Abstract
Background
A variety of skin cancer susceptibility among mouse strains has allowed identification of genes responsible for skin cancer development. Fifteen Skts loci for skin tumour susceptibility have been mapped so far by using the two-stage skin carcinogenesis model [induced by 7.12-dimethylbenz(a)anthracene (DMBA)/12-O-tetradecanoylphorbol-13-acetate (TPA)]. A few responsible genes have been identified using wild-derived dominant resistant Mus spretus mice, and one has been confirmed as a low penetrance cancer susceptibility gene in a variety of human cancers.
Results
In the present study, we found that wild-derived PWK mice developed no tumour by treatment with the two-stage skin carcinogenesis protocol. This phenotype is dominant resistant when crossed with the highly susceptible strain FVB. By analyzing the F1 backcross generation between PWK and FVB, we found empirical evidence of significant linkage at the new loci Skts-fp1 on chromosome 4 and suggestive linkage on chromosomes 1, 3, 11, 12 and 14 for skin tumour susceptibility. Skts-fp1 includes the Skts7 interval, which was previously mapped by a Mus spretus and NIH backcross. We also observed suggestive linkage on chromosomes 1 and 2 in the female population only, while suggestive linkage on chromosomes 14 and 15 only was observed in the male population. A significant genetic interaction was seen between markers of D11Mit339 and D16Mit14.
Conclusion
Analysis of this new cross may facilitate the identification of genes responsible for mouse skin cancer susceptibility and may reveal their biological interactions.
Background
Germ-line mutation has been reported in tumour suppressor genes and in oncogenes that are responsible for the highly penetrated familial cancer syndrome [1]. The majority of cancers, however, are sporadic and often associated with familial aggregation. Therefore, cancer susceptibility in general is explained by multiple low penetrance cancer susceptibility genes [2,3].
Although analysis of the human population is useful in finding the gene related to high penetrance disease, the identification of low penetrance tumour susceptible genes is still difficult. This is mainly due to heterogeneous genetic background and complex environmental effects. Mouse models of cancer have been used extensively for the analysis and identification of the genetic components of tumour susceptibility [4,5]. The susceptibility for the two-stage skin carcinogenesis model varies among mouse strains [6,7], and the genetic approach has been performed to identify genes related to cancer susceptibility [8-11]. Previous studies indicate that the percent contribution of skin cancer susceptibility can be explained by each genetic component consisting of one of multiple loci and their interaction [12]. Using NIH and Mus spretus crosses, in which Mus spretus is dominant resistant against susceptible Mus musculus strains, skin cancer susceptibility loci Skts1~15 were mapped [8,9,12]. Psl1~4 were mapped by using C57BL/6J(B6) and DBA crosses, in which DBA shows semi-dominant susceptibility against resistant B6 [10,13]. Tomaso Dragani's group also mapped Skts1 using Car-R and Car-S outbred. crosses [14], which were generated by balanced intercross of inbred strains, and reported loss of tyrosinase activity conferred increased skin tumor susceptibility [15]. Skts13 was identified as the Aurka gene [16], and Skts14 was identified as Tgfb1 [11]. However, none of the responsible genes have been cloned yet at the remaining loci. Since functional polymorphisms of AURKA were repeatedly reported to be determinants of individual risk for colon, esophageal, skin, lung, breast, ovarian, and prostate cancers in the human population [16-19], identification of genetic components in the mouse skin cancer susceptibility could facilitate the identification of candidate cancer risk factors in the human population.
To narrow down the candidate loci and to identify the corresponding gene for the genetic components of mouse skin cancer susceptibility, information from other strains and crosses are valuable. In this study, we found that the PWK strain, which is an inbred mouse derived from wild Mus musculus musculus mice [20], is dominantly resistant to chemically induced skin carcinogenesis when crossed with a highly susceptible FVB strain. Because of the dominant resistance and high genetic diversity between PWK and FVB strains [21], we expected that there were multiple genetic components involved in skin cancer susceptibility as seen in our previous report of NIH/spretus cross. Therefore, we performed genetic analysis of a large backcross between PWK and FVB strains.
Results
Using the two stage skin tumour induction treatment, all 16 FVB mice developed papillomas with an average multiplicity of 32.56 ± 3.6, at 20 weeks after the initiation. However, none of the 10 PWK/Rbrc mice developed any papillomas on dorsal skin (Table 1). Also, none of the 17 (PWK/Rbrc × FVB/N)F1 mice, abbreviated as PF, developed tumours. Four out of the 25 (FVB/N × PWK/Rbrc)F1 mice, abbreviated as FP, developed tumours, but the average multiplicity was 0.32 ± 0.16 (Table 1). These results demonstrate that the PWK strain has a dominant resistant phenotype to the DMBA/TPA induced skin tumour against a susceptible FVB strain.
Table 1.
Incidence and multiplicity of papilloma in the parental strains, F1 and F1 backcross
| Strain | Animal No. | Incidence (%) | Multiplicity a) | |
| FVB/N | total | 16 | 16 (100) | 32.56 ± 3.60 |
| female | 9 | 9 (100) | 28.22 ± 4.46 | |
| male | 7 | 7 (100) | 38.14 ± 5.36 | |
| PWK/Rbrc | total | 10 | 0 (0) | 0 |
| female | 10 | 0 (0) | 0 | |
| male | 10 | 0 (0) | 0 | |
| FVB × PWK | total | 25 | 4 (16) | 0.32 ± 0.8 |
| female | 14 | 4 (28.6) | 0.57 ± 1.01 | |
| male | 11 | 0 (0) | 0 | |
| PWK × FVB | total | 17 | 0 (0) | 0 |
| female | 8 | 0 (0) | 0 | |
| male | 9 | 0 (0) | 0 | |
| (FxP) × F | total | 208 | 123(56.7) | 4.86 ± 0.56 |
| female | 107 | 60(56.1) | 3.45 ± 0.64 | |
| male | 101 | 63(62.4) | 6.34 ± 0.92 |
a) data are shown as mean ± SEM.
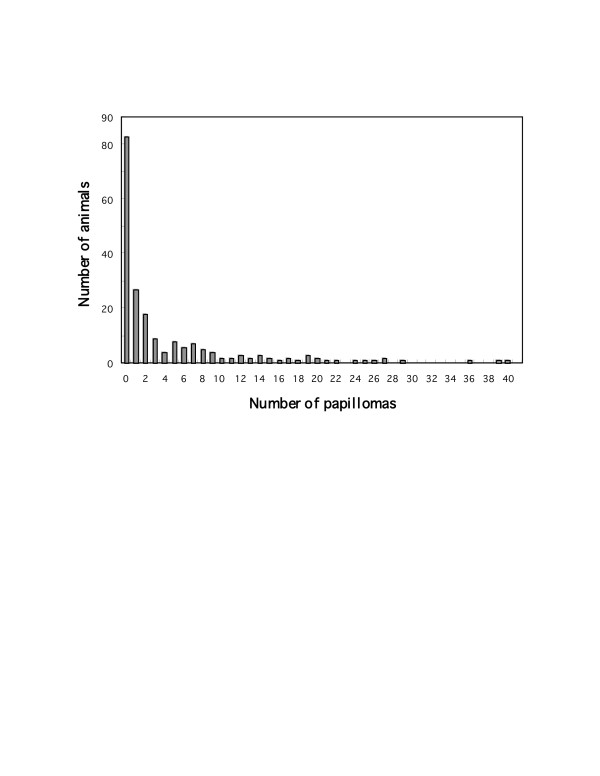
Linkage analysis was performed by genotyping 208 F1 backcross mice (abbreviated as FPxF) for 204 polymorphic markers spread throughout the genome. The number of papillomas 20 weeks after the initial treatment was used as a trait. One hundred twenty three mice (56.7%) developed papillomas, and average multiplicity was 4.86 ± 0.56 (Table 1, Fig. 1). Males developed a significantly higher number of papillomas than females. Even though it was not significantly associated, FVB males also showed higher multiplicity than FVB females. On the contrary, only female mice developed papillomas in the F1 hybrids (Table 1).
Figure 1.
Distribution patterns of papilloma multiplicity. The number of papillomas 20 weeks after the initiation is plotted as histograms.
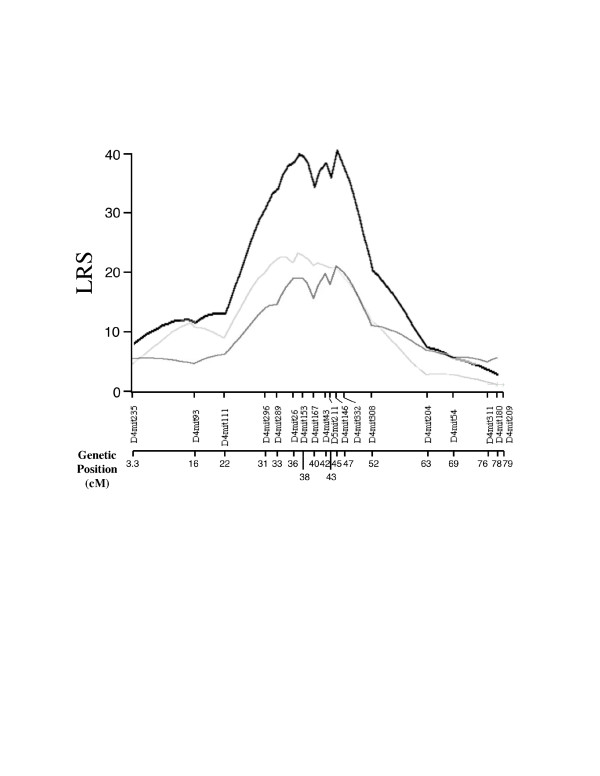
For parametric quantitative trait loci (QTL) regression analysis, tumour multiplicity data are transformed by root-square transformation to improve the fit of a negative binomial distribution pattern of nominal data sets to a normal distribution [22]. By single point marker regression analysis, significant linkage with likelihood ratio statistics (LRS) larger than 12.9 was mapped only on chromosome 4 (Table 2). LRS of at least 6.9 was considered evidence for suggestive linkage and was detected on chromosomes 1, 3, 4, 11, 12 and 14. Interval mapping on chromosome 4 showed highly significant linkage between markers D4Mit111 and D4Mit308 (Skts-fp1). This QTL interval covered almost one third of chromosome 4 and there seem to be at least two peaks close to D4Mit26 and D4Mit146 with values of LRS 38.5 and 40.5, respectively (Fig. 2). It was estimated that approximately 18% of the total trait variance would be explained by a QTL at this locus.
Table 2.
QTL for papilloma multiplicity
| total | female | male | |||||||||
| Number of papillomas c) | |||||||||||
| Marker | Map location a) | LRS | P-value b) | FF (Number of animals) | FP (Number of animals) | LRS | P-value b) | LRS | P-value b) | ||
| Chromosome1 | |||||||||||
| D1Mit18 | 21.4 | 7.4 | 0.00671* | 6.33 ± 0.96 | (103) | 3.37 ± 0.56 | (104) | 8.9 | 0.00288* | 0.9 | >0.01 |
| D1Mit284 | 35.6 | 6.8 | 0.00924 | 9.4 | 0.00219* | 0.9 | >0.01 | ||||
| D1Mit10 | 44.1 | 6.0 | >0.01 | 8.4 | 0.00383* | 0.7 | >0.01 | ||||
| Chromosome2 | |||||||||||
| D2Mit484 | 50 | 1.7 | >0.01 | 8.1 | 0.00445* | 0.2 | >0.01 | ||||
| D2Mit401 | 58.1 | 5.2 | >0.01 | 8.9 | 0.00289* | 0.5 | >0.01 | ||||
| D2Mit423 | 63.7 | 3.2 | >0.01 | 8.8 | 0.00308* | 0.0 | >0.01 | ||||
| Chromosome3 | |||||||||||
| D3mit40 | 36.1 | 7.6 | 0.0058* | 6.39 ± 0.94 | (109) | 3.19 ± 0.54 | (98) | 3.6 | >0.01 | 2.8 | >0.01 |
| Chromosome4 | |||||||||||
| D4Mit235 | 3.3 | 7.4 | 0.00648* | 6.56 ± 0.98 | (98) | 3.33 ± 0.57 | (110) | 4.1 | >0.01 | ||
| D4Mit93 | 16.1 | 11.0 | 0.00092* | 6.55 ± 0.94 | (99) | 3.31 ± 0.60 | (109) | 10.4 | 0.00123* | 3.2 | >0.01 |
| D4Mit111 | 22.1 | 12.5 | 0.00041* | 6.57 ± 0.94 | (99) | 3.29 ± 0.61 | (109) | 8.8 | 0.00354* | 4.8 | >0.01 |
| D4Mit26 | 36.3 | 34.4 | <0.00001*** | 7.94 ± 1.05 | (95) | 2.11 ± 0.39 | (109) | 16.5 | 0.00005** | 18.7 | 0.00002** |
| D4Mit167 | 40 | 34.2 | <0.00001*** | 7.82 ± 1.01 | (100) | 2.11 ± 0.39 | (108) | 21.5 | <0.00001*** | 15.0 | 0.00011** |
| D4Mit308 | 51.9 | 19.9 | 0.00001** | 6.72 ± 0.86 | (100) | 3.12 ± 0.69 | (108) | 11.6 | 0.00067* | 10.1 | 0.00147* |
| Chromosome11 | |||||||||||
| D11Mit339 | 33.8 | 8.3 | 0.00388* | 6.36 ± 0.92 | (99) | 3.48 ± 0.65 | (109) | 3.3 | >0.01 | 5.3 | >0.01 |
| Chromosome12 | |||||||||||
| D12Mit283 | 9.4 | 7.8 | 0.00534* | 6.24 ± 0.92 | (107) | 3.39 ± 0.60 | (101) | 2.5 | >0.01 | 4.6 | >0.01 |
| D12Mit153 | 15.1 | 7.0 | 0.0081* | 6.25 ± 0.93 | (105) | 3.44 ± 0.60 | (102) | 3.0 | >0.01 | 3.3 | >0.01 |
| Chromosome14 | |||||||||||
| D14Mit257 | 24.6 | 0.6 | >0.01 | 9.4 | 0.00222* | ||||||
| D14Mit203 | 44.1 | 8.4 | 0.00382* | 6.18 ± 0.89 | (100) | 3.63 ± 0.68 | (108) | 1.8 | >0.01 | 5.9 | >0.01 |
| D14Mit193 | 48.3 | 12.3 | 0.00046* | 6.43 ± 0.88 | (103) | 3.31 ± 0.67 | (105) | 1.8 | >0.01 | 11.0 | 0.00089* |
| D14Mit195 | 60.3 | 8.0 | 0.00465* | 5.96 ± 0.78 | (107) | 3.68 ± 0.81 | (101) | 1.5 | >0.01 | 6.2 | >0.01 |
| D14Mit197 | 72.7 | 1.0 | >0.01 | 8.0 | 0.00475* | ||||||
| Chromosome15 | |||||||||||
| D15Mit15 | 64.9 | 3.4 | >0.01 | 1.7 | >0.01 | 10.6 | 0.0011* | ||||
| D15Mit161 | 68.8 | 4.8 | >0.01 | 0.2 | >0.01 | 9.1 | 0.0026* | ||||
a) cM from the centrosome
b) P-value for the association at each locus
c) Data are shown as mean ± SEM.
Values in parenthesis are number of samples.
* Suggestive linkage
** Significant linkage
*** Highly Significant linkage
Figure 2.
Interval mapping on chromosome 4. The QTL on chromosome 4 shows significant linkage to multiplicity of papilloma 20 weeks after the initiation. Black line indicates the linkage calculated for whole mice, right gray is for female mice, dark gray is for male mice. Genetic distances of each marker from centrosome are indicated as genetic position (cM) on the bottom.
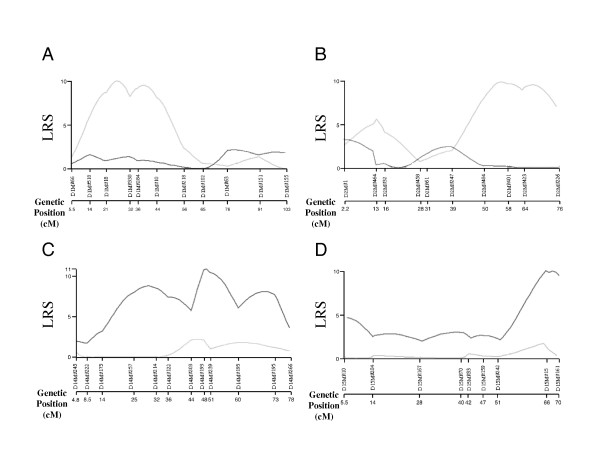
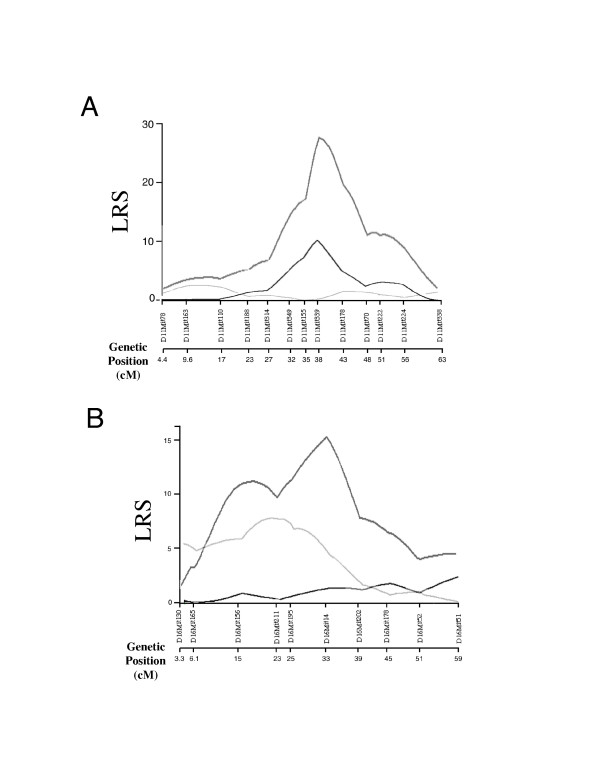
The suggestive linkage on chromosomes 1, 2, 14 and 15 showed gender difference. Linkage on chromosomes 1 and 2 was observed only in female, and linkage on chromosomes 14 and 15 was in males exclusively. Suggestive evidence of linkage at loci on chromosome 1, 2, 14 and 15 was obtained with interval mapping in either male or female population exclusively (Fig. 3A~D).
Figure 3.
Interval mapping on chromosome 1, 2, 14 and 15. Result of interval mapping on (A) chromosome 1, (B) chromosome 2, (C) chromosome 14 and (D) chromosome 15 are shown. Right gray line indicates the linkage for female mice and dark gray line for male mice. Genetic distances of each marker from centrosome are indicated as the genetic position (cM) on the bottom.
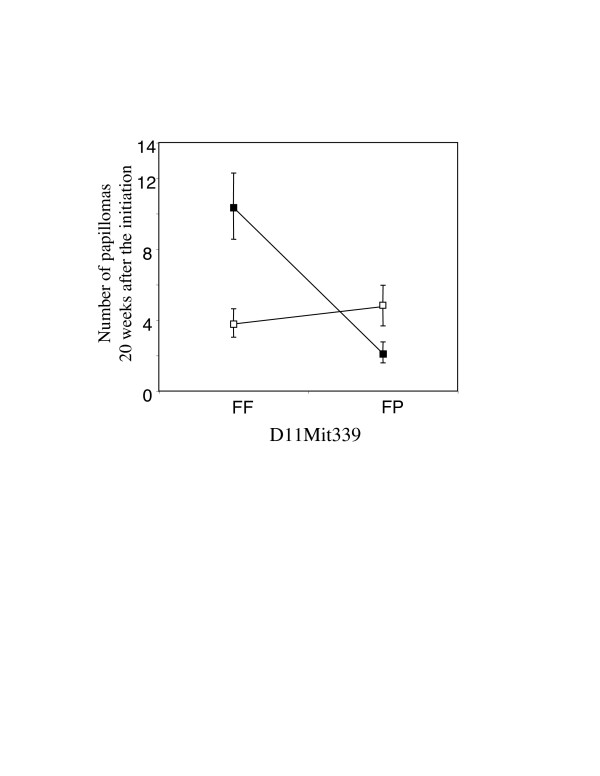
To find interaction components among QTLs, a two-dimensional scan of per-mutation test using J/qtl was performed. With a standard criteria (Joint LOD>5 and [Epistatic LOD > 3 or minimum LOD {locus1, locus2} >3]), possible interactive QTL with epistatic LOD 4.14 was found between markers D11Mit339 and D16Mit14. A plot of the average number of papillomas of each genotype at D11Mit339 and D16Mit14 is shown in Fig. 4. Homozygous FVB allele (FF) at the D16Mit14 marker is associated with susceptibility if the genotype at the interacting locus D11Mit339 is a homozygous (FF) genotype. In the mice with heterozygous FP genotype at D16Mit14, genotype at D11Mit339 had no association with the multiplicity of papillomas. Interval mapping indicated a highly significant linkage Skts-fp2 (LRS 27.6) between markers D11Mit155 and D11Mit178 locus in mice with FF at D16Mit14, but no linkage was observed in mice with FP at D16Mit14 (Fig. 5A). By interval mapping, significant linkage Skts-fp3 (LRS15.3) was also mapped between markers D16Mit202 and D16Mit195 in mice with FF genotype at D11Mit339 (Fig. 5B). Some weak linkage was observed also in mice with FP at D11Mit339, however the additive effect was opposite, i.e. PWK allele on chromosome16 could be susceptible allele for skin tumor development in FP at D11Mit339, while it is resistant in FF at D11Mit339. Approximately 26% of the total trait variance would be explained by Skts-fp2 in mice with FF genotype at D16Mit14 locus, and 14% for Skts-fp3 in mice with FF genotype at D11Mit339. No sexual difference was observed in Skts-fp2 and Skts-fp3.
Figure 4.
Allelic interaction between D11Mit339 and D16Mit14. Association between D11Mit339 genotype and multiplicity of papilloma 20 weeks after the initiation is shown. Black square indicates the mice with FF genotype at the D16Mit14 locus. White square indicates the mice with FP genotype at the D16Mit14 locus. Data are shown as mean ± SEM.
Figure 5.
Interval mapping on chromosome 11. (A) Interval mapping results on chromosome 11 are shown for the mice with FF genotype (dark gray line) and the mice with FP genotype (right gray line) at the D16Mit14 locus. Black line indicates interval mapping for whole mice. Significant linkage is observed only in the mice with FF genotype at the D16Mit14 locus. (B) Interval mapping results on chromosome 16 are shown for the mice with FF genotype (dark gray line) and the mice with FP genotype (right gray line) at the D11Mit339 locus. Black line indicates interval mapping for whole mice. Significant linkage is observed only in the mice with FF genotype at the D11Mit339 locus. Genetic distances of each marker from centrosome are indicated as the genetic position (cM) on the bottom.
Discussion
The present study indicates that PWK is a dominant resistant strain to two-stage skin carcinogenesis, when crossed with highly susceptible FVB mice. Among the 20 mice in each group, no PWK developed papillomas, and only a few FP F1 hybrid mice developed one papilloma. In a 208 backcross mice study, which should detect QTLs with more than 4% variance, a significant QTL of Skts-fp1 has been mapped. The linkage study using genetically and phenotypically diverged strains FVB and PWK can facilitate analysis and understanding the genetic component of this complex cancer susceptibility.
PWK is a wild-derived strain that could help ensure high genetic diversity [21,23]. As seen in NIH and Mus spretus inter-specific crosses, we expected to have a large genetic variation to skin tumour susceptibility between FVB and PWK. However, we mapped a significant locus on chromosome 4 only. Interval mapping indicated that Skts-fp1 showed multiple significant peaks located between D4Mit111 and D4Mit308, which covered almost one third of chromosome 4, suggesting that a linkage group consisting of multiple genes in a chromosome may be involved in Skts-fp1. Contributions of the D4Mit26 locus and the D4Mit146 locus (two major linkage peaks between which recombination are observed in 16 mice) are estimated to explain the approximately 15% and 18% of the variance of PWK resistant phenotype, respectively. These contributions are higher than the contribution of major QTLs in the NIH and Mus spretus backcross study, indicating that contributions on a single chromosome are important in the FVB and PWK cross. Linkage analysis also showed suggestive linkage at loci on chromosomes 1, 3, 11, 12 and 14 in addition to significant linkage at D4Mit26 and D4Mit167 (Skts-fp1). Although additional markers and backcross mice are needed to hypothesize the multiple genetic effects in this cross, it is possible that many other weak loci and epistatic components contribute to the resistant phenotype of PWK.
Skts7, which was mapped as a QTL on chromosome 4 for chemically induced skin tumour susceptibility by the analysis of the NIH/Ola and Mus spretus cross [8,9], is located within Skts-fp1. A common gene may be responsible for the skin susceptibilities of the two crosses, segregating FVB with PWK and NIH with Mus spretus. So far, the SNP information in the Phenome database is not sufficient to find common haplotype blocks between PWK and Mus spretus. However, after narrowing down the locus and carrying out expression and functional analysis of the candidate genes, we can perform a haplotype analysis identifying a common polymorphism in the gene between resistant strains (Mus spretus, PWK) and susceptible strains (NIH, and FVB). The haplotype analysis will help to identify a candidate skin resistant gene.
From the study with Ornithine decarboxylase (Odc) transgenic BALB/cJ and C57BL/6J mice, significant linkage of the skin tumour induced by DMBA and the Odc transgene was detected in the interval between D4Mit31and D4Mit52 (51.3–54.9 cM) [24]. These two strains may also share a common resistant allele and could be used for haplotype mapping to identify the candidate.
On this interval of Skts-fp1, many QTLs for susceptibility to other types of tumours have been mapped [4,5]. Pctr1 and Pctr2 have been mapped as loci for susceptibility to pristine induced plasmacytoma [25,26], and Cdkn2a has been suggested to be a corresponding gene for Pctr1[27,28]. Cdkn2a is also a strong candidate for Papg1, QTL for progression of urethane induced lung tumours [29]. The Cdkn2a locus encodes two tumour suppressors, P16INK4a and P19ARF. There are no non-synonymous polymorphisms between FVB and PWK in ankirin 1 and 2 domains of P16INK4a, which are suggested to be corresponding polymorphisms for Pctr1 and Papg1 (data not shown). Because Figure 2 shows multiple peaks at the Skts-fp1 locus on chromosome 4, as seen with other tumour susceptibility QTLs [30], the locus is complex and may create a linkage group containing two or more overlapping susceptibility and/or resistant regions. Thus, we have initiated the creation of congenic mice containing segments of the donor PWK allele on chromosome 4 to the recipient FVB background.
Skts-fp2 on chromosome 11 was identified as a significant linkage among FVB homozygous mice at marker D16Mit14, but not in mice heterozygous at this locus. Skts-fp3 on chromosome 16 was also identified among FVB homozygous mice at the D11Mit339 locus, but not in mice with heterozygous at this locus. Our results suggest that Skts-fp2 and Skts-fp3 showed the evidence of interaction. An interaction component between two loci has been suggested to play an important role in cancer susceptibility [4,12] and recently Tgfb1 was indicated as a corresponding gene for a skin tumour susceptible locus Skts14. This locus was shown to have interaction with Skts15, which overlaps with the Tgfbm3 locus mapped as Tgfb1 modifier locus [11]. Skts-fp2 could also be functionally related to Skts-fp3. Scc15, the QTL for 1,2-dimethylhydrazine induced colon tumour susceptibility in mice [31] and Sluc4, the QTL for N-ethyl-N-nitrosourea induced lung tumour in mice [32] have been mapped around the Skts-fp2 locus, but no QTL for skin tumor susceptibility is mapped at this locus. No QTL for susceptibility to skin cancer or other types of tumour has been mapped around the Skts-fp3 locus, however Skts9 for DMBA/TPA induced skin cancer [9] and Sluc27 for N-ethyl-N-nitrosourea induced lung tumour [33] are mapped on the same chromosome. If extreme phenotypic differences exist between two strains and dominant interactions are expected, the interaction components can be detected more efficiently using a simple backcross [12]. Although further analysis with an additional number of mice is needed to identify responsible genes at the interactedlocus and significant interaction component between two QTLs, the genetic interaction between Skts-fp2 and Skts-fp3 reveals a biological interaction of genes at two loci.
In this study, we found that linkages at four loci show sexual difference. Suggestive linkage on chromosomes 1 and 2 was exclusively observed in females and suggestive linkage on chromosomes 14 and 15 was observed only in males. Usually this kind of study is performed with either all female or all male animals, and sexual differences in tumour susceptibility genes are not identified. An increased number of mice will enable us to reveal the QTL for tumour susceptibility, which interacts with sex chromosomes directly or indirectly.
The absence of PWK alleles at both markers remarkably increased the average number of papillomas, whereas no effect was seen in animals that retained one PWK allele at either locus. A dominant resistant to cancer development may be redundant regulation. Candidate accessory regulatory genes modify a major effect gene and can be identified as a dominant interacted modifier. Although individual modifying genes may show only subtle quantitative effects on tumourigenesis, combinations of suppressive alleles can completely eliminate neoplasia. The identification of effective combinations of gene activities reflects the general finding of "genetic redundancy", in which the loss of function of a single gene has no obvious phenotype.
Here we present three significant loci related to skin tumour susceptibility from the analysis of a new strain combination of FVB and PWK. Among previous studies using other tumour induction systems, many tumour susceptible loci mapped on the same chromosomal regions where skin cancer susceptibility loci were mapped. For example, Psl2 was mapped close to Skts13, Psl3 was mapped close to Skts8, and the locus found from the analysis of Car-S and Car-R strains was located on the Skts1 locus. Also, Skts-fp1 identified in this study covered the region on which Skts7 and the linkage obtained from ODC transgenic mice have been mapped. As described in the studies for in silico mapping methods using many inbred strains, congenic, recombinant inbred, advanced intercross, chromosome substitution strains or very recently collaborative crosses [34-36], it is obvious that accumulated information from several genetic analyses using many other strains provides an important and rapid route to identification of complex germ-line genetic variants that confer increased cancer risk.
Conclusion
This study shows that PWK is a dominant resistant strain to two-stage skin carcinogenesis when it is crossed with the FVB strain. By linkage analysis using backcross between FVB and PWK, three significant QTLs, Skts-fp1, Skts-fp2 and Skts-fp3, for the skin tumour susceptibility locus were mapped. Studies using this new cross could be an efficient method for finding new genes responsible for skin cancer susceptibility and for examining genetic/biological interaction between the responsible genes.
Methods
Animals
Mice used in this study were bred in the SPF Facility in the Department of Laboratory Animals at Roswell Park Cancer Institute and are treated in accordance with IACUC regulations. FVB/N mice were purchased from TACONIC (Germantown, NY). PWK/Rbrc inbred mice were obtained from RIKEN BRC (Ibaraki, Japan). Twenty five (FVB/N × PWK/Rbrc) F1 and 17 (PWK/Rbrc × FVB/N) F1, abbreviated as FP and PF respectively, were generated from the two strains of mice. FP mice were then backcrossed with FVB/N to generate 208 F1 backcross animals.
Skin cancer induction
7.12-Dimethylbenz(a)anthracene (DMBA) and 12-O-tetradecanoylphorbol-13-acetate (TPA) were purchased from Sigma Chemical Co. (St. Louis, MO). DMBA is used as a carcinogen and TPA as a promoter.
Nine of FVB female and 7 of male mice, each 10 female and 10 male of PWK mice, 14 female and 11 male of FP hybrids, 8 female and 9 male of PF mice, 107 female and 101 male of FPxF backcross mice were applied for skin carcinogen.
At 8–11 weeks of age, the back skin of each mouse was carefully shaved with an electric clipper. Two days after shaving, 200 ul of DMBA (0.125 mg/ml) was dissolved in acetone and applied to the back of each mouse prior to the hair growth cycle. A total of 97.4 nmol of DMBA was applied to each mouse. One week after the treatment, mice received 400 ul of TPA (5 × 10-5 M solution in acetone) as a promoter, i.e., 32.4 nmol of TPA, twice weekly for 20 weeks. Animals are assessed twice weekly for the appearance of papillomas during the promotion phase. The number of papillomas was counted every other week until 20 weeks after tumour initiation. Papilloma incidence during the promotion phase and papilloma multiplicity 20 weeks after the initiation was used for assessment of skin tumour susceptibility in this study.
Genotype
Genomic DNA was isolated from mouse tails using the alkali method. Briefly, 1~2 mm of tails were boiled in 300 ul of 50 mM NaOH for 30 min, then 25 ul of 1 M Tris-HCL (ph8.0) was added. DNAs were then genotyped for 204 polymorphic microsatellite markers spaced at approximately 8-cM intervals through chromosome 1 to 19 and X by polymerase chain reaction (PCR). Information of MIT markers used in this study is available upon request. PCR reactions were carried out using a Biometlar thermocyler, and PCR products were separated by electrophoresis through 4% Nusieve-GTG low melting temperature agarose gels (FMC) in the 0.5 × TBE buffer.
Statistical analysis
MapManager QTX ver1.03 [37,38] was used to analyse phenotype and genotype data to map QTLs related to susceptibility for chemically induced skin tumours. Permutation tests were performed for FPxF F1 backcross generation, to estimate empirical threshold values for quantitative trait mapping. Single-point marker regression was performed for the whole chromosomes to find linkage, and then interval mapping was performed to analyse the chromosome with significant linkage in the single point marker regression. A permutation test (10,000 permutations at a 1 cM interval) was performed, and the significance threshold was calculated to establish the empirical significance levels in QTL mapping experiments. Suggestive (LRS > 6.9), significant (LRS > 12.9), highly significant (LRS > 20.8) thresholds were established and used for present analysis. A two-dimensional genome scan using J/qtl [39,40] was performed to analyse interactions between QTLs.
Abbreviations
DMBA: 7.12-dimethylbenz(a)anthracene, TPA: 12-O-tetradecanoylphorbol-13-acetate, F: FVB/N, P: PWK/Rbrc, FP: (FVB/N × PWK/Rbrc)F1 mice, PF: (PWK/Rbrc × FVB/N)F1 mice, FPxF: (FVB/N × PWK/Rbrc) × FVB/N F1 backcross mice, LRS: likelihood ratio statistics, B6: C57BL/6J, Odc: Ornithine decarboxylase, PCR: polymerase chain reaction
Authors' contributions
JI and KF carried out skin carcinogen experiments. KF analysed the data of skin tumour developed in mice. KF and NI performed genotyping of F1 backcross mice and linkage analysis. HN (corresponding author) is the PI of the program that funded the work and designed this study. This manuscript was written by KF and HN. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This research was supported by the National Institute of Environmental Health Services to HN (ES012249-01), the Roswell Park Alliance Foundation and, in part, by the NCI Cancer Center Support Grant to Roswell Park Cancer Institute (CA016156). We are grateful to Ms. T. Triplet, Ms. D. Tabaczynski and Ms. J. Jackson for technical support; Ms. K. DeRosa, Ms. Y. Harigaya and Mr. E. Morien for genotyping analyses; and Ms. D. Ovak, Ms. E. Sanders-Noonan and Ms. J. Augus for editorial and secretarial assistance.
Contributor Information
Kyoko Fujiwara, Email: kyoko.fujiwara@roswellpark.org.
Jun Igarashi, Email: ijun@med.nihon-u.ac.jp.
Natsumi Irahara, Email: natsumi.irahara@roswellpark.org.
Makoto Kimura, Email: makoto.kimura@roswellpark.org.
Hiroki Nagase, Email: hiroki.nagase@roswellpark.org.
References
- Li FP. Identification and management of inherited cancer susceptibility. Environ Health Perspect. 1995;103 Suppl 8:297–300. doi: 10.2307/3432329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat Genet. 2003;33 Suppl:238–244. doi: 10.1038/ng1107. [DOI] [PubMed] [Google Scholar]
- Houlston RS, Peto J. The search for low-penetrance cancer susceptibility alleles. Oncogene. 2004;23:6471–6476. doi: 10.1038/sj.onc.1207951. [DOI] [PubMed] [Google Scholar]
- Demant P. Cancer susceptibility in the mouse: genetics, biology and implications for human cancer. Nat Rev Genet. 2003;4:721–734. doi: 10.1038/nrg1157. [DOI] [PubMed] [Google Scholar]
- Mao JH, Balmain A. Genomic approaches to identification of tumour-susceptibility genes using mouse models. Curr Opin Genet Dev. 2003;13:14–19. doi: 10.1016/S0959-437X(03)00005-4. [DOI] [PubMed] [Google Scholar]
- Bangrazi C, Mouton D, Neveu T, Saran A, Covelli V, Doria G, Biozzi G. Genetics of chemical carcinogenesis. 1. Bidirectional selective breeding of susceptible and resistant lines of mice to two-stage skin carcinogenesis. Carcinogenesis. 1990;11:1711–1719. doi: 10.1093/carcin/11.10.1711. [DOI] [PubMed] [Google Scholar]
- Ashman LK, Murray AW, Cook MG, Kotlarski I. Two-stage skin carcinogenesis in sensitive and resistant mouse strains. Carcinogenesis. 1982;3:99–102. doi: 10.1093/carcin/3.1.99. [DOI] [PubMed] [Google Scholar]
- Nagase H, Bryson S, Cordell H, Kemp CJ, Fee F, Balmain A. Distinct genetic loci control development of benign and malignant skin tumours in mice. Nat Genet. 1995;10:424–429. doi: 10.1038/ng0895-424. [DOI] [PubMed] [Google Scholar]
- Nagase H, Mao JH, Balmain A. A subset of skin tumor modifier loci determines survival time of tumor-bearing mice. Proc Natl Acad Sci U S A. 1999;96:15032–15037. doi: 10.1073/pnas.96.26.15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel JM, Caballero M, DiGiovanni J. Identification of novel genetic loci contributing to 12-O-tetradecanoylphorbol-13-acetate skin tumor promotion susceptibility in DBA/2 and C57BL/6 mice. Cancer Res. 2003;63:2747–2751. [PubMed] [Google Scholar]
- Mao JH, Saunier EF, de Koning JP, McKinnon MM, Higgins MN, Nicklas K, Yang HT, Balmain A, Akhurst RJ. Genetic variants of Tgfb1 act as context-dependent modifiers of mouse skin tumor susceptibility. Proc Natl Acad Sci U S A. 2006;103:8125–8130. doi: 10.1073/pnas.0602581103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase H, Mao JH, de Koning JP, Minami T, Balmain A. Epistatic interactions between skin tumor modifier loci in interspecific (spretus/musculus) backcross mice. Cancer Res. 2001;61:1305–1308. [PubMed] [Google Scholar]
- Angel JM, Beltran L, Minda K, Rupp T, DiGiovanni J. Association of a murine chromosome 9 locus (Psl1) with susceptibility to mouse skin tumor promotion by 12-O-tetradecanoylphorbol-13-acetate. Mol Carcinog. 1997;20:162–167. doi: 10.1002/(SICI)1098-2744(199710)20:2<162::AID-MC2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Peissel B, Zaffaroni D, Pazzaglia S, Manenti G, Zanesi N, Zedda I, Rebessi S, Covelli V, Dragani TA, Saran A. Use of intercross outbred mice and single nucleotide polymorphisms to map skin cancer modifier loci. Mamm Genome. 2001;12:291–294. doi: 10.1007/s003350010274. [DOI] [PubMed] [Google Scholar]
- Saran A, Spinola M, Pazzaglia S, Peissel B, Tiveron C, Tatangelo L, Mancuso M, Covelli V, Giovannelli L, Pitozzi V, Pignatiello C, Milani S, Dolara P, Dragani TA. Loss of tyrosinase activity confers increased skin tumor susceptibility in mice. Oncogene. 2004;23:4130–4135. doi: 10.1038/sj.onc.1207565. [DOI] [PubMed] [Google Scholar]
- Ewart-Toland A, Briassouli P, de Koning JP, Mao JH, Yuan J, Chan F, MacCarthy-Morrogh L, Ponder BA, Nagase H, Burn J, Ball S, Almeida M, Linardopoulos S, Balmain A. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nat Genet. 2003;34:403–412. doi: 10.1038/ng1220. [DOI] [PubMed] [Google Scholar]
- Kimura MT, Mori T, Conroy J, Nowak NJ, Satomi S, Tamai K, Nagase H. Two functional coding single nucleotide polymorphisms in STK15 (Aurora-A) coordinately increase esophageal cancer risk. Cancer Res. 2005;65:3548–3554. doi: 10.1158/0008-5472.CAN-04-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicioccio RA, Song H, Waterfall C, Kimura MT, Nagase H, McGuire V, Hogdall E, Shah MN, Luben RN, Easton DF, Jacobs IJ, Ponder BA, Whittemore AS, Gayther SA, Pharoah PD, Kruger-Kjaer S. STK15 polymorphisms and association with risk of invasive ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:1589–1594. [PubMed] [Google Scholar]
- Ewart-Toland A, Dai Q, Gao YT, Nagase H, Dunlop MG, Farrington SM, Barnetson RA, Anton-Culver H, Peel D, Ziogas A, Lin D, Miao X, Sun T, Ostrander EA, Stanford JL, Langlois M, Chan JM, Yuan J, Harris CC, Bowman ED, Clayman GL, Lippman SM, Lee JJ, Zheng W, Balmain A. Aurora-A/STK15 T+91A is a general low penetrance cancer susceptibility gene: a meta-analysis of multiple cancer types. Carcinogenesis. 2005;26:1368–1373. doi: 10.1093/carcin/bgi085. [DOI] [PubMed] [Google Scholar]
- Gregorova S, Forejt J. PWD/Ph and PWK/Ph inbred mouse strains of Mus m.musculus subspecies--a valuable resouce of phenotypic variations and genomic polymorphisms. Folia Biol. 2000;46:31–41. [PubMed] [Google Scholar]
- Tsang S, Sun Z, Luke B, Stewart C, Lum N, Gregory M, Wu X, Subleski M, Jenkins NA, Copeland NG, Munroe DJ. A comprehensive SNP-based genetic analysis of inbred mouse strains. Mamm Genome. 2005;16:476–480. doi: 10.1007/s00335-005-0001-7. [DOI] [PubMed] [Google Scholar]
- Drinkwater NR, Klotz JH. Statistical methods for the analysis of tumor multiplicity data. Cancer Res. 1981;41:113–119. [PubMed] [Google Scholar]
- Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George K, Iacobucci A, Uitto J, O'Brien TG. Identification of an X-linked locus modifying mouse skin tumor susceptibility. Mol Carcinog. 2005;44:212–218. doi: 10.1002/mc.20130. [DOI] [PubMed] [Google Scholar]
- Potter M, Mushinski EB, Wax JS, Hartley J, Mock BA. Identification of two genes on chromosome 4 that determine resistance to plasmacytoma induction in mice. Cancer Res. 1994;54:969–975. [PubMed] [Google Scholar]
- Mock BA, Hartley J, Le Tisser P, Wax JS, Potter M. The plasmacytoma resistance gene, Pctr2, delays the onset of tumorigenesis and resides in the telomeric region of chromosome 4. Blood. 1997;90:4092–4098. [PubMed] [Google Scholar]
- Zhang S, Ramsay ES, Mock BA. Cdkn2a, the cyclin-dependent kinase inhibitor encoding p16INK4a and p19ARF, is a candidate for the plasmacytoma susceptibility locus, Pctr1. Proc Natl Acad Sci U S A. 1998;95:2429–2434. doi: 10.1073/pnas.95.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Qian X, Redman C, Bliskovski V, Ramsay ES, Lowy DR, Mock BA. p16 INK4a gene promoter variation and differential binding of a repressor, the ras-responsive zinc-finger transcription factor, RREB. Oncogene. 2003;22:2285–2295. doi: 10.1038/sj.onc.1206257. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang Y, Herzog CR, Liu G, Lee HW, DePinho RA, You M. A strong candidate gene for the Papg1 locus on mouse chromosome 4 affecting lung tumor progression. Oncogene. 2002;21:5960–5966. doi: 10.1038/sj.onc.1205725. [DOI] [PubMed] [Google Scholar]
- Cormier RT, Bilger A, Lillich AJ, Halberg RB, Hong KH, Gould KA, Borenstein N, Lander ES, Dove WF. The Mom1AKR intestinal tumor resistance region consists of Pla2g2a and a locus distal to D4Mit64. Oncogene. 2000;19:3182–3192. doi: 10.1038/sj.onc.1203646. [DOI] [PubMed] [Google Scholar]
- Ruivenkamp CA, Csikos T, Klous AM, van Wezel T, Demant P. Five new mouse susceptibility to colon cancer loci, Scc11-Scc15. Oncogene. 2003;22:7258–7260. doi: 10.1038/sj.onc.1207096. [DOI] [PubMed] [Google Scholar]
- Fijneman RJ, de Vries SS, Jansen RC, Demant P. Complex interactions of new quantitative trait loci, Sluc1, Sluc2, Sluc3, and Sluc4, that influence the susceptibility to lung cancer in the mouse. Nat Genet. 1996;14:371–373. doi: 10.1038/ng1296-465. [DOI] [PubMed] [Google Scholar]
- Tripodis N, Hart AA, Fijneman RJ, Demant P. Complexity of lung cancer modifiers: mapping of thirty genes and twenty-five interactions in half of the mouse genome. J Natl Cancer Inst. 2001;93:1484–1491. doi: 10.1093/jnci/93.19.1484. [DOI] [PubMed] [Google Scholar]
- Grupe A, Germer S, Usuka J, Aud D, Belknap JK, Klein RF, Ahluwalia MK, Higuchi R, Peltz G. In silico mapping of complex disease-related traits in mice. Science. 2001;8:1915–1918. doi: 10.1126/science.1058889. [DOI] [PubMed] [Google Scholar]
- Brockmann GA, Bevova MR. Using mouse models to dissect the genetics of obesity. Trends Genet. 2002;18:367–376. doi: 10.1016/S0168-9525(02)02703-8. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD. The collavorative cross, a community resouce for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Manly KF, Cudmore RHJ, Meer JM. Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome. 2001;12:930–932. doi: 10.1007/s00335-001-1016-3. [DOI] [PubMed] [Google Scholar]
- Website http://www.mapmanager.org/mmQTX.html
- Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Website http://www.jax.org/staff/churchill/labsite/software/Jqtl/Jqtl-install.html