geo, β-gal/neoR.
References
- 1.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular Biology of the Cell. New York: Garland; 1994. [Google Scholar]
- 2.Treisman R, Maniatis T. Nature (London) 1985;315:72–75. doi: 10.1038/315072a0. [DOI] [PubMed] [Google Scholar]
- 3.Weber F, Schaffner W. Nature (London) 1985;315:75–77. doi: 10.1038/315075a0. [DOI] [PubMed] [Google Scholar]
- 4.Moreau P, Hen R, Wasylyk B, Everett R, Gaub M P, Chambon P. Nucleic Acids Res. 1981;9:6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon A M, Ley T J. Proc Natl Acad Sci USA. 1990;87:7693–7697. doi: 10.1073/pnas.87.19.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walters M C, Fiering S, Eidemiller J, Magis W, Groudine M, Martin D I K. Proc Natl Acad Sci USA. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walters M C, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin D I K. Genes Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- 8.Weintraub H. Proc Natl Acad Sci USA. 1988;85:5819–5823. doi: 10.1073/pnas.85.16.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radtke F, Heuchel R, Georgiev O, Hergersberg M, Gariglio M, Dembic Z, Schaffner W. EMBO J. 1993;12:1355–1362. doi: 10.1002/j.1460-2075.1993.tb05780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmiter R. Proc Natl Acad Sci USA. 1994;91:1219–1223. doi: 10.1073/pnas.91.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller P R, Salser S J, Wold B. Genes Dev. 1988;2:412–427. doi: 10.1101/gad.2.4.412. [DOI] [PubMed] [Google Scholar]
- 12.Stuart G W, Searle P F, Palmiter R D. Nature (London) 1985;317:828–831. doi: 10.1038/317828a0. [DOI] [PubMed] [Google Scholar]
- 13.Henikoff S. Curr Opin Genet Dev. 1992;2:907–912. doi: 10.1016/s0959-437x(05)80114-5. [DOI] [PubMed] [Google Scholar]
- 14.Lewis E B. Adv Genet. 1950;3:73–115. doi: 10.1016/s0065-2660(08)60083-8. [DOI] [PubMed] [Google Scholar]
- 15.Nolan G P, Fiering S, Nicolas F F, Herzenberg L A. Proc Natl Acad Sci USA. 1988;85:2603–2607. doi: 10.1073/pnas.85.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiering S, Northrop J P, Nolan G P, Mattila P S, Crabtree G R, Herzenberg L A. Genes Dev. 1990;4:1823–1834. doi: 10.1101/gad.4.10.1823. [DOI] [PubMed] [Google Scholar]
- 17.Fiering S N, Roederer M, Nolan G P, Micklem D R, Parks D R, Herzenberg L A. Cytometry. 1991;12:291–301. doi: 10.1002/cyto.990120402. [DOI] [PubMed] [Google Scholar]
- 18.Friedrich G, Soriano P. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 19.Allshire R C, Javerzat J-P, Redhead N J, Cranston G. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 20.Cattanach B M. Genet Res. 1974;23:291–306. doi: 10.1017/s0016672300014932. [DOI] [PubMed] [Google Scholar]
- 21.Fauvarque M-O, Dura J-M. Genes Dev. 1993;7:1508–1520. doi: 10.1101/gad.7.8.1508. [DOI] [PubMed] [Google Scholar]
- 22.Horn D, Cross G A M. Cell. 1995;83:555–561. doi: 10.1016/0092-8674(95)90095-0. [DOI] [PubMed] [Google Scholar]
- 23.Paro R. Trends Genet. 1995;118:295–297. doi: 10.1016/s0168-9525(00)89081-2. [DOI] [PubMed] [Google Scholar]
- 24.Renauld H, Aparicio O M, Zierath P D, Billington B L, Chablani S K, Gottschling D E. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 25.Rivier D H, Rine J. Curr Opin Genet Dev. 1992;2:286–292. doi: 10.1016/s0959-437x(05)80286-2. [DOI] [PubMed] [Google Scholar]
- 26.Rudenko G, Blundell P A, Dirks-Mulder A, Kieft R, Borst P. Cell. 1995;83:547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- 27.Davies R L, Fuhrer-Krusi S, Kucherlapati R S. Cell. 1982;31:521–529. doi: 10.1016/0092-8674(82)90308-7. [DOI] [PubMed] [Google Scholar]
- 28.Conklin K F, Groudine M. Mol Cell Biol. 1986;6:3999–4007. doi: 10.1128/mcb.6.11.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. Nature (London) 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 30.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 31.Aparicio O M, Gottschling D E. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 32.Tsukiyama T, Becker P B, Wu C. Nature (London) 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 33.Svaren J, Schmitz J, Horz W. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mymryk J S, Archer T K. Genes Dev. 1995;9:1366–1376. doi: 10.1101/gad.9.11.1366. [DOI] [PubMed] [Google Scholar]
- 35.Paranjape S M, Kamakaka R T, Kadonaga J T. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 36.Kornberg R D, Lorch Y. Annu Rev Cell Biol. 1992;8:563–587. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- 37.Wasylyk B, Wasylyk C, Chambon P. Nucleic Acids Res. 1984;12:5589–5608. doi: 10.1093/nar/12.14.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weatherall D J. In: The Molecular Basis of Blood Diseases. Stamatoyannopoulos G, Nienhuis A, Leder P, Majerus P, editors. Philadelphia: Saunders; 1994. pp. 157–206. [Google Scholar]
- 39.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I K, Enver T, Ley T, Groudine M. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 40.Hug B A, Wesselschmidt R L, Fiering S, Bender M A, Epner E, Groudine M, Ley T J. Mol Cell Biol. 1996;16:2906–2912. doi: 10.1128/mcb.16.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin D I K, Fiering S, Groudine M. Curr Opin Genet Dev. 1996;6:488–495. doi: 10.1016/s0959-437x(96)80072-4. [DOI] [PubMed] [Google Scholar]


