Abstract
The SH2 domain of growth factor receptor-bound protein 2 (Grb2) has been the focus of numerous studies, primarily because of the important roles it plays in signal transduction. More recently, it has emerged as a useful protein to study the consequences of ligand preorganization upon energetics and structure in protein-ligand interactions. The Grb2-SH2 domain is known to form a domain-swapped dimer, and as part of our investigations toward correlating structure and energetics in biological systems, we examined the effects that domain-swapping dimerization of the Grb2-SH2 domain had upon ligand binding affinities. Isothermal titration calorimetry was performed using Grb2-SH2 in both its monomeric and domain-swapped dimeric forms and a phosphorylated tripeptide AcNH–pTyr–Val–Asn-NH2 that is similar to the Shc sequence recognized by Grb2-SH2 in vivo. The two binding sites of domain-swapped dimer exhibited a 4- and a 13-fold reduction in ligand affinity compared to monomer. Crystal structures of peptide-bound and uncomplexed forms of Grb2-SH2 domain-swapped dimer were obtained and reveal that the orientation of residues V122, V123, and R142 may influence the conformation of W121, an amino acid that is believed to play an important role in Grb2-SH2 ligand sequence specificity. These findings suggest that domain-swapping of Grb2-SH2 not only results in a lower affinity for a Shc-derived ligand, but it may also affect ligand specificity.
Keywords: Grb2, SH2 domain, domain-swapping, dimerization, crystallography
Introduction
Growth factor receptor-bound protein 2 (Grb2)1 is a 25 kDa protein composed of a single SH2 domain flanked on its N- and C-terminal ends by SH3 domains. This structural motif enables Grb2 to serve as a linker between a phosphotyrosine signal and downstream cellular events, characterizing it as an adaptor protein [1, 2]. It has been shown that Grb2 binds to phosphotyrosine residues on Shc through its SH2 domains, whereas the SH3 domains of Grb2 interact with proline rich regions of SOS. Once bound to Grb2, SOS associates with Ras and exchanges GDP for GTP, thereby activating Raf and initiating the MAP kinase pathway [1, 3, 4]. Because the MAP kinase pathway has been linked to a number of cancers [5], drugs that block the activation of this pathway are of interest as potential anticancer agents. Indeed, and compounds that bind to the Grb2-SH2 domain have been shown to block Ras activation in vivo [6–10].
First coined to describe the structure of diphtheria toxin [11–13], domain-swapping occurs when two protein molecules exchange identical or highly similar regions, usually near the N- or C-terminus. There are two reports showing that the Grb2-SH2 domain can form a homodimer via domain-swapping in which two molecules swap C-terminal helices [14, 15]. However, in both of these cases a truncated form of Grb2-SH2 was expressed as a fusion protein with GST, which has been postulated to facilitate domain-swapping in some cases [16].
That the Grb2-SH2 domain can dimerize by domain swapping in vitro raises the interesting question of whether there is any biological significance to such dimerization. In this regard it is perhaps noteworthy that two Grb2-SH2 binding sites on Shc are required for efficient recruitment and binding of SOS to the Grb2-Shc complex in B-cell antigen receptor-stimulated cells [3]. This observation suggests that Grb2-SOS complex formation occurs only when multiple Grb2-SH3 domains are bound to SOS. Comparing the structures of full length Grb2 (1GRI) [2] and a domain-swapped dimeric Grb2-SH2/peptide complex (1FYR) led to a model for a full length domain-swapped Grb2 dimer suggesting that both SH2 domains and each of the four SH3 domains would be available to interact with other cellular proteins [15]. Even though binding of two Grb2 molecules to Shc may be required for SOS recruitment in vivo, it is unknown whether this process involves two monomers or one dimer or whether two full length molecules of Grb2 associate through domain-swapping.
During the course of studies of complexes of the Grb2-SH2 domain with ligands derived from a phosphorylated tyrosine [17], we observed domain-swapping of a Grb2-SH2 domain that was not part of a GST fusion protein. Isothermal titration calorimetry (ITC) data indicated that this dimeric form of the Grb2-SH2 domain exhibited a reduction in affinity for a Shc-derived ligand compared to the monomeric domain. We solved the crystal structures of domain-swapped dimeric Grb2-SH2 in its uncomplexed form and complexed with a Shc-derived ligand. Comparison of these structural data with structures available from the protein data bank, reveal differences in the interactions to residues within the binding sites of monomeric and domain-swapped dimeric Grb2-SH2, allowing for a plausible explanation for the differences in ligand affinities.
Materials and Methods
Chemicals
All chemicals used were purchased from Fisher, Sigma, or Fluka. E. coli cells (SG13009, Qiagen) transformed with the PEQ vector encoding Grb2 residues 53–163, corresponding to the SH2 domain, were kindly provided by Dr. Andrew Prongay (Schering-Plough Research Institute, Kenilworth, NJ). Gel filtration, ion exchange chromatography, and CNBr-activated Sepharose 4B beads were purchased from G.E. Healthcare.
Preparation of Phosphotyrosine Column
In a slight modification of a procedure developed by Dr. Patrick J. Finerty, Jr. (The Hospital for Sick Children and Department of Biochemistry, University of Toronto), CNBr-activated Sepharose 4B beads were swelled in 1 mM HCl (5 mL/g resin); the beads were then washed extensively with 1 mM HCl. Phosphotyrosine coupling solution (10 mM O-phospho-L-tyrosine, 0.5 M NaCl, 0.1 M NaHCO3 at pH 8.3) was added to the activated resin (5 mL/g), and the solution was mixed with gentle agitation for 3 h at room temperature. The excess phosphotyrosine was removed by thoroughly rinsing the resin with coupling buffer (0.5 M NaCl, 0.1 M NaHCO3 at pH 8.3). The resin was suspended in 5 gel volumes of blocking solution (0.1 M Tris-HCl at pH 8.0) and gently agitated for 3 h at room temperature. The resin was washed with 5 cycles of alternating pH (0.1 M sodium acetate, 0.5 M NaCl at pH 4.0 and 0.1 Tris-HCl, 0.5 M NaCl at pH 8.0). The resin was then suspended in 30 mM Tris, 1 mM EDTA at pH 7.5, packed into a column, and equilibrated with 30 mM Tris, 1 mM EDTA at pH 7.5 prior to use.
Synthesis of Ligands
The known tripeptide ligand AcNH-pTyr-Val-Asn-NH2 (Ac–pYVN) [18] was prepared in solution by modifying literature procedures for solid phase synthesis. The ligand was purified using reverse phase HPLC and characterized using proton and carbon NMR along with high-resolution mass spectrometry.
Preparation of Recombinant Grb2-SH2
Cultures (1 L) were grown at 30 °C in L.B. from a single colony, and expression was induced with 1.0 mM IPTG when an O.D.λ600 of 0.5–0.9 was reached. Cultures were allowed to express protein for roughly 15 h. Pellets were lysed in 30 mM Tris, 1 mM EDTA, pH 7.5. The lysate was centrifuged, and the supernatant was applied to a phosphotyrosine column. Grb2-SH2 was eluted from the phosphotyrosine column with 30 mM Tris and 250 mM NaCl at pH 7.5 and dialyzed (100-fold, twice) in 30 mM Tris and 1mM EDTA at pH 7.5. The sample was applied to a Q-Sepharose column (10–15 mL), and the flow through contained >95% pure Grb2-SH2 as visualized by commassie-stained SDS-PAGE. Dimeric and monomeric Grb2-SH2 were separated on a Superose 12 prep grade column (100 mL) that had been pre-equilibrated with 30 mM Tris and 150 mM NaCl at pH 7.5. Two peaks eluted, corresponding to the molecular weights of monomer (~13 kDa) and dimer (~26 kDa) Grb2-SH2 as confirmed by a gel filtration molecular weight standard (Bio-Rad). Using this protocol, roughly 80% of the Grb2-SH2 domain eluted from the column as monomer.
The extinction coefficient of dimer at 280 nm was determined by the method described by Edelhoch [19]. Briefly, an initial calculated extinction coefficient of 30,440 M−1 was determined for fully denatured protein in 6.0 M guanidinium hydrochloride, 0.2 M phosphate buffer, pH 6.5. This was done by employing the known sequence of Grb2 SH2 and the measured contributions that tryptophan and tyrosine make to the overall extinction coefficient. Each of those values was in turn determined by Gill and Hippel [20], by approximating each residue in the environment of a fully extended protein with model tripeptides Gly-X-Gly, in which X represents either tryptophan or tyrosine. In our laboratory, we have measured the extinction coefficient of dimer to be 30,120 M−1, whereas a value of 15,000 M−1 for the pure monomeric form has been measured. Our value for monomer compares reasonably well with that obtained by McNemar and coworkers (15,600 M−1) [21].
Protein Behavior
Separated monomer and dimer were found to be surprisingly kinetically stable, as very little interconversion was seen over a course of months at 4 °C. However, the dimer to monomer ratio changed dramatically by binding the protein to an SP Sepharose column in 30 mM Tris at pH 7.5 and 1 mM EDTA; roughly 60% of the Grb2-SH2 domain eluted from this column as dimer. Furthermore, concentrating either dimer or monomer using an YM-10 Centriprep Centrifugal Filter Device (Millipore) resulted in some interconversion to monomer or dimer, respectively. Protein intended for use in the ITC experiments was directly obtained from the gel filtration column, because concentrating the dimeric or monomeric Grb2-SH2 resulted in interconversion.
Isothermal Titration Calorimetry
Monomeric and dimeric Grb2-SH2 were isolated from the gel filtration column at 50–100 μM and were at least 95% pure. The domain was dialyzed (three times, 500-fold) in 50 mM HEPES at pH 7.5 and 150 mM NaCl. Tripeptide Ac–pYVN was dissolved in the third dialysis buffer to a concentration of 1.0–3.0 mM. Experiments were performed in duplicate using a Microcal VP-ITC microcalorimeter. A solution of ligand was injected (typically 30–50 injections of 4–10 μL each) into the sample cell containing the Grb2-SH2 domain. Raw data were integrated and analyzed using Microcal Origin software, and values for the stoichiometry of the binding (n), the association constant (Ka), the change in enthalpy of binding (ΔH), the change in Gibbs free energy upon binding (ΔG), and the change in entropy (ΔS) were determined (Table 1 and Supplementary Figure 1). Blank titrations were performed by injecting ligand into buffer (50 mM HEPES at pH 7.5, 150 mM NaCl).
Table 1.
Isothermal titration calorimetry data for the binding of AcNH–pTyr–Val–Asn-NH2 (Ac–pYVN) to monomeric and domain-swapped dimeric Grb2-SH2. ITC experiments were conducted in duplicate at 25 °C with same batch of ligand and Grb2 SH2 domain in HEPES at pH 7.5. Uncertainties in ΔG, ΔH, and ΔS represent deviations from the average.
| Monomer | Ka(M−1) 6.1 (±0.3) × 105 | ΔG (kcal mol−1) −7.9 (±0.1) | ΔH (kcal mol−1) −6.6 (±0.2) | ΔS (cal mol−1 K−1) 4.3 (±0.7) | TΔS (kcal mol−1) 1.3 (±0.2) |
|---|---|---|---|---|---|
| Dimer | |||||
| Site 1: | 1.5 (±0.3) × 105 | −7.1 (±0.1) | −3.3 (±0.1) | 12.5 (±0.4) | 3.7 (±0.1) |
| Site 2: | 4.7 (±0.4) × 104 | −6.4 (±0.1) | −2.3 (±0.1) | 13.8 (±0.2) | 4.1 (±0.1) |
Protein Crystallization
All crystals were grown in 7 μL drops via vapor diffusion using the hanging drop method. Crystals of uncomplexed domain-swapped dimer Grb2-SH2 domain (PDB 2H46) grew in the I422 space group and formed within a few days at 25 °C by mixing equal volumes of 15 mg/mL Grb2-SH2 in 50 mM HEPES at pH 7.5 with 100 mM MES and 2.1 M NH4SO4 at pH 6.0. Crystals of Grb2-SH2 domain-swapped dimer that were dissolved in 30 mM Tris and 500 mM NaCl at pH 7.5 and loaded on a size exclusion column eluted at a volume corresponding to a molecular weight of ~28 kDa. Uncomplexed domain-swapped dimer crystals were cryoprotected by adjusting the crystallization drop to 25% glycerol.
The crystal structure of the complex between domain-swapped dimer Grb2-SH2 and tripeptide Ac–pYVN (PDB 2H5K) crystallized at 4 °C by mixing equal volumes of protein-ligand solution (25 mM sodium cacodylate at pH 6.0, 20 mg/mL Grb2-SH2, with tripeptide Ac–pYVN added to a 1.5 M excess) and 0.1 M sodium cacodylate, 0.1 M calcium acetate, 18% PEG 8000, pH 6.0. Crystals grew in the P6222 space group and were allowed to grow for at least 1 week. Prior to data collection, the crystals were cryoprotected by slowly making the drops 25% (v/v) ethylene glycol.
Data Collection and Refinement
All data were collected under liquid N2 on either a Mar345 or RAXIS 4++ detector using CuKα radiation at 1.5418 Å and images were processed using HKL2000 [22] (See Supplemental Figure 2 for data collection and refinement statistics). A molecular replacement solution, using the PDB deposited structure 1JYU [14] for both the uncomplexed and peptide bound structures, was determined using the CNS and CCP4 program suites [23, 24]. Ligand building and structure manipulation was achieved using CORINA, PRODRG, and O [25–27]. The quality of the final models was assessed using Procheck from CCP4. Images were created with the aid of Pymol [28].
Results
ITC of Monomeric and Domain-Swapped Dimeric Grb2-SH2
Thermodynamic data were acquired in duplicate for the binding of the tripeptide AcNH-pTyr-Val-Asn-NH2 (Ac–pYVN) to domain-swapped dimeric and monomeric forms of Grb2-SH2 at 25 °C (Table 1). Blank titrations were performed, and the corresponding heats from these injections were subtracted from the protein-ligand titrations prior to data fitting. In contrast to previous reports [15], we found that dimeric and monomeric Grb2-SH2 domains bind the Ac–pYVN tripeptide with significantly different affinities. The Ka for binding of Ac–pYVN to each of the peptide-binding sites of the domain-swapped dimer of Grb2-SH2 was 1.5 × 105 and 4.7 × 104 M−1 (Kd = 6.7 and 21.2 μM, respectively). Hence, the affinity of the dimeric form of Grb2-SH2 for tripeptide Ac–pYVN is about 4 to 13-fold less than for the monomeric Grb2-SH2 domain (Ka = 6.1 × 105 M−1; Kd = 1.6 μM). The reduction in affinity of Ac–pYVN for dimeric Grb2 SH2 domain arises from a significantly less favorable (ca 3–4.3 kcal mol−1) enthalpy of binding for the dimer relative to monomer. Conversely, the entropic term, TΔS, contributes significantly more (i.e., 2.4–2.8 kcal mol−1) to the binding free energy for complex formation involving the dimeric form of the domain than it does for the monomer. Namely, the enthalpic term dominates the entropic term in the binding energetics for complexes of the monomeric Grb2 SH2 domain, whereas the entropic term assumes equal or greater importance in binding to the dimer. However, this increase in the entropy of binding is not sufficient to override the loss in enthalpy of binding. It is perhaps noteworthy that the dissociation constant reported herein for the binding of tripeptide Ac–pYVN to monomeric Grb2-SH2 (Kd = 1.6 μM) is somewhat greater than that reported for Ac-PSpYVNVQN-NH2 (Kd = ~0.3 μM) [29] and comparable to the ELSIA-derived IC50 value (Kd = 4.3 μM) for Ac-pTyr-Val-Asn-NH2 reported by Garcia-Echeverria and colleagues [30].
Uncomplexed Domain-Swapped Dimer Grb2-SH2 Crystal Structure
Uncomplexed, domain-swapped dimeric Grb2-SH2 crystallized with one molecule of Grb2-SH2 in the asymmetric unit, and applying crystallographic symmetry generated the full domain-swapped dimer. According to the nomenclature for SH2 domains proposed by Eck [31], the asymmetric unit comprises two α-helices, α A and αB, and seven β-strands, βA–βG. The swapped portion of the dimer includes residues 122–152 and perhaps additional residues, although there was no observable density beyond residue 152. A glycerol molecule from the cryoprotectant was found in the phosphotyrosine-binding site. The hydroxyl moieties of the glycerol molecule serve as hydrogen bond donors to residues within αA and the BC loop, which comprises residues between β-strands B and C.
The major differences between uncomplexed forms of full length Grb2 monomer [2] and the Grb2-SH2 domain-swapped dimer are found in the orientations of residues W121, V122, V123 and R142. Residues 121–123 undergo a major conformational change upon domain-swapping and adopt an extended conformation in the domain-swapped dimer that has several notable structural consequences (Figure 1a). For example, the carbonyl oxygen atom of V122 in dimeric Grb2-SH2 is now positioned so that it forms a hydrogen bond with the guanidino group of R142 (Figure 1b). The orientations and solvent accessible area of the side chain of W121 in the two structures differ significantly. For example, the χ1 and χ2 angles for this residue in uncomplexed full length Grb2 are −51° and −68°, respectively, whereas the corresponding angles in the domain swapped dimer are 63° and −78°. In the uncomplexed domain-swapped dimer, the side chain of R142 and the side chain of V123 bury most of the side chain of W121 so the non-polar solvent accessible area of W121 (11.6 Å2) is significantly smaller than it is in the full length, uncomplexed monomeric form of Grb2 (41.1 Å2). It is interesting to note that the structure published by Nioche and colleagues [14] of uncomplexed domain-swapped dimeric Grb2-SH2 is very similar to the one reported here. However, in this structure R142 is not within hydrogen bonding distance of the carbonyl oxygen atom of V122.
Fig. 1.
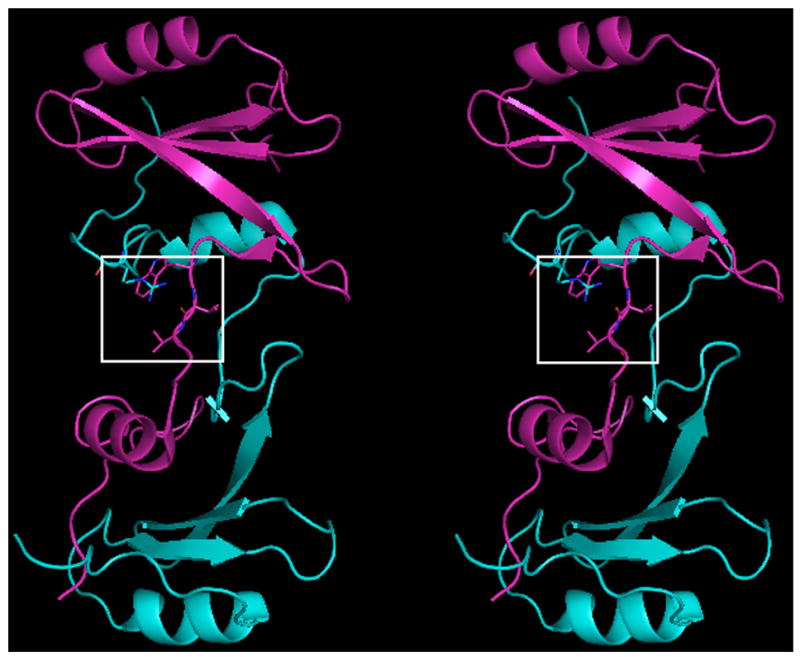
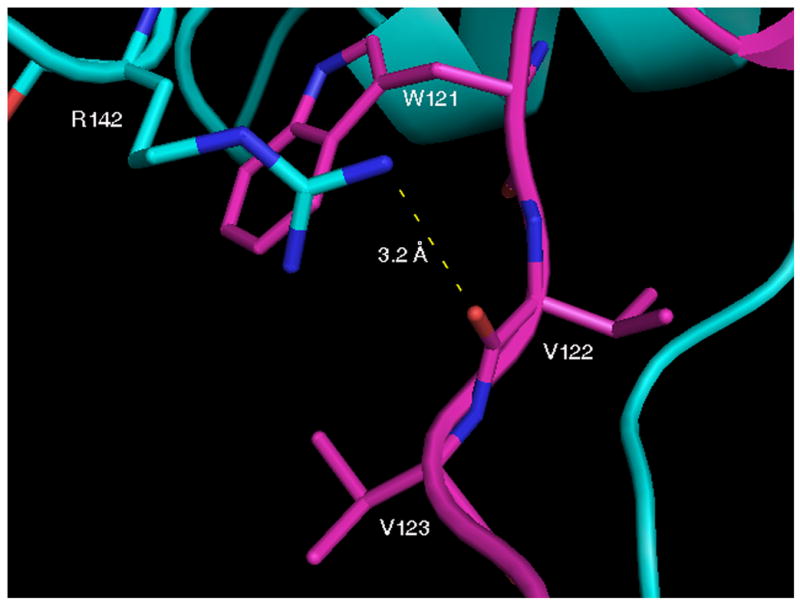
Crystal structure of native domain-swapped dimeric Grb2-SH2. A, Cross-eyed stereo view of native domain-swapped dimeric Grb2-SH2, generated by applying crystallographic symmetry. The two molecules composing the dimer, shown in magenta and cyan, swap the C-terminus, composed of residues 124–152. B, In domain-swapped dimeric Grb2-SH2 residues 121–123 are in an extended conformation, which allows for a hydrogen bond (dashed yellow line) to form between the □guanidine group of R142 and the carbonyl oxygen atom of V122. The side chains of R142 and V123 mostly bury W121.
Crystal structure of domain-swapped dimer Grb2-SH2 complexed to tripeptide Ac–pYVN
The complex of Grb2-SH2 with tripeptide Ac–pYVN crystallized with one domain-swapped dimer in the asymmetric unit. One of the two binding sites in the domain-swapped dimer was occupied by the tripeptide Ac–pYVN (site A), whereas what appears to be a cacodylate ion, which was a component of the crystallization buffer, was bound to the other ligand binding site (site B). Residues from the two molecules comprising the domain-swapped dimer Grb2-SH2/Ac–pYVN complex adopt nearly identical conformations as the backbone atoms align with an RMS deviation of 0.5 Å. Temperature factors are generally two-fold higher for residues that form binding site B compared to site A, but such variations are not unexpected given the dramatic dissimilarities in the structures of tripeptide Ac–pYVN and cacodylate ion.
Within binding site A, the pY-1 carbonyl oxygen atom of tripeptide Ac–pYVN engages in hydrogen bonds to the guanidino group of R67, whereas the phosphate moiety of the phosphotyrosine residue is stabilized by hydrogen bonding interactions with the side chains of residues R67, R86, S88, S90, and S96. The pY+1 Val side chain of tripeptide Ac–pYVN lies within a hydrophobic pocket and is involved in hydrophobic contacts with the phenyl group of F108 and the Cβ atom of Q106. The tripeptide Ac–pYVN adopts a β-turn about the Val residue with an intramolecular hydrogen bond between the C–terminal amide N–H group and the carbonyl carbon atom of the tyrosine residue. The pY+2 Asn Nδ2 and Oδ1 atoms are involved in hydrogen bonds with the carbonyl oxygen atom of K109 and the amide N–H of L120 in the A binding site. As in the structure of the uncomplexed form of the domain-swapped dimer of Grb2-SH2, the W121 side chain is mostly buried by the side chains of R142 and V123 at both ligand binding sites A and B. The side chain of W121 thus does not interact with the pY+2 Asn residue of tripeptide Ac–pYVN. Residues R86, S88, E89, and S96 form hydrogen bonds to the cacodylate ion at the B site. It is important to note that despite the somewhat low resolution of this structure (3.2 Å) the location of the ligand tripeptide Ac–pYVN is clearly visible in the |Fo-Fc| omit map (Figure 2a). Temperature factors for the tripeptide Ac–pYVN were relatively low, with an average value of 20 Å2.
Fig. 2.
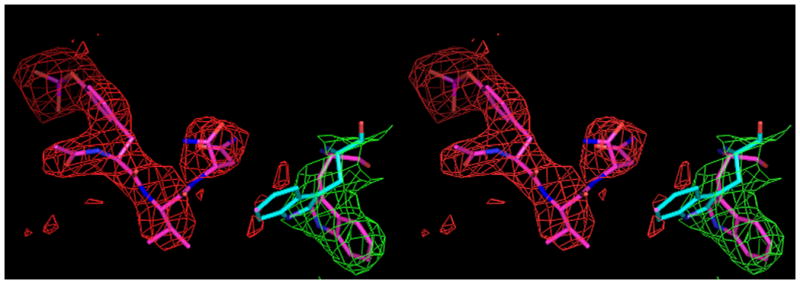
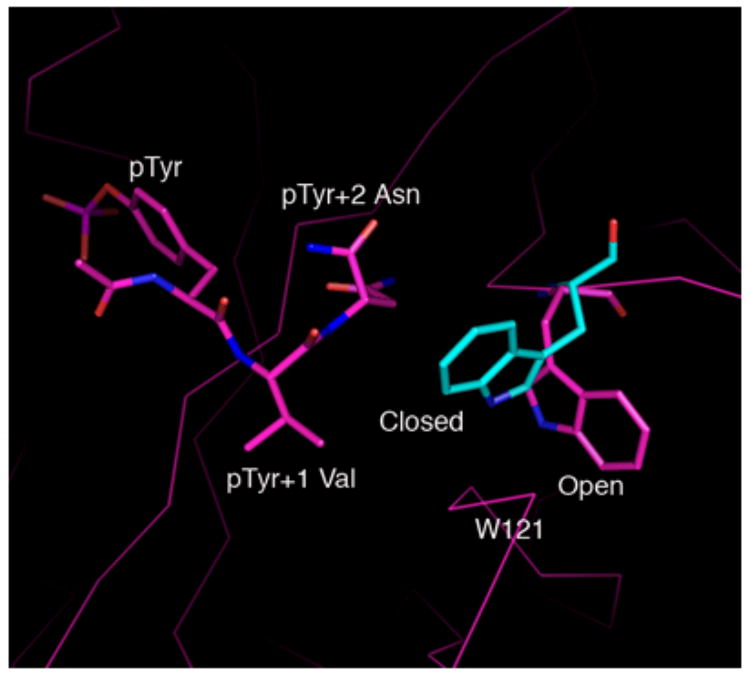
A, Shown in cross-eyed stereo are the 2Fo-Fc (green) and |Fo-Fc| (red) electron density maps contoured at 1.0 and 2.5 σ, respectively. The latter maps were calculated with tripeptide Ac–pYVN omitted. The |Fo-Fc| omit map clearly shows the location of the ligand despite the relatively low resolution data (3.2 Å). For information regarding the quality of electron density maps at this resolution see reference [37]. B, Open and closed conformations of W121. Shown is W121 from a monomeric Grb2-SH2/ligand complex (cyan) (PDB code 1JYR [14]) aligned to the Grb2-SH2 domain-swapped dimer/Ac–pYVN structure (magenta). In crystal structures of monomeric Grb2-SH2 W121 adopts the closed conformation (cyan), bringing the idole ring within close contact to the ligand pTyr+2 residue. W121 from the domain-swapped dimer/Ac–pYVN complex adopts the open conformation and points away from the ligand binding site (magenta). W121 also adopts the open conformation in uncomplexed domain-swapped dimer.
Discussion
The uncomplexed and ligand-complexed structures of domain-swapped dimeric Grb2-SH2 are similar with the backbone atoms in the two structures aligning with an rms deviation of 0.6 Å; all atoms overlay with an rmsd of 0.9 Å. The significant differences between the complex of domain-swapped dimeric Grb2-SH2 with Ac–pYVN and the eight known crystal structures of monomericGrb2 SH2/ligand complexes [2, 14, 32–34] are found in the conformations of residues 121–123 and 142 and their interactions with the domain and the side chain of the pY+2 Asn residue. In the domain-swapped dimer complex with Ac–pYVN, residues 121–123 are in an extended conformation that allows for formation of a hydrogen bond between the guanidino group of R142 and the carbonyl oxygen atom of V122. The side chain of W121 thus forms contacts with the side chains of V123 and R142 rather than with the pY+2 residue of Ac–pYVN. Conversely, in monomeric Grb2-SH2 complexes, residues 121–123 form part of a loop, precluding formation of the aforementioned hydrogen bond between V122 and R142. The indole ring of W121 is thus not in proximity to the V123 and R142 side chains; rather it forms part of the ligand binding site, making van der Waals contact with the side chain of the pY+2 Asn residue. These data suggest that the side chain of W121 can adopt two different conformations upon ligand binding, and these are herein denoted as open and closed (Figure 2b). In the closed conformation, the side chain of W121 is oriented toward the pTyr+2 residue, and in the open conformation it is positioned away from the ligand binding site so it cannot interact with the ligand.
Besides the structures reported here, three other structures of ligand complexes of the domain-swapped dimer of Grb2-SH2 are known (PDB codes 1FYR, 2AOA, 2AOB) [15, 35]. In each of those structures the side chain of W121 adopts both open and closed conformations within the different molecules that comprise the asymmetric units. In our structure of uncomplexed domain-swapped dimeric Grb2-SH2, electron density is only observed for the open conformation of W121, whereas in the complex with Ac–pYVN there appears to be some |Fo-Fc| electron density in the area corresponding to the closed conformation (Figure 2a). Taken together the data suggests that both open and closed conformations likely exists in solution.
Positioning the side chain of W121 in the open and closed conformations does not significantly change the amount of solvent accessible surface area of the W121 side chain itself upon complexation. On the other hand, the orientation of the W121 side chain does have a considerable affect upon the accessible surface area for the pY+2 Asn residue of the bound Ac–pYVN. For example, the calculated solvent accessible surface area (ASAp = 48.5 Å2 and ASAnp = 14.6 Å2) of W121 in the complex of Ac–pYVN with the domain-swapped dimer is similar (ASAp = 44.7 Å2 and 2, ASAnp = 22.5 Å2) to that calculated using the monomeric/ligand complex reported by Nioche (PDB code 1JYR) [14]. However, the pY+2 Asn residue of the ligand makes close contact with the indole ring of W121 in this and all other reported peptide complexes with monomeric Grb2-SH2. Hence, less polar and non-polar surface area of the pY+2 Asn residue is solvent accessible (ASAp = 16.5 Å2 and ASAnp = 0.7 Å2) in monomer compared to that of the complex between Ac–pYVN and the domain-swapped dimer of Grb2-SH2 (ASAp = 19.2 Å2 and ASAnp = 17.9 Å2). The burial of additional non-polar surface area upon ligand binding to monomeric Grb2-SH2 could explain its higher affinity for the latter. In this context, Marengere observed an approximately 14-fold reduction in the affinity for pTyr-Val-Asn-Val, a peptide similar to Ac–pYVN, when W121 was mutated to a threonine [36]. The crystallographic evidence presented herein clearly shows that the open conformation may be populated in ligand complexes. Therefore, it appears that ligands, other than Ac-pYVN, have the potential to bind to domain-swapped dimeric Grb2 in an extended conformation.
Previous reports on the domain-swapping of the Grb2-SH2 domain fragment have prompted questions about its biological significance. A model proposed by Schiering suggests that if full length Grb2 does form a domain-swapped dimer, the SH2 domains and flanking SH3 domains would be fully accessible [15]. Therefore, it is at the least feasible that Grb2 may form domain-swapped dimers in vivo. The ITC data presented here suggest Ac–pYVN binds sequentially to the two binding sites of domain-swapped dimer. Compared to the monomeric form, the observed lower affinity and noncooperative binding for domain-swapped dimeric Grb2-SH2 could have physiological relevance. For example, cellular events which lead to the formation of domain-swapped dimer from monomer would likely down regulate the signals that Grb2 is involved. It may also be the case that the SH2 domain of domain-swapped dimeric Grb2 exhibits different sequence specificity than the monomeric form.
In summary, the thermodynamic data indicate that monomeric Grb2-SH2 domain binds the phosphotyrosine peptide Ac–pYVN with significantly higher affinity than the domain-swapped dimeric form of Grb2-SH2. Interestingly, the binding free energy of Ac–pYVN to the monomeric domain is dominated by the ΔH term, which is substantially more favorable than the enthalpy for its binding to the dimeric domain, whereas TΔS contributes more then ΔH to the free energy for Ac–pYVN binding to the dimeric domain. That ligand binding to the monomeric domain is enthalpy driven while binding to the dimeric domain is entropy driven is intriguing. The side chain of W121 forms close contacts with the pY+2 residue of pYVN-derived ligands in all reported monomeric Grb2-SH2/ligand complexes, but there is no such interaction in the complex of the domain-swapped dimer of the Grb2-SH2 domain with Ac–pYVN that is reported herein. These structural data are consistent with the hypothesis that the lack of a van der Waals contact between W121 and the Asn residue at the pY+2 position of phosphotyrosine-derived ligands may be at least partly responsible for the reduced affinity and less favorable enthalpy for the binding of Ac–pYVN to the domain-swapped dimer of Grb2-SH2. Other studies correlating energetics and structure in protein-ligand interactions involving the Grb2-SH2 domain and other proteins are ongoing, and the results of these investigations will be reported in due course.
Supplementary Material
Acknowledgments
We are grateful to the Robert A. Welch Foundation and the Texas Advanced Research Program for their support of this work. We also thank Dr. Andrew Prongay (Schering-Plough Research Institute, Kenilworth, NJ) for E. coli cells (SG13009, lon−) containing the GE-60 plasmid. We also thank Dr. John Pascal (Thomas Jefferson University) for help in data collection and processing, Professor John Tesmer (Professor of Life Sciences Institute and Pharmacology, Medical School, University of Michigan) for valuable discussions, and Laura Millspaugh (The University of Texas at Austin) and Dr. Hilary Plake (The University of Texas at Austin) for preparing the ligands used in these studies. We also thank Dr. Patrick Finerty, Jr. (The Hospital for Sick Children and Department of Biochemistry, University of Toronto) for advice on preparing the phosphotyrosine column.
The abbreviations used are
- ASA
accessible solvent area
- Grb2-SH2
growth factor receptor-bound protein 2-src homology domain 2
- GST
glutathione S-transferase
- ITC
isothermal titration calorimetry
- PAGE
polyacrylamide gel electrophoresis
- pTyr
phosphotyrosine
- Ac–pYVN
AcNH–pTyr–Val–Asn-NH2
- Shc
Src Homologous and Collagen
- SOS
son of sevenless
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chardin P, Cussac D, Maignan S, Ducruix A. The Grb2 adaptor. FEBS Lett. 1995;369:47–51. doi: 10.1016/0014-5793(95)00578-w. [DOI] [PubMed] [Google Scholar]
- 2.Maignan S, Guilloteau JP, Fromage N, Arnoux B, Becquart J, Ducruix A. Crystal structure of the mammalian Grb2 adaptor. Science. 1995;268:291–293. doi: 10.1126/science.7716522. [DOI] [PubMed] [Google Scholar]
- 3.Harmer SL, DeFranco AL. Shc contains two Grb2 binding sites needed for efficient formation of complexes with SOS in B lymphocytes. Mol Cell Biol. 1997;17:4087–4095. doi: 10.1128/mcb.17.7.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harmer SL, DeFranco AL. The src homology domain 2-containing inositol phosphatase SHIP forms a ternary complex with Shc and Grb2 in antigen receptor-stimulated B lymphocytes. J Biol Chem. 1999;274:12183–12191. doi: 10.1074/jbc.274.17.12183. [DOI] [PubMed] [Google Scholar]
- 5.Segawa N, Nakamura M, Nakamura Y, Mori I, Katsuoka Y, Kakudo K. Phosphorylation of mitogen-activated protein kinase is inhibited by calcitonin in DU145 prostate cancer cells. Cancer Res. 2001;61:6060–6063. [PubMed] [Google Scholar]
- 6.Gay B, Suarez S, Caravatti G, Furet P, Meyer TJ. Selective GRB2 SH2 inhibitors as anti-Ras therapy. Int J Cancer. 1999;83:235–241. doi: 10.1002/(sici)1097-0215(19991008)83:2<235::aid-ijc15>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 7.Lung FD, Tsai JY. Grb2 SH2 domain-binding peptide analogs as potential anticancer agents. Biopolymers. 2003;71:132–140. doi: 10.1002/bip.10396. [DOI] [PubMed] [Google Scholar]
- 8.Shakespeare WC. SH2 domain inhibition: a problem solved? Curr Opin Chem Biol. 2001;5:409–515. doi: 10.1016/s1367-5931(00)00222-2. [DOI] [PubMed] [Google Scholar]
- 9.Bradshaw JM, Waksman G. Molecular recognition by SH2 domains. Adv Protein Chem. 2002;61:161–210. doi: 10.1016/s0065-3233(02)61005-8. [DOI] [PubMed] [Google Scholar]
- 10.Machida K, Mayer BJ. The SH2 domain: versatile signaling module and pharmaceutical target. Biochim Biophys Acta. 2005;1747:1–25. doi: 10.1016/j.bbapap.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Bennett MJ, Choe S, Eisenberg D. Refined structure of dimeric diphtheria toxin at 2.0 A resolution. Protein Sci. 1994;3:1444–1463. doi: 10.1002/pro.5560030911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett MJ, Choe S, Eisenberg D. Domain swapping: entangling alliances between proteins. Proc Natl Acad Sci U S A. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nioche P, Liu WQ, Broutin I, Charbonnier F, Latreille MT, Vidal M, Roques B, Garbay C, Ducruix A. Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor. J Mol Biol. 2002;315:1167–1177. doi: 10.1006/jmbi.2001.5299. [DOI] [PubMed] [Google Scholar]
- 15.Schiering N, Casale E, Caccia P, Giordano P, Battistini C. Dimer formation through domain swapping in the crystal structure of the Grb2-SH2-Ac-pYVNV complex. Biochemistry. 2000;39:13376–13382. doi: 10.1021/bi0012336. [DOI] [PubMed] [Google Scholar]
- 16.Newcomer ME. Protein folding and three-dimensional domain swapping: a strained relationship? Curr Opin Struct Biol. 2002;12:48–53. doi: 10.1016/s0959-440x(02)00288-9. [DOI] [PubMed] [Google Scholar]
- 17.Benfield AP, Teresk MG, Plake HR, DeLorbe JE, Milspaugh LE, Martin SF. Ligand Preorganization May Be Accompanied by Entropic Penalties in Protein–Ligand Interactions. Angew Chem Int Ed. 2006;45:6830–6835. doi: 10.1002/anie.200600844. [DOI] [PubMed] [Google Scholar]
- 18.Furet P, Gay B, Garcia-Echeverria C, Rahuel J, Fretz H, Schoepfer J, Caravatti G. Discovery of 3-aminobenzyloxycarbonyl as an N-terminal group conferring high affinity to the minimal phosphopeptide sequence recognized by the Grb2-SH2 domain. J Med Chem. 1997;40:3551–3556. doi: 10.1021/jm9702185. [DOI] [PubMed] [Google Scholar]
- 19.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 20.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 21.McNemar C, Snow ME, Windsor WT, Prongay A, Mui P, Zhang R, Durkin J, Le HV, Weber PC. Thermodynamic and structural analysis of phosphotyrosine polypeptide binding to Grb2-SH2. Biochemistry. 1997;36:10006–10014. doi: 10.1021/bi9704360. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst D Biol Cryst. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.N. Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Cryst D Biol Cryst. 1994;1:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Gasteiger J, Sadowski J, Schuur J, Selzer P, Steinhauer L, Steinhauer V. Chemical Information in 3D-Space. J Chem Inf Comput Sci. 1996;36:1030–1037. [Google Scholar]
- 26.van Aalten DM, Bywater R, Findlay JB, Hendlich M, Hooft RW, Vriend G. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comp Mol Des. 1996;10:255–262. doi: 10.1007/BF00355047. [DOI] [PubMed] [Google Scholar]
- 27.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Delano WL. Delano Scientific LLC; San Carlos: 2004. [Google Scholar]
- 29.de Mol NJ, Dekker FJ, Broutin I, Fischer MJE, Liskamp RMJ. Surface plasmon resonance thermodynamic and kinetic analysis as a strategic tool in drug design. Distinct ways for phosphopeptides to plug into Src- and Grb2 SH2 domains. J Med Chem. 2005;48:753–763. doi: 10.1021/jm049359e. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Echeverria C, Gay B, Rahuel J, Furet P. Mapping the X(+1) binding site of the Grb2-SH2 domain with alpha, alpha-disubstituted cyclic alpha-amino acids. Bioorg Med Chem Lett. 1999;9:2915–2920. doi: 10.1016/s0960-894x(99)00501-6. [DOI] [PubMed] [Google Scholar]
- 31.Eck MJ, Shoelson SE, Harrison SC. Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature. 1993;362:87–91. doi: 10.1038/362087a0. [DOI] [PubMed] [Google Scholar]
- 32.Ettmayer P, France D, Gounarides J, Jarosinski M, Martin MS, Rondeau JM, Sabio M, Topiol S, Weidmann B, Zurini M, Bair KW. Structural and conformational requirements for high-affinity binding to the SH2 domain of Grb2(1) J Med Chem. 1999;42:971–980. doi: 10.1021/jm9811007. [DOI] [PubMed] [Google Scholar]
- 33.Rahuel J, Gay B, Erdmann D, Strauss A, Garcia-Echeverria C, Furet P, Caravatti G, Fretz H, Schoepfer J, Grutter MG. Structural basis for specificity of Grb2-SH2 revealed by a novel ligand binding mode. Nat Struct Biol. 1996;3:586–589. doi: 10.1038/nsb0796-586. [DOI] [PubMed] [Google Scholar]
- 34.Rahuel J, Garcia-Echeverria C, Furet P, Strauss A, Caravatti G, Fretz H, Schoepfer J, Gay B. Structural basis for the high affinity of amino-aromatic SH2 phosphopeptide ligands. J Mol Biol. 1998;279:1013–1022. doi: 10.1006/jmbi.1998.1790. [DOI] [PubMed] [Google Scholar]
- 35.Phan J, Shi ZD, Burke TRJ, Waugh DS. Crystal structures of a high-affinity macrocyclic peptide mimetic in complex with the Grb2 SH2 domain. J Mol Biol. 2005;353:104–115. doi: 10.1016/j.jmb.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 36.Marengere LE, Songyang Z, Gish GD, Schaller MD, Parsons JT, Stern MJ, Cantley LC, Pawson T. SH2 domain specificity and activity modified by a single residue. Nature. 1994;369:502–505. doi: 10.1038/369502a0. [DOI] [PubMed] [Google Scholar]
- 37.Minichino A, Habash J, Raftery J, Helliwell JR. The properties of (2Fo - Fc) and (Fo - Fc) electron-density maps at medium-to-high resolutions. Acta Crystallogr D Biol Crystallogr. 2006;59:843–849. doi: 10.1107/s0907444903004219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


