Abstract
Transformation-associated recombination (TAR) can be exploited in yeast to clone human DNAs. TAR cloning was previously accomplished using one or two telomere-containing vectors with a common human repeat(s) that could recombine with human DNA during transformation to generate yeast artificial chromosomes (YACs). On basis of the proposal that broken DNA ends are more recombinogenic than internal sequences, we have investigated if TAR cloning could be applied to the generation of circular YACs by using a single centromere vector containing various human repeats at opposite ends. Transformation with these vectors along with human DNA led to the efficient isolation of circular YACs with a mean size of ≈150 kb. The circular YACs are stable and they can be easily separated from yeast chromosomes or moved into bacterial cells if the TAR vector contains an Escherichia coli F-factor cassette. More importantly, circular TAR cloning enabled the selective isolation of human DNAs from monochromosomal human–rodent hybrid cell lines. Although <2% of the DNA in the hybrid cells was human, as much as 80% of transformants had human DNA YACs when a TAR cloning vector contained Alu repeats. The level of enrichment of human DNA was nearly 3000-fold. A comparable level of enrichment was demonstrated with DNA isolated from a radiation hybrid cell line containing only 5 Mb of human DNA. A high selectivity of human DNA cloning was also observed for linear TAR cloning with two telomere vectors. No human–rodent chimeras were detected among YACs generated by TAR cloning. The results with a circular TAR cloning vector or two vectors differed from results with a single-telomere vector in that the latter often resulted in a series of terminal deletions in linear YACs. This could provide a means for physical mapping of cloned material.
Keywords: selective cloning, human chromosomes
A critical step in the characterization of large genomes, including the genome of humans, has been the cloning of large chromosomal fragments. This has been fulfilled through the development of artificial chromosomes in the yeast Saccharomyces cerevisiae (YACs) and has led to the large-scale physical map of the human genome derived from several YAC libraries (1, 2). The recombinational properties of yeast and the opportunities to manipulate YACs (3) provide important additional benefits for characterizing genomes. Recently we developed an alternative approach for cloning human DNA in yeast as large linear YACs that omits the in vitro ligation step (4). The approach is based on transformation-associated recombination (TAR) between a repeat within transformed human DNA fragments (such as an Alu or LINE) and a human repeat sequence on a cotransformed linearized plasmid that also contains a yeast centromere and a telomere.
It is apparent that the ability to isolate specific chromosomal regions would greatly benefit positional cloning and studies of various human diseases as well as fill the gaps in existing maps. Many chromosome-specific libraries have been generated by cloning directly from monochromosomal human–rodent hybrid lines, with the clones being screened for human inserts by hybridization (5).
On the basis of our proposal that repeats at broken ends might be preferred sites of recombination (4), we have exploited TAR cloning to directly isolate human DNA from monochromosomal hybrid cells as large circular YACs. The circular TAR cloning system was also applied to the rapid isolation of human DNAs from radiation hybrids containing only a small fragment of a human chromosome. The circular YACs greatly facilitate subsequent physical isolation and analysis of the cloned material.
MATERIALS AND METHODS
TAR Cloning Vectors.
Centromeric and acentric vectors pVC1 (Alu-CEN6-HIS3-TEL), and pVL27 (Alu-URA3-TEL) used for generation of linear YACs have been described earlier (4). New TAR circularizing vectors containing two Alus or Alu plus a medium reiteration (MER) frequency human repetitive sequence or a LINE sequence were constructed (Fig. 1A) from the vector pVCX0 (Alu-CEN6-HIS3-TEL). pVCX0 was obtained from pVC1 by replacement of a 297-bp BamHI fragment containing BLUR13 Alu sequence with a 320-bp BamHI fragment containing an Alu consensus sequence (6). Another targeting sequence (Alu, LINE, or MER) was included in pVCX0 by replacement of a 0.4-kb EcoRV telomere-containing fragment (TEL). Three constructed vectors, pVC39-AAH2, pVC39-AAH4, and pVC39-AAT3, contain Alu targeting sequences (i.e., an Alu consensus at one end and a Eco-BLUR13.R1 Alu sequence at another end). Eco-BLUR13.R1 differs from BLUR13.R1 in that the order of two Alu halves is reversed. Reversal arose when the Alu family repeat was isolated as an EcoRI fragment from a tandem duplication of a BamHI BLUR13.R1 fragment (7). In pVC39-AAT3 (Alu-CEN6-TRP1-Alu) and pVC39-AAH2 (Alu-CEN6-HIS3-Alu) the targeting sequences were cloned in inverted orientation. pVC39-AAT3 is marked by TRP1 and contains a tandem repeat of Eco-BLUR13.R1. The vector was cut with PstI (the site is located in the polylinker) before transformation to yield a molecule bounded by complete Alu sequences. pVC39-AAH2 is marked by HIS3 and contains one copy of Eco-BLUR13.R1. pVC39-AAH2 was cut with SmaI before transformation. Since a SmaI site is present in both targeting sequences, the linearized form of pVC39-AAH2 contains 207 bp from 320 bp of the Alu consensus at one end and 52 bp from 297 bp of BLUR13.R1 Alu sequence at the other end. In the linearized vector, 45 bp of the 52 bp of BLUR13.R1 are identical to the terminal region of the Alu consensus. It is worth noting that both targeting sequences in pVC39-AAH2 lack an 82-bp fragment of 3′ Alu repeat. This fragment was used as a specific probe for detection of human YACs generated by pVC39-AAH2. pNKBAC39 is a derivative of pVC39-AAH2 containing pBeloBAC11 (8). The plasmid was cut by SalI before transformation. pVC39-AAH4 contains Alu consensus and BLUR13.R1 sequences cloned in direct orientation. Linearization of pVC39-AAH4 with SmaI before transformation was accompanied by a partial deletion of Alu repeats. The linearized form of the vector contains 207 bp of Alu consensus at one end and 246 bp of Eco-BLUR13.R1 at the other end. pVC39-MAH8 (MER8-CEN6-HIS3-Alu) and pVC39-MAH10 (MER10-CEN6-HIS3-Alu) contain MER8 and MER10 human repeats (9) and an Alu consensus as targeting sequences. pVC39-MAH8 and pVC39-MAH10 were cut with EcoRI (the site is located in the polylinker) before transformation to yield molecules bounded by complete Alu and MER sequences. pVC39-LAH2 (LINE1-CEN6-HIS3-Alu) contains a human repeat LINE1.1 from pVC3 (4) at one end and an Alu consensus repeat at the other end. The plasmid was cut by SmaI before transformation. All the plasmids were purified by CsCl/ethidium bromide (EtdBr) centrifugation for TAR cloning experiments.
Figure 1.
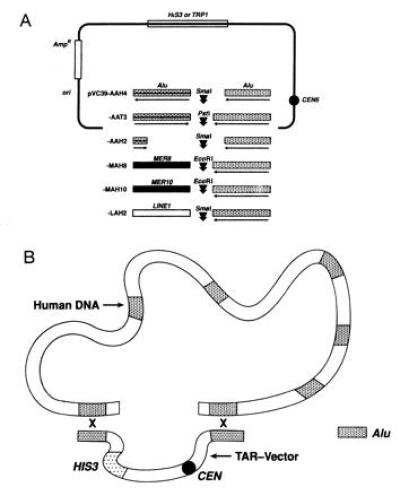
Isolation of human DNAs as circular YACs by using the TAR cloning method. (A) TAR cloning vectors. Presented are pVC39-AAH4, -AAT3, -AAH2, -MAH8, -MAH10, and -LAH2 vectors containing Alu, LINE, or MER sequences. CEN6, centromere; AmpR, ampicillin-resistance gene. (B) The TAR cloning scheme. Yeast spheroplasts are transformed with human DNA along with a TAR cloning vector. Recombination between repeats in the vector and human DNA leads to the establishment of a circular YAC. The filled-in blocks in human DNA and vectors identify repeated Alu, LINE, or MER sequences.
Yeast Strains and Mammalian Cell Lines.
S. cerevisiae strain YPH857 with the HIS3 gene deleted (MATα, his3-Δ200, trp1-Δ1, ura3-52, leu2-Δ1, lys2-801, ade2-101) was used (10). Human (HL-60), mouse (NIH 3T3), hamster (CHOK1), and chromosome 22-containing hamster–human hybrid (GM10888) cells were obtained from the University of North Carolina Tissue Culture Facility. Mouse–human monochromosomal somatic cell hybrid CY18 with human chromosome 16 was provided by L. Deaven (Los Alamos National Laboratory). Mouse–human monochromosomal somatic cell hybrid GM10926D with human chromosome 10 was provided by M. Cancilla (The Murdoch Institute for Research into Birth Defects, Melbourne, Australia). Radiation hybrid hamster–human cell line D2-X-38 that contained an approximately 5-Mb fragment from chromosome 2 with the XRCC5 (Ku80) DNA repair gene (11) was obtained from P. Jeggo (University of Sussex, Sussex, U.K.). Yeast cells were grown on complete medium (YEPD) or synthetic standard selective media (14).
Yeast and Escherichia coli Transformation.
A protocol for spheroplasting cells was used that results in efficient transformation (4). Agarose plugs (100 μl) containing approximately 5 μg of gently prepared DNAs from monochromosomal cell lines were used for transformation (4). Linearized vector(s) (1 μg) was added to DNA-containing plugs before treating with agarase, and the vector/DNA mixture was presented to spheroplasts. DH10B cells (GIBCO/BRL) were used for transfer of circular YACs into E. coli. A Bio-Rad Gene Pulser was used for electroporation with the setting 2.5 kV, 200 Ω, and 25 μF. Yeast DNA for YAC transfer was prepared by standard procedure. Colonies were selected on plates containing chloramphenicol at 12.5 μg/ml.
Identification and Characterization of YAC Clones.
Chromosomal size DNAs from transformants were separated by transverse alternating field electrophoresis (TAFE), blotted, and hybridized with human and/or rodent DNA as previously described (4, 12). Human, hamster, and mouse probes were labeled by random priming. To estimate the size of circular YACs, agarose DNA plugs were exposed to a low dose of γ-rays [30 krad (300 Gy)] before TAFE analysis.
A specific Alu probe for detection of human YACs generated by the TAR circularizing vector pVC39-AAH2 was developed. This probe is the 82-bp fragment of 3′ Alu consensus sequence that is omitted in the linearized pVC39-AAH2 as described above. Two primers were used to amplify this fragment from pPD39 plasmid containing Alu consensus sequence (6): 5′-CCCGGGAGGCGGAGCTTGCAGTGA-3′ and 5′-TTTGAGACGGAGTCTCGCTCTGTCGCCCAG-3′. The Alu probe was labeled with [32P]dCTP during PCR. Alu profiles of YACs were done as previously described (4).
Genetic analysis of YAC mitotic stability was described previously (4).
RESULTS
Rationale for Generation of Circular YACs.
The TAR cloning method is based on recombination between human DNA fragments transformed into yeast and cotransformed plasmids containing a repeat commonly represented in human DNA. On the basis of efficiency of recombination during transformation between diverged DNAs and the likelihood that repeats near the ends of DNA fragments might be more likely to undergo recombination compared with internal repeats (4), we reasoned that TAR could also be applied to the generation of circular YACs. In the proposed scheme (Fig. 1B), a linear plasmid is generated that contains commonly occurring repeats, such as Alu or LINE, at each end, a yeast selectable marker, and a centromere. Recombination between the plasmid and homologous or diverged repeats in cotransformed human DNA can result in circular YAC molecules. As proposed for linear TAR cloning (4), the capability for replication is provided by one of the frequent sequences present in human DNA that can function as replication origins (ARS) in yeast.
TAR Vectors Containing Human Repeats Can Generate Circular YACs.
The generation of circular YACs by TAR cloning was investigated in a mixture of human DNA and vectors with terminal Alu sequences. Total human DNA that was gently isolated by lysing cells in low-melting agarose was combined (after melting the agarose) with a linearized Alu-TRP1-CEN6-Alu plasmid pVC39-AAT3 and presented to yeast spheroplasts. The full-size Alus were in inverted orientation. Between 300 and 1000 tryptophan-independent (Trp+) transformants were typically obtained when a mixture containing 1 μg of plasmid and 5 μg of human DNA was used (Table 1). Since fewer than 10 transformants were obtained when only plasmid DNA was presented to spheroplasts, the high levels of transformation were due to interactions between the human DNAs and the plasmid molecules. These frequencies were comparable to the frequency observed when a single Alu-containing TAR vector, pVC1, that generates linear YACs (Table 1) (see also ref. 4) was used. High frequencies of transformation were also obtained when a plasmid, pVC39-AAH2, containing parts of Alu sequences was used (Table 1). This TAR-circularizing vector had a 207-bp 5′-truncated Alu fragment at one end and a 52-bp Alu internal sequence at the other end in opposite orientation; the Alus were 10% diverged and shared a common region of 45 bp (see Materials and Methods). No or few transformants were obtained with the pVC39-AAH4 vector, which contained Alu repeats in direct orientation (Table 1); presumably, intraplasmid recombination between targeting sequences competes with recombinational interaction with human DNA.
Table 1.
Efficiency of transformation by Alu-containing TAR vectors when various DNAs are included
| TAR vector* | Alu orientation | Mammalian DNA | No. of His+ or Trp+ transformants† |
|---|---|---|---|
| pVC39-AAT3 | → ← | None | 1 –3 |
| -AAH2 | → ← | None | 0 –7 |
| -AAH4 | → → | None | 0 –7 |
| -AAT3 | → ← | Human | 300 –1000 |
| -AAH2 | → ← | Human | 400 –1000 |
| -AAH4 | → → | Human | 5 –20 |
| -AAH2 | → ← | Hamster | 3 –10 |
| -AAH2 | → ← | Mouse | 1 –20 |
| -LAH2 | LINE ← | Human | 200 –500 |
| -MAH8 | MER ← | Human | 50 –100 |
| -MAH10 | MER ← | Human | 100 –200 |
| pVC1 | TEL-Alu | Human | 300 –1000 |
| pNKBAC39 | → ← | Human | 200 –500 |
Yeast spheroplasts were transformed with 1 μg of linearized plasmids along with 5 μg of human or rodent DNA.
Three to 20 independent transformations were carried out for each condition.
The presence and nature of YACs generated by the circularizing vectors were determined. Chromosomal-size DNA of the Trp+ or His+ transformants obtained with pVC39-AAT3 or pVC39-AAH2 vectors was analyzed by TAFE in a gel. Large circular DNA molecules are expected to be retained in the starting wells under the TAFE conditions employed. Among 100 transformants analyzed (60 obtained with pVC39-AAH2 and 40 obtained with pVC39-AAT3 vectors), no new chromosome bands were detected by EtdBr staining. However, when the gels were probed with a radioactively labeled human DNA (under conditions in which hybridization to Alus was prevented) strong signals were located at the positions of the starting wells for 93 of 100 clones, which were presumed to be circular DNAs (Fig. 2). In addition there was usually a faint single band characteristic of each of the 93 transformants. A second set of plugs was, therefore, exposed to a low dose of radiation to produce breaks in the molecules. If YAC molecules were circular, radiation-induced breaks would result in the material appearing within the gel, with those molecules having only a single break resulting in strong bands (discussed in ref. 13). Multiple breaks would result in a smear. As shown in Fig. 2, irradiation of the plugs resulted in broad bands of material that hybridized with the human probe. The upper position of broad bands corresponded to the position of the faint single bands found with unirradiated DNA. We conclude that nearly all the YACs were circular and that the single faint band obtained with unirradiated DNA corresponded to broken molecules arising during DNA isolation. These results also suggest that each transformant colony typically had only one YAC (see below). Over 50% of YACs were greater than 150 kb. (Specific analysis of size distribution for chromosome 16 YACs is presented below.) While the size of the YACs is smaller than reported previously for linear TAR cloning (4), we note that the system has not been optimized for the development of large circular YACs.
Figure 2.
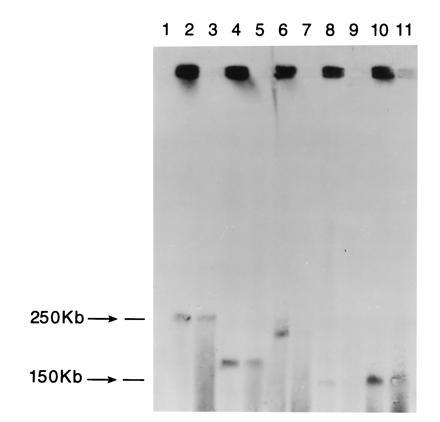
Physical characterization of five randomly selected circular human YACs developed by TAR cloning from total human DNA. Chromosomal-size DNA was separated by TAFE in a gel and blot-hybridized with a human DNA probe. Strong signals located at the positions of the starting wells correspond to circular YACs (lanes 2, 4, 6, 8, and 10). A band corresponding to linear molecules was also detected. Irradiation of the plugs with human YACs resulted in appearance of the bands corresponding to linear forms of the YACs (lanes 3, 5, 7, 9, and 11).
Among 100 transformants 7 contained new chromosome bands corresponding to linear human YACs with sizes between 120 and 350 kb. Since they were mitotically unstable, the bands could not be detected by EtdBr staining but could be visualized by blot-hybridization.
We also investigated TAR circular cloning after introducing into pVC39-AAH2 the E. coli F-factor-based cassette from a bacterial artificial chromosome (BAC) vector. As shown in Table 1, the shuttle pNKBAC39 vector generates circular human YACs in yeast. These YACs can subsequently be propagated in E. coli as BACs. Total DNA was gently isolated from three transformants carrying circular YACs (≈120 kb each) and used for electroporation of E. coli cells. Large circular DNAs were isolated by the standard alkaline method from five E. coli chloramphenicol-resistant transformants obtained with each YAC. The size of the circular YACs obtained from all the E. coli transformants corresponded to the original circular YACs. Moreover, the restriction patterns for the individual transformants were identical for a given YAC (data not shown).
The stability of the large circular YACs containing human DNA was investigated, since (i) linear human YACs are known to be less stable than natural yeast chromosomes and (ii) the circular YACs might undergo sister chromatid exchange leading to dicentrics. In most (110/120) primary transformant colonies obtained with circularizing vectors pVC39-AAH2 and pVC39-AAT3, the YACs were stable. Less than 10% of cells in a colony grown without selection for the YAC lacked the YAC marker, similar to what was observed with linear YACs generated either by a standard ligation method or by linear TAR cloning with two telomere-containing vectors (4, 14). The YACs also appeared to be structurally stable in that changes in size were generally not seen. For example, in most transformants only one faint band corresponding to a linear form of the YAC was observed for unirradiated DNA, arguing against the presence of several YACs in the transformant or the development of various-sized YACs during propagation. The structural stability of circular YACs from five primary transformant colonies was examined further. Cells were inoculated into YEPD medium and grown for 20 generations. The cells were plated and the sizes of YACs in five subclones of each transformant were determined after exposure to γ-rays. No changes in size from the original ≈150- to 200-kb YACs were observed for subclones of four transformants. For one of the transformants 40-kb and 80-kb deletions were detected in two of the five subclones analyzed. While these results demonstrate that human DNA cloned in circular YACs may undergo rearrangements during mitotic propagation, the frequency of rearrangements is comparable to that observed for linear YACs (14, 15).
We also investigated the use of other classes of repeat elements, the LINE and the MER moderate repeats, which are less than 1/10th as frequent as Alus. One end of the circularizing TAR vectors contained a MER or LINE element and the other end had an Alu consensus. As shown in Table 1, both MER8 and MER10 and LINE1 led to an increase in numbers of TAR-derived transformants as compared with the control. The lower efficiency of transformation as compared with Alu-containing vectors presumably reflects the reduced frequency of these repeats in the genome. Among 60 transformants analyzed (20 for each vector) all contained human DNA as circular YACs. The sizes were comparable to those obtained with vectors having only Alus (data not shown).
We conclude that TAR cloning can be applied to the generation of large, stable circular YACs from human genomic DNA. Once generated, the circular YACs can be easily separated from yeast chromosomes by TAFE or can be transferred into E. coli for further analysis.
Selective Cloning of Human DNA with a TAR-Circularizing Vector from Rodent–Human Monochromosomal Hybrid Cells.
We investigated the use of circular TAR cloning for the specific isolation of human DNA. Total genomic DNA from the hybrid line CY18 containing human chromosome 16 was presented to yeast spheroplasts along with linearized vector pVC39-AAH2 (Alu-CEN6-HIS3-Alu). The efficiency of transformation was 100–200 colonies per 5 μg of genomic DNA (Table 2). Transformation with mouse DNA plus vector yielded only a few transformants (Table 1). Among 200 His+ transformants (from three independent transformations), nearly 80% (161/200) contained human DNA, as identified by hybridization of chromosome-size DNA in TAFE gels with an 82-bp Alu probe. Since the probe had no homology to the linearized pVC39-AAH2 (see Materials and Methods), it was diagnostic for human DNA. Among 39 transformants that lacked human DNA, 3 had mouse DNA. Since chromosome 16, which is 100 Mb, represents only 1.5% of the total cellular DNA, circular TAR cloning can provide a highly efficient means for isolating human DNA from a hybrid cell line. The enrichment of human DNA relative to mouse DNA is greater than 3000-fold, an estimate based on the isolation of 161 human YAC clones versus 3 mouse YAC clones and the fraction of human DNA in the hybrid. Most of the clones analyzed by TAFE (154/161) contained circular YACs that were retained in the wells. The size distribution of YACs was determined after exposure to γ-rays. As shown in Fig. 3 most of the YACs were between 100 and 200 kb; 18% were greater than 200 kb. The Alu profiles of 40 randomly selected YACs (size range from 70 to 300 kb) indicated that there were no identical clones (data not shown).
Table 2.
Specific isolation of human DNAs by TAR cloning from rodent–human monochromosomal hybrid cell lines
| Hybrid cell line | TAR vector | No. of His+ or His+ Ura+ transformants | % clones with human DNA |
|---|---|---|---|
| Circular | |||
| Mouse–human 16 | pVC39-AAH2 (Alu-ulA)* | 100–200 | 80 (161/200) |
| Mouse–human 10 | pVC39-AAH2 (Alu-ulA) | 130–180 | 76 (74/98) |
| Hamster–human 22 | pVC39-AAH2 (Alu-ulA) | 90–170 | 85 (51/60) |
| Linear | |||
| Hamster–human 22 | pVC1 (TEL-Alu) | 20–70 | 25 (24/98) |
| Hamster–human 22 | pVC1 + pVL27 (TEL-Alu) + (Alu-TEL) | 85–190† | 55 (55/100) |
Alu-ulA indicates that the repeats are in opposite orientation.
Approximately half of the His+ transformants contained the unselectable URA3 marker, which in all cases (152 examined) was linked to HIS3.
Figure 3.
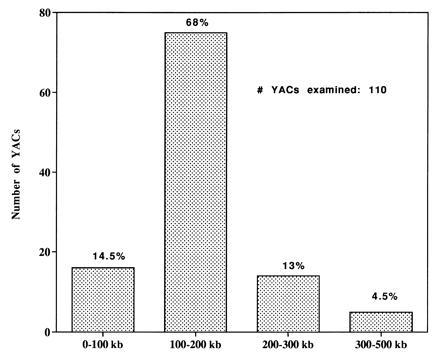
Size distribution of circular YACs generated by the TAR vector pVC39-AAH2 from a human hybrid cell line containing chromosome 16. Presented are the results for 110 YACs.
Comparable transformation efficiencies and frequencies of YACs containing human DNA were found for two other hybrid cell lines containing human chromosome 10 or 22 (Table 2). On the basis of TAFE analyses of unirradiated and irradiated DNAs nearly 80% of the transformants for each hybrid cell line (125/158) contained circular YACs with sizes from 70 kb to 350 kb. The Alu profiles of 20 YACs generated from each hybrid line were determined. No identical patterns were observed (data not shown). Among 33 transformants analyzed that lacked human DNA, none had rodent DNA. These transformants presumably arose by illegitimate recombination between vector and yeast chromosome(s). Selective cloning of human DNA was also observed with the F-factor-based pNKBAC39 vector (data not shown). We conclude that circular TAR cloning vectors containing Alu sequences are highly selective for isolation of human DNA from hybrid cell lines.
Selective TAR Cloning of Human DNA from a Radiation Hybrid Line Containing a Small Fragment of Human DNA.
The above results suggested that the TAR cloning approach could be applied to isolation of DNAs from small fragments of chromosomal DNA. We, therefore, examined the ability of TAR cloning to isolate human DNA from a radiation hybrid containing a 5-Mb region of chromosome 2 that includes the Ku80 gene required for double-strand break and VDJ recombination (11). Among 113 isolates obtained with pVC39-AAH2, there were 20 containing human DNA and 5 with hamster DNA. Among the 20 human YACs, 15 were circular with sizes between 70 kb and 200 kb. In the 5 remaining transformants the YACs were small (about 70 kb) and linear. Since the Alu profiles of the YACs were different, it appears that the cloned regions correspond to different regions of the human chromosomal fragment (Fig. 4). These results indicate that there was a nearly 5000-fold enrichment of human DNA by the circular TAR cloning.
Figure 4.
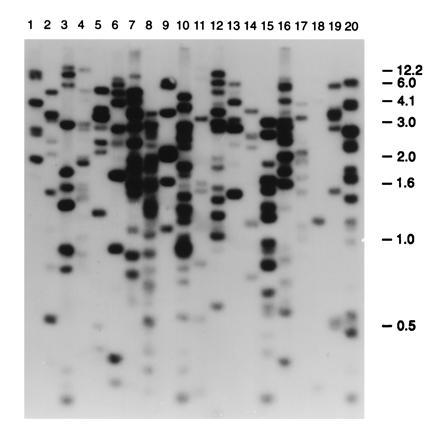
Alu profiles of 20 YACs generated by the TAR cloning method from hamster radiation hybrid cells containing a 5-Mb human DNA fragment. The profiles were produced by hybridization of an 82-bp Alu probe with TaqI-digested DNA isolated from the transformants. Size markers are kb.
Selective Linear TAR Cloning of Human DNA from a Hybrid Cell Line.
We compared linear and circular TAR cloning in terms of efficiency and the nature of the cloned material. The combination of vectors Alu-CEN6-HIS3-TEL (pVC1) and Alu-URA3-TEL (pVL27) containing full-size Alus led to the selective cloning of human DNA from a hamster–human chromosome 22 hybrid line (Table 2). Among His+ Ura+ transformants 55% (55/100) contained linear human YACs (on the basis of hybridization with human DNA). The size of the YACs varied from 70 kb to >600 kb. More than 50% of the YACs were >200 kb. Since only 3% of transformants had hamster DNA, there was a nearly 1100-fold enrichment of human DNA. Thus, linear TAR cloning with two vectors containing Alu targeting sequences also provides a means for the selective isolation of human DNA as YACs from hybrid cell lines.
We also examined linear TAR cloning of human DNA from a hybrid cell line using a single, telomere-containing Alu-CEN6-HIS3-TEL vector, pVC1, to compare the present results with those obtained previously (4). Typically, from 20 to 100 His+ transformants were obtained with the vector (Table 2). On the basis of about 25% of transformants having human DNA and 4% having mouse DNA, there was a nearly 400-fold enrichment of human DNA among the mammalian DNAs isolated.
Unlike the circular YACs, linear YACs in the primary transformants obtained with the pVC1 vector were lost with a high frequency. However, they were mitotically stable in the subclones of the transformants (4). The YACs in the subclones were different sizes. However, the Alu profiles found in the smallest YAC were present in the YACs of increasing size (data not shown). On the basis of this and restriction analysis, we conclude that the size differences in YACs in the subclones was the result of a progenitor YAC that was degraded prior to establishment of a telomere. Thus, the pVC1 vector provides the opportunity to generate a set of YACs with terminal deletions that can be isolated by simply restreaking the transformant colony.
Human–Rodent Chimeras Are Not Observed During TAR Cloning.
Previously we demonstrated that chimeric human–mouse YACs are not generated during TAR cloning with a single-telomere TAR vector (4). We examined 80 circular human YACs generated from the chromosome 16 hybrid cell line for the presence of mouse DNA. Chromosome-size DNAs from these clones were separated by TAFE and hybridized with a mouse DNA probe to identify human–mouse chimeras. There were no YACs that hybridized to a mouse probe. Similarly, among 38 circular YACs containing chromosome 22 DNA, none hybridized to hamster DNA. We also examined linear human YACs and found no chimeras among 24 generated with one- and 55 generated with two-telomere-containing vectors (data not shown). Thus we conclude that few, if any, chimeras are developed during circular or linear TAR cloning of human DNA from hybrid cell lines, suggesting that recombination between the two DNAs is excluded because of the lack of homology.
DISCUSSION
The yeast S. cerevisiae is highly efficient at recombining broken homologous or highly diverged DNA molecules through a RAD52-dependent pathway. The high efficiency also extends to intermolecular recombination during yeast transformation (16, 17, 18). In our TAR cloning approach, we have exploited the frequent copenetration of large and/or small molecules along with efficient recombination to isolate human DNAs as large size YACs. When a circularizing plasmid containing a commonly occurring human repeat at each end (i.e., Alu-CEN-Marker-Alu) along with human DNA was used to transform yeast spheroplasts, there was a high yield of large circular YACs; nearly half were greater than 150 kb. Since the average distance between Alu sequences is about 3 kb, the TAR cloning of human DNA fragments appears to occur by nonrandom recombination. These results support our previous proposal that recombinational interactions between the repeats of the plasmid(s) occur preferentially with repeats near the ends of the human cotransformed DNA (4). Surprisingly, there was efficient cloning when the cloning vector contained only a 52-bp Alu fragment, suggesting that the region of homology required for TAR cloning is small.
The application of TAR cloning to the development of circular YACs has provided several new utilities in the cloning of human DNAs or the DNAs of any organism in yeast. Most prominent is the ease of isolation of YACs from total yeast DNA by using pulsed-field gel electrophoresis, since the circular molecules are trapped in the starting well. Circular YACs are more resistant to shear than linear YACs and can be easily manipulated for further characterization of the cloned material. Furthermore, the presence of an F-factor origin in a TAR vector provides the opportunity for transfer of a YAC to E. coli. Thus, TAR cloning using BAC type vectors provides many of the genetic utilities of yeast along with the capability for rapid isolation from E. coli.
Although there have been several reports on the cloning of DNA as large circular molecules in yeast (19, 20), their stability has not been well characterized. We have established that TAR-cloned circular YACs exhibit structural and segregational stabilities that are comparable to those observed for linear YACs. Moreover the structural stability of circular YACs can be further enhanced in a recombination-repair-deficient rad52 strain similar to that for linear YACs (unpublished data).
An important utility of TAR cloning is the opportunity to specifically isolate human DNA from rodent–human hybrid cell lines. Previous approaches involved either cloning of DNA from physically separated specific chromosomes or the random cloning of DNA from hybrid cell lines and identification of human clones (5, 21). Both methods are very laborious. Previously we showed that TAR cloning with a single-telomere-containing vector led to a 400-fold enrichment of human DNA among the linear mammalian YACs that were isolated (4). In the present experiments using TAR-circularizing vectors with Alus at each end, the enrichment was nearly 3000-fold. The selectivity extended even to small human chromosome fragments, since we were able to demonstrate that there was a comparable level of enrichment when circular TAR cloning was used for the isolation of human DNA from a 5-Mb fragment in a radiation hybrid.
Selective isolation of human DNA from mouse–human hybrid cells was also observed with two telomere-containing vectors. In this case up to 55% of the transformants contained human YACs and the enrichment of human to mouse DNA among the YACs was approximately 1000-fold. The YACs size varied from 70 kb to more than 600 kb. The difference in levels of human DNA enrichment observed with circular and linear TAR vectors systems may be due to circular TAR cloning being essentially a bimolecular reaction, whereas for linear TAR cloning the reaction is trimolecular, involving two telomere vectors plus the human DNA. Once an initial recombinant is formed with the circular TAR cloning vector, the second recombination event with the other target sequence on the vector would seem more likely.
For the case of TAR cloning with two vectors, the YACs in primary transformants were stable, as was also found for circular TAR cloning. However, for TAR cloning with a single vector, the sizes of YACs were often different in subclones of the primary transformant. On the basis of physical analysis of YACs in the subclones, we propose that the degradation occurs from the end that initially lacked a telomere. Eventually the YACs are stabilized, presumably by telomere formation at various yeast telomere-like sequences. These results suggest that a human YAC DNA with only one protected end can pass through several generations before healing of the broken end. While the nature of the stabilizing sequences has not been established, a wide spectrum of YAC derivatives observed for each transformant suggests that such sequences are frequent in human DNA. We suggest that this feature of isolating human DNAs with a single TAR vector can be useful for physical mapping of the cloned material.
No rodent–human chimeras were observed during TAR cloning of human DNA from monochromosomal hybrid cell lines. It seems that the lack of homology between human and mouse (hamster) DNAs prevents recombinational interactions. The lack of chimeras is consistent with the previous conclusion that recombination is an important source of chimeras (15, 22). Human–human chimeras are unlikely during TAR cloning of DNA from monochromosomal hybrid cells because of the low probability of copenetration of more than one human DNA molecule from a transformation mixture in which the concentration of human DNA is low.
Although our results demonstrate that the TAR method is highly efficient in the selective cloning of human DNA from hybrid cell lines as either circular or linear YACs, conditions remain to be optimized for the cloning of larger human fragments. At present most of the YACs cloned are in the size range of 100–200 kb. For example, size selection of molecules prior to transformation might increase the likelihood of obtaining larger YACs. The size distribution of TAR-cloned YACs could depend on the distribution of repeats in chromosomal DNA. For example, use of moderately repeated sequences such as MERs (9) in circularizing TAR vectors or pairs of telomere-containing vectors could result in the isolation of larger YACs. We note that many MER sequences are not present in the rodent genome. Thus, TAR vectors containing MERs can provide a great selectivity in the cloning of human DNA from human–rodent hybrid cell lines.
It remains to be established whether the TAR cloned fragments are representative of the total DNA being examined. Although the overall density of Alu sequences in chromosomes should be high enough to generate random clones by Alu-containing TAR vectors, there may be limitations, such as Alu clustering and the likelihood that a cloned fragment contains a yeast ARS-like sequence. Possibly a set of TAR vectors containing various repeats would ensure that most regions of the genome are isolated. While ARS-like sequences are frequent (nearly 1 per 20 kb in the human genome (ref. 23 and references in ref. 4), there could be long regions lacking such elements, thereby limiting the isolation of a small fraction of the genome.
In conclusion, TAR cloning greatly expands the usefulness of YACs in that it provides the possibility for direct cloning of DNA fragments through recombination. It provides opportunity for the simple isolation of specific chromosome sequences and it is likely to lead to the isolation of gene families, and possibly single-copy genes. To this end, we have been able to selectively isolate ribosomal DNA repeats from total human DNA (unpublished results).
Acknowledgments
We thank N. Nikolaishvili for help in experiments with the Ku radiation hybrid cell line. P. Jeggo’s contribution in providing DNA from the D2-X-38 radiation hybrid cells, which were pretested for radiation resistance, is greatly appreciated. Support was provided in part by an interagency grant (1-YO2-HG-60021-01) from the National Institutes of Health Human Genome Center.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
Abbreviations: YAC, yeast artificial chromosome; BAC, bacterial artificial chromosome; TAR, transformation-associated recombination; EtdBr, ethidium bromide; TAFE, transverse alternating field electrophoresis.
References
- 1.Burke D T, Carle G F, Olson M V. Science. 1987;236:806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- 2.Guyer M S, Collins F S. Proc Natl Acad Sci USA. 1995;92:10841–10848. doi: 10.1073/pnas.92.24.10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reeves R H, Pavan W J, Hieter P. Methods Enzymol. 1992;216:584–603. doi: 10.1016/0076-6879(92)16051-k. [DOI] [PubMed] [Google Scholar]
- 4.Larionov V, Kouprina N, Graves J, Chen X-N, Korenberg J, Resnick M A. Proc Natl Acad Sci USA. 1996;93:491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gingrich J C, Lowry S R, Kuo W L, Gray J, Smith C L, Cantor C R. Genomics. 1993;15:228–230. doi: 10.1006/geno.1993.1043. [DOI] [PubMed] [Google Scholar]
- 6.Batzer M A, Alegria-Hartman M, Deininger P L. Genet Anal Tech Appl. 1994;11:34–38. doi: 10.1016/1050-3862(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 7.Pavan W J, Hieter P, Reeves R H. Mol Cell Biol. 1990;10:4163–4169. doi: 10.1128/mcb.10.8.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri Y, Simon M. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jurka J, Kaplan D J, Duncan C H, Walichiewicz J, Milosavljevic A, Murali G, Solus J F. Nucleic Acids Res. 1993;21:1273–1279. doi: 10.1093/nar/21.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blunt T, Taccioli G E, Priestley A, Hafezparast M, Mcmillan T, Liu J, Cole C C, White J, Alt F W, Jackson S P, Schurr E, Lehmann A R, Jeggo P A. Genomics. 1995;30:320–328. doi: 10.1006/geno.1995.9871. [DOI] [PubMed] [Google Scholar]
- 12.Carle G, Olson M. Nucleic Acids Res. 1984;12:5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Game J C, Sitney K C, Cook V E, Mortimer R K. Genetics. 1989;123:695–713. doi: 10.1093/genetics/123.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouprina N, Eldarov M, Moyzis R, Resnick M, Larionov V. Genomics. 1994;21:7–17. doi: 10.1006/geno.1994.1218. [DOI] [PubMed] [Google Scholar]
- 15.Larionov V, Kouprina N, Nikolaishvili N, Resnick M. Nucleic Acids Res. 1994;22:4154–4161. doi: 10.1093/nar/22.20.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erickson J R, Johnston M. Genetics. 1993;134:151–157. doi: 10.1093/genetics/134.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ketner G, Spencer F, Tugendreich S, Connelly C, Hieter P. Proc Natl Acad Sci USA. 1994;91:6186–6190. doi: 10.1073/pnas.91.13.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Degryse E, Dumas B, Dietrich M, Laruelle L, Achstetter T. Yeast. 1995;11:629–640. doi: 10.1002/yea.320110704. [DOI] [PubMed] [Google Scholar]
- 19.Featherstone T, Huxley C. Genomics. 1993;17:267–278. doi: 10.1006/geno.1993.1321. [DOI] [PubMed] [Google Scholar]
- 20.McGonigal T, Bodelle P, Schopp C, Sarthy A V. Gene. 1995;155:267–271. doi: 10.1016/0378-1119(94)00887-x. [DOI] [PubMed] [Google Scholar]
- 21.McCormick M K, Buckler A, Bruno W, Campbell E, Shera K, Torney D, Deaven L, Moyzis R. Genomics. 1993;18:553–558. doi: 10.1016/s0888-7543(05)80355-6. [DOI] [PubMed] [Google Scholar]
- 22.Green E D, Riethman H C, Dutchik J E, Olson M V. Genomics. 1991;11:658–669. doi: 10.1016/0888-7543(91)90073-n. [DOI] [PubMed] [Google Scholar]
- 23.Stinchomb D T, Thomas M, Kelly I, Selker E, Davis R W. Proc Natl Acad Sci USA. 1980;77:4559–4563. doi: 10.1073/pnas.77.8.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]


