Abstract
Duramycin is a 19-amino-acid tetracyclic lantibiotic closely related to cinnamycin (Ro09-0198), which is known to bind phosphatidylethanolamine (PE). The lipid specificity of duramycin was not established. The present study indicates that both duramycin and cinnamycin exclusively bind to ethanolamine phospholipids (PE and ethanolamine plasmalogen). Model membrane study indicates that the binding of duramycin and cinnamycin to PE-containing liposomes is dependent on membrane curvature, i.e., the lantibiotics bind small vesicles more efficiently than large liposomes. The binding of the lantibiotics to multilamellar liposomes induces tubulation of membranes, as revealed by electron microscopy and small-angle x-ray scattering. These results suggest that both duramycin and cinnamycin promote their binding to the PE-containing membrane by deforming membrane curvature.
INTRODUCTION
Duramycin is a 19-amino-acid tetracyclic peptide produced by Streptoverticillium cinnamoneus and is closely related to cinnamycin (Ro09-0198) (Fig. 1 A) (1–5). Both compounds belong to the lantibiotics. Lantibiotics are bacteriocins that are characterized by the presence of a high proportion of unusual amino acids. Cinnamycin is unique in that it specifically binds phosphatidylethanolamine (PE) (6–8). Because of this characteristic, cinnamycin has been employed to study the distribution and metabolism of PE (9–14). Duramycin is also suggested to interact with PE (15–18). However, the lipid specificity of duramycin is not well established. Previously it was proposed that duramycin recognizes a particular membrane conformation determined by the presence of PE or monogalactosyl diglyceride (MGDG) (15). Analysis of the membranes of the duramycin-resistant Bacillus subtilis mutants revealed that they had little or no PE and cardiolipin (15,16). In contrast, mutation of alkalophilic Bacillus firmus to duramycin resistance resulted in a substantial replacement of PE by its plasmalogen form (17).
FIGURE 1.
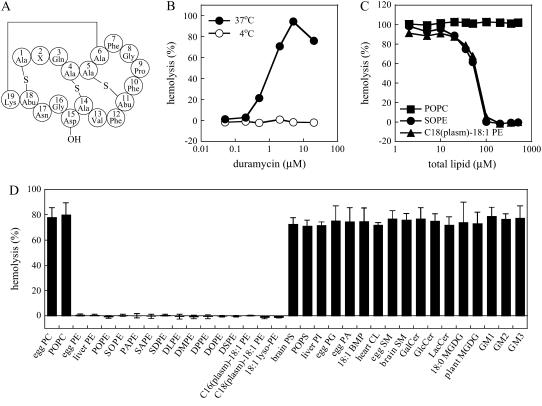
Ethanolamine phospholipids inhibit hemolytic activity of duramycin. (A) Structure of duramycin and cinnamycin (Ro09-0198). Abbreviations: Abu, α-aminobutyric acid; Ala, alanine; Asn, asparagine; Asp, aspartic acid; Gln, glutamine; Gly, glycine; Lys, lysine; Phe, phenylalanine; Pro, proline; Val, valine. Ala6 is linked to Lys19 as lysinoalanine. Ala-S-Ala: lanthionine, Ala-S-Abu: β-methyllanthionine, X2: lysine (duramycin) or arginine (cinnamycin) (4,5). (B) Rabbit erythrocytes (final 3 × 107 cells/ml) were incubated with various concentrations of duramycin for 30 min at 4°C or 37°C. Hemolysis was measured as described in Materials and Methods. (C) Duramycin was preincubated with various concentrations of MLVs composed of POPC, 90 mol % POPC and 10 mol % SOPE, or 90 mol % POPC and 10 mol % C18(plasm)-18:1 PE for 1 h at 37°C. The mixtures (final concentration of duramycin was 5 μM) were then added to rabbit erythrocytes (final 3 × 107 cells/ml) and further incubated for 30 min at 37°C, followed by the measurement of hemolysis. Horizontal axis indicates the final concentration of the total lipids in MLVs. (D) Duramycin was preincubated with MLVs containing 90 mol % POPC and 10 mol % of indicated lipids, followed by the measurement of hemolysis, as described in panel C. Final concentrations of duramycin and total lipids were 5 μM and 500 μM, respectively. Data are means ± SD of at least three independent experiments.
In eukaryotic cells, PE is mainly restricted to the inner leaflet of the plasma membrane (19–21). Recently we showed that cinnamycin induces transbilayer phospholipid movement of target cells in a PE-dependent manner (8). This causes exposure of the inner leaflet PE to the peptide and promotes binding of cinnamycin. When the surface concentration of PE is high, cinnamycin induces membrane reorganization such as membrane fusion and the alteration of the membrane gross morphology (8). However, the detailed membrane ultrastructure induced by cinnamycin binding is not well determined. Although duramycin was known to alter the membrane permeability of mammalian cells (18,22,23), the precise mechanism(s) of duramycin-induced membrane damage is not yet determined.
In this study, we examined the interaction of duramycin and cinnamycin with model membranes. The results indicate that both duramycin and cinnamycin selectively bind ethanolamine phospholipids, irrespective of whether they are of diacyl- or plasmalogen type. The binding of the lantibiotics induces reorganization of the membrane into highly curved tubular structures as revealed by electron microscopy and small-angle x-ray scattering (SAXS). In addition, we found that the binding of duramycin and cinnamycin to PE-containing liposomes is dependent on the curvature of the membrane, and the lantibiotics preferentially bind PE in the highly curved membranes. Thus, both duramycin and cinnamycin promote their binding to the membrane by inducing transbilayer movement and by changing membrane curvature.
MATERIALS AND METHODS
Materials
The following were purchased from Avanti Polar Lipids (Alabaster, AL):
L-α-phosphatidylcholine (egg, chicken; egg PC).
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC).
1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC).
1-palmitoyl-2-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]-sn-glycero-3-phosphocholine (C6-NBD-PC).
L-α-phosphatidylethanolamine (egg, chicken; egg PE). L-α-phosphatidylethanolamine (liver, bovine; liver PE).
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE).
1-stearoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (SOPE).
1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphoethanolamine (PAPE).
1-stearoyl-2-arachidonoyl-sn-glycero-3-phosphoethanolamine (SAPE).
1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphoethanolamine (SDPE).
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE).
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE).
1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE).
1-O-1′-(Z)-hexadecenyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (C16(plasm)-18:1 PE).
1-O-1′-(Z)-octadecenyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (C18(plasm)-18:1 PE).
1-oleoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine (18:1 lyso-PE).
1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (N-NBD-PE).
L-α-phosphatidylserine (brain, porcine; brain PS).
1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-L-serine] (POPS).
L-α-phosphatidylinositol (liver, bovine; liver PI).
L-phosphatidyl-DL-glycerol (egg, chicken; egg PG).
L-α-phosphatidic acid (egg, chicken; egg PA).
sn-(3-oleoyl-2-hydroxy)-glycerol-1-phospho-sn-3′-(1′-oleoyl-2′-hydroxy)-glycerol (18:1 BMP).
cardiolipin (heart, bovine; heart CL).
(2S,3R,4E)-2-acylaminooctadec-4-ene-3-hydroxy-1-phosphocholine (egg, chicken; egg SM).
(2S,3R,4E)-2-acylaminooctadec-4-ene-3-hydroxy-1-phosphocholine (brain, porcine; brain SM).
Total cerebrosides (brain, porcine; GalCer).
From Matreya (Pleasant Gap, PA):
Glucosylceramide (human; GlcCer), lactosylceramide (LacCer), and monogalactosyl diglyceride (plant, hydrogenated; 18:0 MGDG).
From Larodan Fine Chemicals (Malmö, Sweden):
Monogalactosyl diglyceride (plant MGDG).
From Wako Pure Chemical Industries (Osaka, Japan):
Ganglioside GM1 (bovine brain; GM1), ganglioside GM2 (NeuAc) (bovine brain; GM2), and ganglioside GM3 (NeuAc) (bovine; GM3).
From Sigma (St. Louis, MO):
1,2-didodecanoyl-sn-glycero-3-phosphoethanolamine (DLPE), 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine (DMPE), and duramycin and cinnamycin.
From Nacalai Tesque (Kyoto, Japan):
Sodium hydrosulfite (sodium dithionite), o-phenylenediamine, and Dulbecco's phosphate-buffered saline (−) (PBS). (The PBS, pH 7.4, contained 200 mg/l potassium chloride, 200 mg/l potassium dihydrogenphosphate, 8000 mg/l sodium chloride, and 1150 mg/l di-sodium hydrogenphosphate.)
From CovalAb (Lyon, France):
Rabbit polyclonal antisera against duramycin.
Methods
Preparation of lipid vesicles
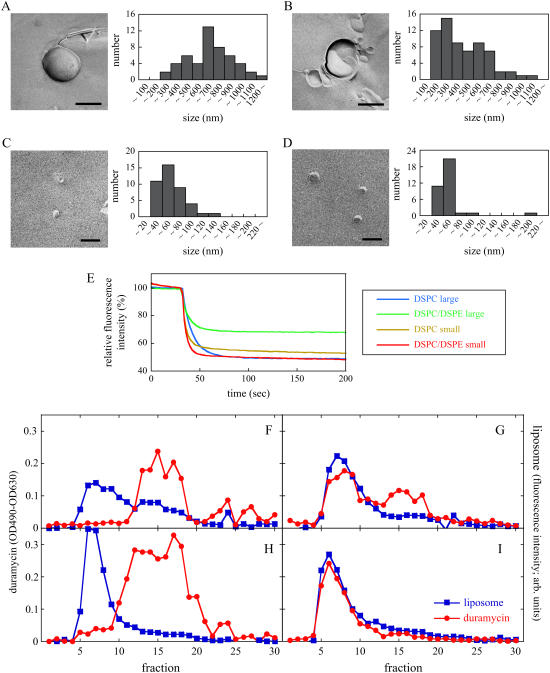
Multilamellar vesicles (MLVs) were prepared by hydrating a lipid film with 20 mM HEPES-NaOH (pH 7.4) and 100 mM NaCl, unless otherwise indicated, and vortex mixing. To prepare large vesicles, MLVs were subjected to extrusion through polycarbonate filters (Nucleopore, Maidstone, UK) for 25 times using a two-syringe extruder (Avanti Polar Lipids, Alabaster, AL). To prepare small vesicles, MLVs were subjected to sonication using a XL-2020 sonicator (Misonix, Farmingdale, NY) until peak diameters (nm) of the size distribution by number became <50 nm. The vesicle size was examined by dynamic light scattering measurements at 37°C using a Zetasizer Nano ZS (Malvern, Worcestershire, UK). In Fig. 5, the vesicles were also examined by freeze-fracture electron microscopy.
FIGURE 5.
Curvature-dependent binding of duramycin to PE-containing membranes. Small and large vesicles (1 mM total phospholipids) containing 1 mol % N-NBD-PE were prepared by sonication and by extrusion through polycarbonate filters with 1.0-μm pore size, respectively. (A–D) Representative freeze fracture images and the size distribution of large (A and B) and small (C and D) vesicles composed of DSPC (A and C) and DSPC/DSPE (9:1) (B and D) containing 1 mol % N-NBD-PE are shown. Bar, 500 nm (A and B) and 100 nm (C and D). (E) Sodium hydrosulfite (final 10 μM) was added to 50 μM (total lipids) vesicles prepared above and the fluorescence was monitored while stirring at 25°C, as described in Materials and Methods. The percentage of the phospholipid residing in the outer leaflet of the most external layer of liposomes was estimated from the decrease of fluorescence. (F–I) Large (F and G) and small (H and I) vesicles composed of DSPC (F and H) and DSPC/DSPE (9:1) (G and I) containing 1 mol % N-NBD-PE were incubated with 30 μM duramycin for 30 min at 37°C. In this assay, duramycin/(phospholipids in the outer leaflet of most external layer of liposomes) ratio was adjusted to 1:10, using the results of panel E. After incubation, the liposome-bound duramycin was separated from free duramycin by gel filtration as described in Materials and Methods. The amounts of liposomes and duramycin in each fraction are shown. Data are representatives of three independent experiments.
Measurement of the amount of phospholipids residing in the outer leaflet of the most external layer of liposomes
Two milliliters of the liposome suspensions (total 50 μM phospholipids) containing 1 mol % N-NBD-PE were mixed with 20 μl of 1 mM sodium hydrosulfite at 25°C and the fluorescence was measured using an FP-6500 spectrofluorometer (Jasco, Tokyo, Japan) with excitation and emission wavelengths at 475 and 535 nm, respectively. Sodium hydrosulfite selectively quenches N-NBD-PE, which is localized in the outer leaflet of the most external layer of the liposomes (24). The fluorescence was monitored until it reached equilibrium. The percentage of the phospholipid residing in the outer leaflet of the most external layer of liposomes was estimated from the decrease of fluorescence.
Measurement of hemolysis
Rabbit erythrocytes were prepared by washing rabbit whole blood (Nippon Bio-Supply Center, Tokyo, Japan) with PBS. Measurement of hemolysis of rabbit erythrocytes was performed as described previously (25). Zero-percent hemolysis was determined by incubating rabbit erythrocytes (3 × 107 cells/ml) on ice for 30 min, whereas 100% hemolysis was measured after three freeze-thaw cycles of erythrocytes. Inhibitory effects of various lipid vesicles on hemolytic activity of duramycin were examined as described previously (25) with some modifications. In brief, 40 μl of duramycin solution in PBS and 40 μl of liposome suspensions in 20 mM HEPES-NaOH (pH 7.4) with or without 100 mM NaCl were mixed and incubated for 1 h at 37°C or at 4°C. After the addition of 160 μl of rabbit erythrocyte suspensions in PBS, the resultant mixtures were incubated for 30 min at 37°C, and then the hemolysis was measured. Final concentrations of erythrocytes, duramycin and liposomes in the mixture are indicated in the figure legends. In the case of using large vesicles or small vesicles, the amount of phospholipids in liposomes was determined by measuring phosphorus content (26).
High-sensitivity titration calorimetry
Isothermal titration calorimetry (ITC) was performed using a Microcal VP-ITC titration calorimeter (MicroCal, Northampton, MA) as described previously (27) with some modifications. Duramycin was titrated with vesicles at 37°C. Injection volumes were 8 μl. The calorimeter cell had a reaction volume of 1.4034 ml. As duramycin was known to give a positive Biuret test (2), the concentrations of duramycin were measured by using BCA Protein Assay Reagent (Pierce Biotechnology, Rockford, IL) and/or by determining dry weight of duramycin. The lipid concentrations of liposomes were determined by measuring phosphorus content by phosphorus assay (26). The heats of dilution were determined in the control experiments using the buffer (20 mM HEPES-NaOH (pH 7.4) and 100 mM NaCl) in place of duramycin solutions, and were subtracted from the heats determined in the corresponding duramycin-lipid binding experiments. Binding constants (Ka) of duramycin and ethanolamine phospholipids were calculated as
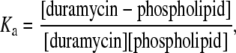 |
assuming duramycin and ethanolamine phospholipid form 1:1 complex (see Results). Binding constants were estimated from ITC results by curve-fitting analysis with “One Set of Sites” model from Origin, Ver. 5.0 (MicroCal, Northampton, MA).
Liposome binding assay using gel filtration
Various liposomes were incubated with duramycin for 30 min at 37°C in 20 mM HEPES-NaOH (pH 7.4), 100 mM NaCl buffer. After incubation, liposome-bound duramycin was separated from free duramycin by gel filtration as described previously (29,30) with some modifications. Five milliliters polypropylene column (Pierce Biotechnology) was filled with 3 ml of Bio-Gel A-15m Gel (Bio-Rad, Hercules, CA) equilibrated with the buffer. After applying 100 μl of the reaction mixture, 200 μl of the buffer was added and the eluent was collected. This step was repeated until majority of duramycin was eluted from the column. Each fraction was analyzed for the amounts of liposomes and duramycin. For quantification of the liposomes, 50 μl of each fraction was diluted with 50 μl of the buffer, followed by the addition of 10 μl 10% Triton X-100. The fluorescence was measured using an ARVO SX Multilabel Counter (Wallac, Turku, Finland) with excitation and emission wavelengths at 485 and 535 nm, respectively. Duramycin was quantified by enzyme-linked immunosorbent assay.
Enzyme-linked immunosorbent assay of duramycin
A fifty-microliter sample was added to each well of an Immulon 2HB (Thermo Fisher Scientific, Waltham, MA) microtiter plate. After overnight absorption at 4°C, the unbound material was washed with Tris-buffered saline (TBS; 10 mM Tris-HCl, pH7.4, 150 mM NaCl). Two-hundred microliters of 30 mg/ml bovine serum albumin (Fraction V; Sigma, St. Louis, MO) in TBS was then added to each well. After 2-h incubation at room temperature, the wells were washed with TBS. The bound duramycin was detected by adding anti-duramycin antiserum followed by incubation with ECL anti-rabbit IgG, horseradish peroxidase-linked species-specific whole antibody (from donkey; GE Healthcare UK, Buckinghamshire, UK). The intensity of the color developed with o-phenylenediamine as a substrate was measured with a Microplate Reader model 680 (Bio-Rad), reading the absorption at 490 nm with reference at 630 nm.
Electron microscopy
Negative staining was performed as reported previously (8) with some modifications. MLVs containing 2 mM total lipids were incubated with various concentrations of duramycin for 30 min at 37°C. After centrifugation at 19,000 × g for 30 min at 4°C, the pellet was suspended in the same volume of the buffer containing 20 mM HEPES-NaOH (pH 7.4) and 100 mM NaCl buffer, fixed with 2.5% glutaraldehyde for 30 min at room temperature and washed three times with the same buffer by centrifugation at 19,000 × g for 30 min at 4°C. For negative staining images, the suspensions were adsorbed onto poly-D-lysine-treated formvar-coated grids and negatively stained with 2% sodium phosphotungstic acid. For freeze fracture images, the samples were frozen in liquid propane cooled by liquid nitrogen, fractured in a freeze-etching machine (Balzers BAF400T, Balzers, Liechtenstein) at −110°C, and replicated by platinum/carbon. Replicated samples were immersed in household bleach to dissolve the lipids, washed in water, and then mounted on formvar-coated copper grids. Both specimens for negative staining and freeze fracture images were examined under transmission electron microscope (Tecnai 12, Philips, Eindhoven, The Netherlands, or 1200EX-II, JEOL, Tokyo, Japan). Electron micrographs recorded on imaging plates were scanned and digitized by an FDL 5000 imaging system (Fuji Photo Film, Tokyo, Japan).
SAXS measurements
SAXS measurements were carried out at RIKEN Structural Biology Beamline I (BL45XU) at SPring-8, 8 GeV synchrotron radiation source (Hyogo, Japan) (31). The x-ray wavelength used was 0.9 Å and the beam size at the sample position was ∼0.4 × 0.7 mm2. The distance of sample-to-detector was 968 mm. Samples were measured in a sample cell with a path length of 1.5 mm and a pair of thin quartz windows (30 μm thickness). The sample temperature was controlled to 37 ± 0.01°C with a high precision thermoelectric device. The samples were allowed to equilibrate for at least 5 min and the diffraction collected with 2 min exposure. Buffer profiles were also taken for background subtraction purposes. The SAXS patterns were recorded with an imaging plate (30 × 30 cm2) system of RIGAKU R-Axis IV2+ (32). The two-dimensional scattering patterns were circularly averaged and reduced to one-dimensional profiles using FIT2D, Ver. 12.012 (http://www.esrf.fr/computing/scientific/FIT2D/), a two-dimensional data reduction and analysis program. The reciprocal spacing (s) and scattering vectors (q),
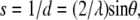 |
(1) |
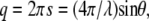 |
(2) |
where d is the lattice spacing, 2θ is the scattering angle, and λ is the wavelength of x ray, were calibrated with silver behenate by the long-period spacing of 5.838 nm (34).
Modeling analysis of SAXS data
Small-angle scattering intensity, I(q), can be described as
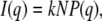 |
(3) |
where k is an instrument constant, N is the number density of particles, and P(q) is the particle scattering function. Of possible structural types tested, the SAXS data of duramycin-membrane complexes was well fitted by a concentric cylindrical shell model. The scattering function of a randomly oriented cylindrical particle composed of ith shells is expressed as
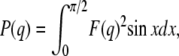 |
(4) |
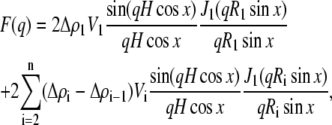 |
(5) |
where F(q) is the form factor of the cylinder with ith shells and 2H is the length of the cylinder. The values Δρi, Vi, and Ri correspond to the average excess electron density (contrast), volume, and radius of ith shell, respectively. The value x is the angle between the longest particle axis and the scattering vector q, and J1 corresponds to the first-order Bessel function. Based on the observation by the electron microscopy, the length of the straight part of the duramycin-membrane rod was estimated to be ∼150 nm, which is much larger than the rod radius (∼10 nm) and is out of limit of our experimental resolution. Therefore, we set the 2H value with 150 nm for this fitting analysis. In case of such large H, the axial factor in Eq. 4 drops to zero very rapidly, unless those orientations where x is very small. This means that the rods make a contribution to the scattering only when they are nearly perpendicular to the scattering vector q. That is, the I(q) remains nearly unaltered with a slight change of H (35,36) in q that we observed.
RESULTS
Duramycin specifically binds ethanolamine phospholipids
Whereas the specific binding of cinnamycin to PE is established, the lipid specificity of duramycin is obscure. Previously it was proposed that duramycin recognizes a particular membrane conformation determined by the presence of PE or MGDG (15). Studies on duramycin-resistant bacteria suggest that duramycin binds to PE (15–17), but not to ethanolamine plasmalogen (17). In the present study, we examined the lipid specificity of duramycin by 1), examination of the inhibitory effects of liposomes from various lipids on the toxicity of duramycin; and 2), heat measurement during interaction of duramycin and the liposomes by ITC. Similar to cinnamycin (6), duramycin exhibited temperature-dependent hemolytic activity against rabbit erythrocytes (Fig. 1 B). When duramycin was preincubated with POPC MLVs, the hemolytic activity was not affected (Fig. 1, C and D). In contrast, preincubation with PE-containing MLVs inhibited the hemolytic activity, suggesting that duramycin binds PE (Fig. 1, C and D). Similar inhibitory effects were observed with ethanolamine plasmalogen-containing MLVs (Fig. 1, C and D). Similar to duramycin, cinnamycin-induced hemolysis was inhibited by PE- and ethanolamine plasmalogen-containing liposomes (Supplementary Material, Fig. S1), suggesting that cinnamycin also binds the plasmalogen form of ethanolamine phospholipids. In Fig. 1 D, we examined the effects of MLVs containing 90 mol % POPC and 10 mol % of various lipids on duramycin-induced hemolysis. Hemolysis was inhibited by the presence of PEs with different fatty acids, ethanolamine plasmalogens, and lyso-PE. However, other lipids, including MGDG, did not affect the hemolysis, suggesting that duramycin specifically binds ethanolamine phospholipids.
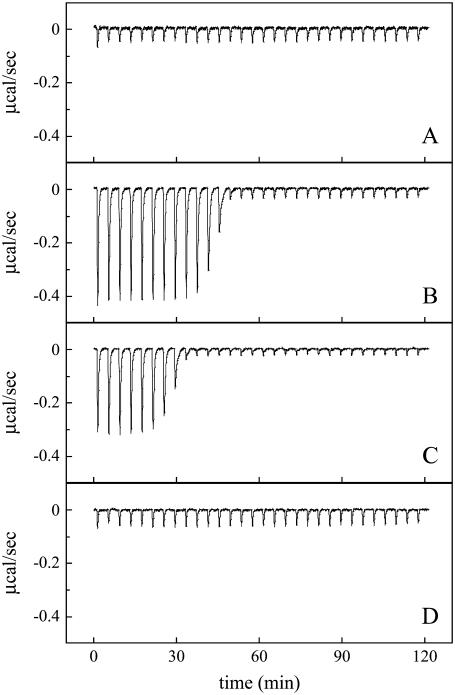
The interaction of duramycin with ethanolamine phospholipids was then examined by ITC. Whereas injections of POPC (Fig. 2 A) or POPC/MGDG (9:1) (Fig. 2 D) liposomes to duramycin solution exhibited only slight exothermic reactions, each injection of POPC/POPE (9:1) (Fig. 2 B) or POPC/C16(plasm)-18:1 PE (9:1) (Fig. 2 C) liposome suspension caused a distinct exothermic reaction. The reaction enthalpy ΔH° was calculated from the ITC profile. In Fig. 2 B, the total amount of duramycin in the sample cell was 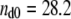 nmol and the total heat measured until the equivalent point was
nmol and the total heat measured until the equivalent point was 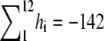 μcal. The reaction enthalpy was thus calculated as ΔH° =
μcal. The reaction enthalpy was thus calculated as ΔH° = 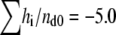 kcal/mol duramycin. Similarly, the reaction enthalpy ΔH° was calculated as ΔH° =
kcal/mol duramycin. Similarly, the reaction enthalpy ΔH° was calculated as ΔH° = 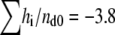 kcal/mol duramycin for POPC/C16(plasm)-18:1 PE liposome suspension (Fig. 2 C). In Fig. 2 B, the amount of PE injected in the first 12 steps was 53.3 nmol. Thus, duramycin/PE ratio was 1:1.89 when the peptide was consumed. Cinnamycin is reported to form 1:1 complex with PE (37,38). Assuming duramycin also forms 1:1 complex, our results suggest that only the PE in the outer leaflet is accessible to duramycin when the PE content was 10 mol %. When the PE content of POPC/POPE liposomes exceeded 20 mol %, duramycin bound more than 50 mol % of the total PE (data not shown), as reported on cinnamycin (38). The binding constant of duramycin and POPE and that of duramycin and C16(plasm)-18:1 PE were estimated as Ka = (2.1 ± 0.4) × 108 M−1 and Ka = (1.1 ± 0.2) × 108 M−1, respectively.
kcal/mol duramycin for POPC/C16(plasm)-18:1 PE liposome suspension (Fig. 2 C). In Fig. 2 B, the amount of PE injected in the first 12 steps was 53.3 nmol. Thus, duramycin/PE ratio was 1:1.89 when the peptide was consumed. Cinnamycin is reported to form 1:1 complex with PE (37,38). Assuming duramycin also forms 1:1 complex, our results suggest that only the PE in the outer leaflet is accessible to duramycin when the PE content was 10 mol %. When the PE content of POPC/POPE liposomes exceeded 20 mol %, duramycin bound more than 50 mol % of the total PE (data not shown), as reported on cinnamycin (38). The binding constant of duramycin and POPE and that of duramycin and C16(plasm)-18:1 PE were estimated as Ka = (2.1 ± 0.4) × 108 M−1 and Ka = (1.1 ± 0.2) × 108 M−1, respectively.
FIGURE 2.
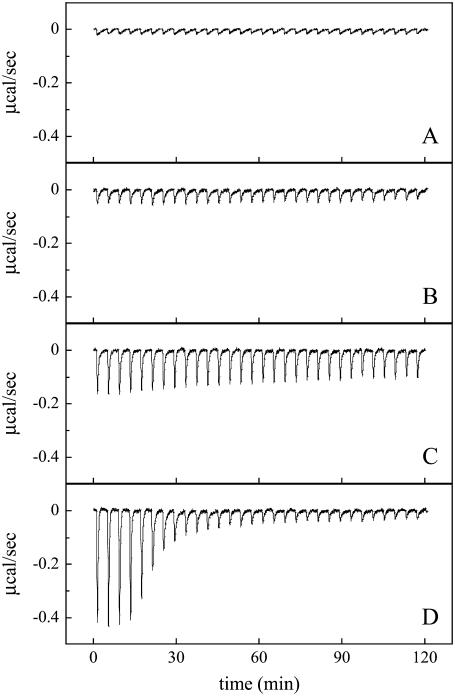
Duramycin specifically interacts with ethanolamine phospholipids. Lipid vesicles were prepared by extrusion through polycarbonate filters with 100-nm pore size. ITC was performed as described in Materials and Methods. The values 20.1 μM (A, B, and D) or 13.7 μM (C) duramycin in the reaction cell (1.4034 ml) was titrated with 5.30 mM POPC (A), 5.55 mM POPC/POPE (9:1) (B), 5.05 mM POPC/C16(plasm)-18:1 PE (9:1) (C) or ∼6 mM POPC/plant MGDG (9:1) (D) at 37°C. Each peak corresponds to the injection of 8 μl of liposomes. Data are representatives of three independent experiments (A and B).
The binding of duramycin and cinnamycin to PE is dependent on the physical properties and the curvature of the membrane
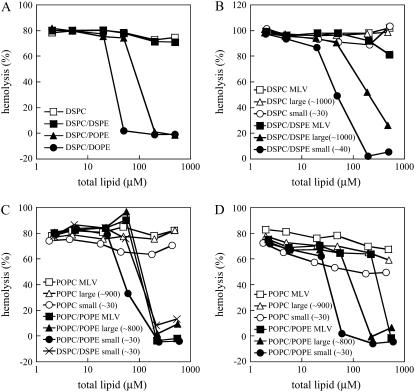
We examined the effect of physical properties of PE-containing liposomes on the inhibition of duramycin-induced hemolysis. As shown in Fig. 1 D, POPC/DSPE MLVs inhibited the hemolytic activity of duramycin. In contrast, DSPC/DSPE (9:1) MLVs did not inhibit duramycin-induced hemolysis (Fig. 3 A). DSPC/POPE (Tm = 20°C (39)) and DSPC/DOPE (Tm = −16°C (39)) inhibited hemolysis (Fig. 3 A). It is speculated that DSPC (Tm = 54.5°C)/DSPE (Tm = 74°C) (39) provides tightly packed surfaces at 37°C. These results suggest that the binding of duramycin to PE is dependent on the physical properties of the membrane. In Fig. 3 B, the effect of membrane curvature of DSPC/DSPE liposomes on the inhibition of duramycin-induced hemolysis was examined. The parentheses in the figure legend indicate the diameters of the liposomes examined by dynamic light scattering. In contrast to MLVs, both small vesicles and large vesicles inhibited hemolysis, small vesicles being more effective than large vesicles. Similar results were obtained with cinnamycin (Supplementary Material, Fig. S2).
FIGURE 3.
Inhibition of duramycin-induced hemolysis by PE-containing liposomes is dependent on the physical properties of the membrane. Large and small vesicles were prepared by extrusion through polycarbonate filters with 1.0-μm pore size and by sonication, respectively. Duramycin was preincubated with various concentrations of liposomes for 1 h at 37°C (A–C) or at 4°C (D). The liposomes were composed of indicated lipids and contained 10 mol % PE when included. Peak diameters (nm) of the vesicles evaluated by dynamic light scattering are given in parentheses. After the preincubation, the mixtures (final concentration of duramycin was 5 μM) were then added to rabbit erythrocytes (final 3 × 107 cells/ml) and incubated for 30 min at 37°C. Hemolysis was measured as described in Materials and Methods. The horizontal axis indicates the final concentrations of total lipids in liposomes.
The curvature-dependent binding of duramycin to DSPC/DSPE membranes was then measured by ITC. Injections of DSPC/DSPE (9:1) large vesicles of ∼700 nm diameters into duramycin solution resulted in slight exothermic reactions (Fig. 4 B). In contrast, injections of DSPC/DSPE small vesicles of ∼40 nm diameters revealed distinct exothermic reactions until the sixth injection (Fig. 4 D). We also found that injections of DSPC small vesicles revealed, to some extent, exothermic reactions, whereas those of DSPC large vesicles did not (Fig. 4, A and C). We next investigated whether duramycin preferentially binds highly curved membranes when lipids contain unsaturated fatty acid. Fig. 3 C shows the inhibition of duramycin-induced hemolysis by liposomes with different sizes. POPC/POPE small vesicles were slightly more effective than large vesicles and MLVs when the liposomes were preincubated with duramycin at 37°C. When POPC/POPE liposomes were preincubated with duramycin at 4°C, a clear difference was observed in the inhibitory effects of small vesicles, large vesicles, and MLVs with the small vesicles showing the most prominent effect (Fig. 3 D). When duramycin was titrated with POPC/POPE (9:1) large vesicles, the peak diameter of which was evaluated as ∼700 nm by dynamic light scattering, the binding constant of duramycin to POPE was estimated to be Ka = (4.0 ± 1.9) × 107 M−1 from the ITC result (data not shown). This value is five times smaller than that of the titration with POPC/POPE vesicles in Fig. 2 B. The peak diameter of the POPC/POPE vesicles used in Fig. 2 B was evaluated as ∼100 nm by dynamic light scattering. These results suggest that duramycin preferentially binds PE in fluid and highly curved membranes.
FIGURE 4.
Curvature-dependent interaction between duramycin and PE-containing membranes. Small and large vesicles were prepared by sonication and by extrusion through polycarbonate filters with 1.0-μm pore size, respectively. ITC was performed as described in Materials and Methods. The values 22.1 μM (A–C) or 26.0 μM (D) duramycin in the reaction cell (1.4034 ml) was titrated with 6.64 mM DSPC large vesicles (A), 6.21 mM DSPC/DSPE (9:1) large vesicles (B), 6.70 mM DSPC small vesicles (C), or 6.50 mM DSPC/DSPE small vesicles (D) at 37°C. Peak diameters (nm) of the liposomes evaluated by dynamic light scattering were ∼800 nm (A), ∼700 nm (B), ∼30 nm (C), and ∼40 nm (D), respectively. Each peak corresponds to the injection of 8 μl of liposome suspension. Data are representatives of two independent experiments.
In Fig. 5, A–D, large vesicles and small vesicles were further characterized by freeze-fracture electron microscopy. Freeze fracture replica indicates that most of the small vesicles are unilamellar (Fig. 5, C and D), whereas large vesicles sometimes contain several layers (Fig. 5, A and B). The average of diameter of DSPC/DSPE small vesicles was estimated to be 49.8 nm whereas that of large vesicles was 410 nm (Supplementary Material, Table S1). Dynamic light scattering estimated the diameters of the same samples as 34.1 nm (small vesicles) and 797 nm (large vesicles), respectively (Supplementary Material, Table S2). Although the sizes of small vesicles were comparable between two methods, freeze fracture replica gave smaller size of large liposomes than dynamic light scattering. This could be because specimens were not always fractured in the plane of the center. In Fig. 5 E, the amount of phospholipids in the outer leaflet of most external layer of liposomes was estimated by measuring the quenching of N-NBD-PE incorporated into liposomes with sodium hydrosulfite. N-NBD-PE becomes nonfluorescent when reduced with sodium hydrosulfite. Since sodium hydrosulfite penetrate membranes very slowly, it is possible to estimate the amount of N-NBD-PE on the outer leaflet of most external layer of liposomes using this method. Fig. 5 E indicates that almost 50% of N-NBD-PE was quenched by sodium hydrosulfite in small vesicles, indicating that most of small vesicle preparations were unilamellar as shown in freeze fracture replica. In contrast, only 30% of N-NBD-PE fluorescence was quenched in DSPC/DSPE large vesicles. This result indicates that DSPC/DSPE large vesicles are in average 1.5 layers in these liposome preparations. In Fig. 5, F–I, the binding of duramycin to small and large liposomes was directly measured. In this assay, duramycin/(phospholipids in the outer leaflet of most external layer of liposomes) ratio was adjusted to 1:10. After incubation with liposomes, the mixture was separated by a Bio-Gel A-15m gel filtration column (BioGel Company, Cleveland, OH). Both small and large liposomes were not retained in the column and recovered in the void fraction whereas free duramycin was retained in the column (data not shown). When duramycin was incubated with DSPC liposomes, duramycin and liposomes were segregated after gel filtration, irrespective of the size of liposomes (Fig. 5, F and H). In contrast, when duramycin was incubated with DSPC/DSPE small vesicles, duramycin eluted as a single peak together with the liposomes, indicating the binding of duramycin to the small liposomes (Fig. 5 I). The incubation of duramycin with DSPC/DSPE large vesicles resulted in the recovery of duramycin in both bound and unbound fraction (Fig. 5 G). These results suggest that duramycin preferentially bind to highly curved membrane.
Duramycin alters the organization of ethanolamine phospholipid-containing model membranes
Previously, we showed that cinnamycin alters the structure of PE-containing liposomes and induces leakage of the contents and exposure of the lipids of the inner leaflet of the liposomes (8). Using duramycin, we confirmed that duramycin also exhibits these properties (Supplemental Fig. S3). These results indicate that duramycin alters the structure of ethanolamine phospholipid-containing membranes, as observed for cinnamycin. Duramycin-induced alteration of membranes was further studied using negative staining and freeze fracture electron microscopy. Whereas duramycin did not alter the structure of POPC MLVs (Fig. 6, C and E), it dramatically deformed POPC/POPE (9:1) MLVs (Fig. 6, D and F) and POPC/C16(plasm)-18:1 PE (9:1) MLVs (data not shown) into a rodlike structure with branching and inflection points. In addition, sometimes small (diameter 20–25 nm) round shape images were obtained in freeze fracture micrograph (Fig. 6 F). Although the image in Fig. 6 D could be interpreted as flattened disks or vesicles, Fig. 6 F suggests the possibility that duramycin induces the tubules with 20–25 nm diameter. This structure was further supported by SAXS measurement (see below). When POPC/POPE and POPC/C16(plasm)-18:1 PE MLVs were treated with lower concentrations of duramycin, i.e., 100 and 40 μM, the rodlike structure was also observed, but at a lower frequency (data not shown). In contrast, when treated with 500 μM duramycin, the rodlike shape was the major structure. The average length of the rod structure between inflection points was 150 nm (n = 193) and the maximum length observed was >1 μm. The average width was 21 nm (n = 177). A similar structure was observed when PE-containing liposomes were treated with cinnamycin (8). However, detailed analysis of the rodlike structure had yet to be performed.
FIGURE 6.
Duramycin deforms PE-containing membranes into a rodlike structure. Two millimolar (total lipids) MLVs composed of POPC (A, C, and E) and POPC/POPE (9:1) (B, D, and F) were incubated in the absence (A and B) or the presence (C–F) of 500 μM duramycin for 30 min at 37°C. (A–D) Negative staining images, and (E, F) freeze fracture replica. Arrows indicate round-shape images, whose diameters were 20–25 nm. Bar, 100 nm.
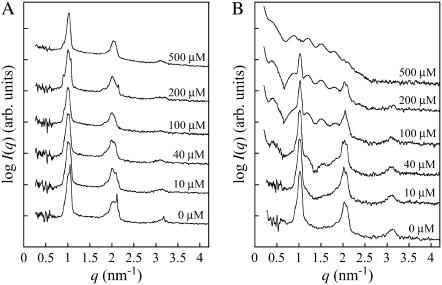
To further explore the ultrastructure of the duramycin-induced rodlike structure, we measured SAXS of MLV suspensions that were treated with various concentrations of duramycin. The SAXS of POPC and POPC/POPE (9:1) MLVs displayed prominent periodic peaks (Fig. 7) corresponding to lamellar distances of 6.28 and 5.90 nm, respectively. The lamellar pattern of POPC MLVs was not affected by duramycin up to at least 500 μM (Fig. 7 A). In contrast, that of POPC/POPE MLVs was dramatically altered by duramycin treatment, i.e., the lamellar structure-derived peaks decreased with increases in the duramycin concentration and, moreover, new scattering curves appeared (Fig. 7 B). These scattering curves, which have well-defined maxima, are attributable to the form factor from the rodlike structure observed in electron microscopy. Similar alteration of the SAXS pattern by duramycin treatment was also observed in POPC/SAPE, POPC/SDPE, POPC/C16(plasm)-18:1 PE, POPC/1-O-1′-(Z)-octadecenyl-2-arachidonoyl-sn-glycero-3-phosphoethanolamine (C18(plasm)-20:4 PE), POPC/1-O-1′-(Z)-octadecenyl-2-docosahexaenoyl-sn-glycero-3-phosphoethanolamine (C18(plasm)-22:6 PE), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)/DOPE, and DOPC/C18(plasm)-18:1 PE MLVs (data not shown). The SAXS pattern of cinnamycin-treated POPC/POPE membrane also gave similar results (data not shown).
FIGURE 7.
Duramycin alters SAXS pattern of PE-containing membranes. Two millimolar (total lipids) MLVs composed of POPC (A) and POPC/POPE (9:1) (B) were incubated with indicated concentrations (0–500 μM) of duramycin for 30 min at 37°C before SAXS measurements. All measurements were performed at 37°C. SAXS measurement was performed as described in Materials and Methods. The splitting of the lamellar structure-derived peak in panel A could be the cation-induced phase separation of POPC in liquid crystalline Lα phase (52). While the lamellar peak pattern of POPC MLVs was not affected by duramycin, the pattern of POPC/POPE MLVs was dramatically altered by duramycin treatment.
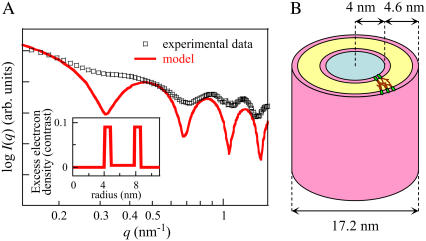
To obtain quantitative insight into the organization of the complexes of duramycin and the POPC/POPE membrane, we analyzed the SAXS data with a model fitting technique. Based on the observation by electron microscopy, it was assumed that the duramycin-membrane complexes form rodlike structure with a ∼20 nm diameter. Moreover, the SAXS patterns in Fig. 7 B exhibit well-defined maxima, suggesting that the size distribution of the rod diameter is narrow. Fig. 8 A shows the result of the fitting and the radial excess electron density profile of the best fit model. Although neither homogeneous solid rod nor core/shell type hollow cylinder models agreed with the experimental data (data not shown), the model of a hollow cylinder with multishells simulated the data very well (Fig. 8 A). This model fitting suggests that the structure formed by duramycin and the POPC/POPE membrane is not a solid rod with a homogeneous electron density, but rather, a hollow tubule consisting of a POPC/POPE bilayer (Fig. 8 B).
FIGURE 8.
Model-fitting analysis of SAXS data. (A) The experimental SAXS data of POPC/POPE (9:1) membrane incubated with 500 μM duramycin were fitted with the theoretical model function. The inset shows the radial excess electron density (contrast, Δρ) map of the best-fit model. The zero level corresponds to the solvent electron density (ρwater = 0.33 electrons/Å3). (B) A schematic tubular model formed by duramycin and POPC/POPE membrane. The core region (light blue), which has zero contrast, corresponds to water. The two high contrast regions (pink) and the intermediate region (yellow) correspond to the hydrophilic and hydrophobic regions of the lipid bilayer, respectively. Duramycin would penetrate into the hydrophobic region (yellow) (see Discussion).
DISCUSSION
Duramycin and cinnamycin specifically bind ethanolamine phospholipids
Duramycin was reported to induce aggregation of PE or MGDG-containing liposomes (15). Duramycin also induces high membrane conductance of PE-containing black lipid membranes (18). These results suggest that the target of duramycin is PE and/or MGDG. Consistent with this, several independently isolated duramycin-resistant mutants of Bacillus strains revealed remarkable reduction of PE and cardiolipin (15–17). One of the mutants, instead, contained plasmenylethanolamine (ethanolamine plasmalogen), which was not detected in the parental strain, B. firmus (17). Plasmalogen is a glycerophospholipid that has an alk-1′-enylether bond at position sn-1. These reports raise the possibility that duramycin binds to PE, but not to the plasmalogen form of PE. In this study, we examined the binding of duramycin to various lipids indirectly by examination of the inhibitory effects of lipids on duramycin-induced hemolysis and by measuring the heat using ITC. The results indicate that duramycin binds both the diacyl- and plasmalogen-form of ethanolamine phospholipids. We did not observe the interaction of duramycin with MGDG. It has not been examined whether cinnamycin binds ethanolamine plasmalogen. Our results indicate that cinnamycin also binds the diacyl- and plasmalogen-form of ethanolamine phospholipids.
The binding of the lantibiotics to PE is dependent on the curvature of the membrane
Whereas duramycin-induced hemolysis was inhibited by small vesicles composed of DSPC/DSPE, MLVs with the same lipid composition did not affect the hemolysis. ITC studies revealed that duramycin interacts with ∼40 nm DSPC/DSPE vesicles, but not with ∼700 nm vesicles. Curvature-dependent inhibition by PE-containing liposomes was observed both in duramycin- and cinnamycin-induced hemolysis. These results suggest that the binding to PE-containing liposomes is dependent on membrane curvature and the lantibiotics preferentially bind to highly curved membrane. This idea was further supported by the direct measurement of the binding of duramycin to different liposomes. Previously it was shown that both the hydrophilic headgroup and hydrophobic tail of PE are required for the recognition by cinnamycin (6,7,40). It is speculated that the hydrophobic region of PE is more easily exposed to duramycin and cinnamycin in small vesicles than in large vesicles.
The titration of DSPC small vesicles to duramycin revealed, to some extent, an exothermic pattern, whereas that of DSPC large vesicles did not. These results imply that duramycin weakly interacts with the highly curved lipid bilayer irrespective of the lipid composition. Since DSPC small vesicles did not exhibit inhibitory effects on duramycin-induced hemolysis, this weak interaction is reversible and is thus distinct from the interaction between duramycin and PE.
Duramycin and cinnamycin induce membrane tubulation
The lantibiotics not only bind to PE in high curvature membranes but also induce high curvature upon binding. According to the obtained model in Fig. 8, the radius of the core region, which has zero contrast, corresponding to water, is 4 nm and the thickness of the three shells is in total 4.6 nm (two 0.8-nm headgroup regions and a 3-nm hydrophobic region). These obtained dimensions of the hydrophilic and hydrophobic regions on the membrane are fairly consistent with the dimensions previously reported on the POPC and POPE membranes (41–43). The estimated diameter of the tubular structure of the duramycin-membrane complex is 17.2 nm. This value is smaller than the value observed with electron microscopy (21 nm). However, considering a widening deformation of the duramycin-membrane tubules adsorbed on the grid during the sample preparation in electron microscopy, this value (17.2 nm) may be reasonable for the actual diameter in the aqueous solution. For duramycin and cinnamycin, the electron density estimated based on the partial molar volumes of the amino acids (44,45) was ∼0.43 electrons/Å3. Since the electron density of the lantibiotics is close to that of the POPC/POPE headgroups (∼0.42 electrons/Å3) (46–48), the presence of the lantibiotic in the hydrophilic region is difficult to evaluate in the present analysis. On the other hand, the contrast in the hydrophobic region differs significantly from the expected value for lipid acyl chains. Generally, the hydrophobic region of the membrane shows negative contrast since the electron density of hydrocarbon chains is lower than that of water. However, in the present analysis, the model which has a contrast corresponding to the electron density of the acyl chains of phospholipids (∼0.16–0.3 electrons/Å3) (46–48) in the hydrophobic region did not fit to the experimental data (data not shown). The best-fit model in this analysis showed a higher density in the hydrophobic region, which is even higher than that of water (0.33 electrons/Å3) (Fig. 8 A). The high contrast in the hydrophobic region can be interpreted as a penetration of duramycin, which has a high electron density, into the hydrophobic region of the membrane. Recent results of Machaidze and Seelig (40) indicate that both the PE headgroup and hydrocarbon chains are important in the binding of cinnamycin to PE. In both cinnamycin and duramycin, lipophilic amino acids are positioned at one side of the peptide, whereas the hydrophilic ones are located on the opposite side (3,4). The results of Machaidze and Seelig suggest that the first 8–10 segments of each hydrocarbon chain of PE are in direct contact with a hydrophobic peptidic surface of cinnamycin. Since the hydrophobic amino acids are identical between cinnamycin and duramycin, one can expect the penetration of duramycin to the hydrophobic region of PE membrane.
Duramycin and cinnamycin promote membrane binding by inducing transbilayer lipid movement and by changing membrane curvature
PE mainly resides in the inner layer of the plasma membrane (19,21,49). To induce cell lysis, duramycin and cinnamycin must bind PE, which is only present in very small amounts on the cell surface. Previously we showed that cinnamycin promotes cell binding by inducing transbilayer lipid movement (8). Transbilayer lipid movement in the plasma membrane causes the exposure of PE to the outer leaflet. The present study indicates that, in addition to inducing transbilayer lipid movement, duramycin and cinnamycin alter the membrane to highly curved tubular structures. Since duramycin and cinnamycin prefer high curvature, the lantibiotics promote further binding of the peptides by inducing tubulation. The mechanism of the lantibiotics-induced membrane tubulation is not clear. It is conceivable that the membrane tubulation is accompanied by the transbilayer lipid movement, and the resultant high curvature may be accounted for by a biased outward-directed transbilayer lipid movement.
Exposure or high curvature?
Our results indicate that the binding of both duramycin and cinnamycin to PE is dependent on the curvature of the membrane. Cinnamycin has been used to study the cellular localization of PE. It is reported that PE is exposed at restricted sites of the cell surface. Using an amino-reactive probe, trinitrobenzene sulfonic acid, it has been shown that in steady-state fibroblasts, 2–2.5 mol % of total PE is exposed on the cell surface (50,51). From the present results, one cannot exclude the possibility that PE is evenly distributed on the cell surface and cinnamycin recognizes the high curvature, instead of the exposure of PE. Further studies are required to understand cellular distribution of PE.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We are grateful to H. Iwase (Japan Atomic Energy Agency (JAEA)) and H. Takahashi (Gunma University) for fruitful discussions on the analysis of SAXS data, to A. Yamaji-Hasegawa (RIKEN) for the technical help in measurement of hemolysis and helpful discussions, to T. Zimmer (Friedrich Schiller Univ. Jena) for valuable discussions, to R. Ishitsuka, H. Shogomori, Y. Ueda, K. Ishii, M. Abe, and K. Tamada for their help in SAXS measurements at SPring-8, to K. Tamada, Y. Ueda, F. Hullin-Matsuda, and R. Ishitsuka for critically reading the manuscript, and to all members of the Kobayashi labs for valuable discussions.
This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan (Nos. 17390025 and 18050040 to T.K., No. 17659058 to M.M.), grants from RIKEN Frontier Research System, Chemical Biology Project of RIKEN, RIKEN Presidential Research Grant for Intersystem Collaboration (to T.K.), and a grant from the Hayashi Memorial Foundation for Female Natural Scientists (to A.M.). K.I. is a Special Postdoctoral Researcher of RIKEN.
Editor: Michael Edidin.
References
- 1.Lindenfelser, L. A., T. G. Pridham, O. L. Shotwell, and F. H. Stodola. 1957. Antibiotics against plant disease. IV. Activity of duramycin against selected microorganisms. Antibiot. Annu. 5:241–247. [PubMed] [Google Scholar]
- 2.Shotwell, O. L., and T. G. Pridham. 1958. Antibiotics against plant disease. III. Duramycin, a new antibiotic from Streptomyces cinnamoneus forma azacoluta. J. Am. Chem. Soc. 80:3912–3915. [Google Scholar]
- 3.Kessler, H., S. Steuernagel, and M. Will. 1988. The structure of the polycyclic nonadecapeptide Ro 09–0198. Helv. Chim. Acta. 71:1924–1929. [Google Scholar]
- 4.Hayashi, F., K. Nagashima, Y. Terui, Y. Kawamura, K. Matsumoto, and H. Itazaki. 1990. The structure of PA48009: the revised structure of duramycin. J. Antibiot. (Tokyo). 43:1421–1430. [DOI] [PubMed] [Google Scholar]
- 5.Fredenhagen, A., G. Fendrich, F. Marki, W. Marki, J. Gruner, F. Raschdorf, and H. H. Peter. 1990. Duramycins B and C, two new lanthionine containing antibiotics as inhibitors of phospholipase A2. Structural revision of duramycin and cinnamycin. J. Antibiot. (Tokyo). 43:1403–1412. [DOI] [PubMed] [Google Scholar]
- 6.Choung, S. Y., T. Kobayashi, J. Inoue, K. Takemoto, H. Ishitsuka, and K. Inoue. 1988. Hemolytic activity of a cyclic peptide Ro09–0198 isolated from Streptoverticillium. Biochim. Biophys. Acta. 940:171–179. [DOI] [PubMed] [Google Scholar]
- 7.Choung, S. Y., T. Kobayashi, K. Takemoto, H. Ishitsuka, and K. Inoue. 1988. Interaction of a cyclic peptide, Ro09–0198, with phosphatidylethanolamine in liposomal membranes. Biochim. Biophys. Acta. 940:180–187. [DOI] [PubMed] [Google Scholar]
- 8.Makino, A., T. Baba, K. Fujimoto, K. Iwamoto, Y. Yano, N. Terada, S. Ohno, S. B. Sato, A. Ohta, M. Umeda, K. Matsuzaki, and T. Kobayashi. 2003. Cinnamycin (Ro 09–0198) promotes cell binding and toxicity by inducing transbilayer lipid movement. J. Biol. Chem. 278:3204–3209. [DOI] [PubMed] [Google Scholar]
- 9.Emoto, K., T. Kobayashi, A. Yamaji, H. Aizawa, I. Yahara, K. Inoue, and M. Umeda. 1996. Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis. Proc. Natl. Acad. Sci. USA. 93:12867–12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emoto, K., O. Kuge, M. Nishijima, and M. Umeda. 1999. Isolation of a Chinese hamster ovary cell mutant defective in intramitochondrial transport of phosphatidylserine. Proc. Natl. Acad. Sci. USA. 96:12400–12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emoto, K., and M. Umeda. 2000. An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J. Cell Biol. 149:1215–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato, U., K. Emoto, C. Fredriksson, H. Nakamura, A. Ohta, T. Kobayashi, K. Murakami-Murofushi, and M. Umeda. 2002. A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J. Biol. Chem. 277:37855–37862. [DOI] [PubMed] [Google Scholar]
- 13.Iwamoto, K., S. Kobayashi, R. Fukuda, M. Umeda, T. Kobayashi, and A. Ohta. 2004. Local exposure of phosphatidylethanolamine on the yeast plasma membrane is implicated in cell polarity. Genes Cells. 9:891–903. [DOI] [PubMed] [Google Scholar]
- 14.Emoto, K., H. Inadome, Y. Kanaho, S. Narumiya, and M. Umeda. 2005. Local change in phospholipid composition at the cleavage furrow is essential for completion of cytokinesis. J. Biol. Chem. 280:37901–37907. [DOI] [PubMed] [Google Scholar]
- 15.Navarro, J., J. Chabot, K. Sherrill, R. Aneja, S. A. Zahler, and E. Racker. 1985. Interaction of duramycin with artificial and natural membranes. Biochemistry. 24:4645–4650. [DOI] [PubMed] [Google Scholar]
- 16.Dunkley, E. A., Jr., S. Clejan, A. A. Guffanti, and T. A. Krulwich. 1988. Large decreases in membrane phosphatidylethanolamine and diphosphatidylglycerol upon mutation to duramycin resistance do not change the protonophore resistance of Bacillus subtilis. Biochim. Biophys. Acta. 943:13–18. [DOI] [PubMed] [Google Scholar]
- 17.Clejan, S., A. A. Guffanti, M. A. Cohen, and T. A. Krulwich. 1989. Mutation of Bacillus firmus OF4 to duramycin resistance results in substantial replacement of membrane lipid phosphatidylethanolamine by its plasmalogen form. J. Bacteriol. 171:1744–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheth, T. R., R. M. Henderson, S. B. Hladky, and A. W. Cuthbert. 1992. Ion channel formation by duramycin. Biochim. Biophys. Acta. 1107:179–185. [DOI] [PubMed] [Google Scholar]
- 19.Devaux, P. F. 1991. Static and dynamic lipid asymmetry in cell membranes. Biochemistry. 30:1163–1173. [DOI] [PubMed] [Google Scholar]
- 20.Cerbon, J., and V. Calderon. 1991. Changes of the compositional asymmetry of phospholipids associated to the increment in the membrane surface potential. Biochim. Biophys. Acta. 1067:139–144. [DOI] [PubMed] [Google Scholar]
- 21.Zachowski, A. 1993. Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem. J. 294:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Racker, E., C. Riegler, and M. Abdel-Ghany. 1984. Stimulation of glycolysis by placental polypeptides and inhibition by duramycin. Cancer Res. 44:1364–1367. [PubMed] [Google Scholar]
- 23.Roberts, M., S. B. Hladky, R. J. Pickles, and A. W. Cuthbert. 1991. Stimulation of sodium transport by duramycin in cultured human colonic epithelia. J. Pharmacol. Exp. Ther. 259:1050–1058. [PubMed] [Google Scholar]
- 24.Kobayashi, T., B. Storrie, K. Simons, and C. G. Dotti. 1992. A functional barrier to movement of lipids in polarized neurons. Nature. 359:647–650. [DOI] [PubMed] [Google Scholar]
- 25.Yamaji, A., Y. Sekizawa, K. Emoto, H. Sakuraba, K. Inoue, H. Kobayashi, and M. Umeda. 1998. Lysenin, a novel sphingomyelin-specific binding protein. J. Biol. Chem. 273:5300–5306. [DOI] [PubMed] [Google Scholar]
- 26.Rouser, G., A. N. Siakotos, and S. Fleisher. 1966. Quantitative analysis of phospholipids by thin-layer chromatography and phosphorus analysis of spots. Lipids. 1:85–86. [DOI] [PubMed] [Google Scholar]
- 27.Ishitsuka, R., A. Yamaji-Hasegawa, A. Makino, Y. Hirabayashi, and T. Kobayashi. 2004. A lipid-specific toxin reveals heterogeneity of sphingomyelin-containing membranes. Biophys. J. 86:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reference deleted in proof.
- 29.Chonn, A., S. C. Semple, and P. R. Cullis. 1991. Separation of large unilamellar liposomes from blood components by a spin column procedure: towards identifying plasma proteins which mediate liposome clearance in vivo. Biochim. Biophys. Acta. 1070:215–222. [DOI] [PubMed] [Google Scholar]
- 30.Ishitsuka, R., and T. Kobayashi. 2007. Cholesterol and lipid/protein ratio control the oligomerization of a sphingomyelin-specific toxin, lysenin. Biochemistry. 46:1495–1502. [DOI] [PubMed] [Google Scholar]
- 31.Fujisawa, T., K. Inoue, T. Oka, H. Iwamoto, T. Uraga, T. Kumasaka, Y. Inoko, N. Yagi, M. Yamamoto, and T. Ueki. 2000. Small-angle x-ray scattering station at the SPring-8 RIKEN beamline. J. Appl. Crystallogr. 33:797–800. [Google Scholar]
- 32.Fujisawa, T., Y. Nishikawa, H. Yamazaki, and Y. Inoko. 2003. Evaluation and improvements of the Rigaku imaging plate reader (R-Axis IV++) for the use in synchrotron x-ray solution scattering. J. Appl. Crystallogr. 36:535–539. [Google Scholar]
- 33.Reference deleted in proof.
- 34.Huang, T., H. Toraya, T. Blanton, and Y. Wu. 1993. X-ray powder diffraction analysis of silver behenate, a possible low-angle diffraction standard. J. Appl. Crystallogr. 26:180–184. [Google Scholar]
- 35.Glatter, O., and O. Kratky. 1982. Small Angle X-Ray Scattering. Academic Press, London.
- 36.Guinier, A., and G. Fournet. 1955. Small Angle Scattering Of X-Rays. Wiley, New York.
- 37.Wakamatsu, K., S. Y. Choung, T. Kobayashi, K. Inoue, T. Higashijima, and T. Miyazawa. 1990. Complex formation of peptide antibiotic Ro09–0198 with lysophosphatidylethanolamine: 1H NMR analyses in dimethyl sulfoxide solution. Biochemistry. 29:113–118. [DOI] [PubMed] [Google Scholar]
- 38.Machaidze, G., A. Ziegler, and J. Seelig. 2002. Specific binding of Ro 09–0198 (cinnamycin) to phosphatidylethanolamine: a thermodynamic analysis. Biochemistry. 41:1965–1971. [DOI] [PubMed] [Google Scholar]
- 39.Marsh, D. 1990. CRC Handbook of Lipid Bilayers. CRC Press, Boca Raton, FL.
- 40.Machaidze, G., and J. Seelig. 2003. Specific binding of cinnamycin (Ro 09–0198) to phosphatidylethanolamine. Comparison between micellar and membrane environments. Biochemistry. 42:12570–12576. [DOI] [PubMed] [Google Scholar]
- 41.Pabst, G., M. Rappolt, H. Amenitsch, and P. Laggner. 2000. Structural information from multilamellar liposomes at full hydration: full q-range fitting with high quality x-ray data. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 62:4000–4009. [DOI] [PubMed] [Google Scholar]
- 42.Rappolt, M., A. Hickel, F. Bringezu, and K. Lohner. 2003. Mechanism of the lamellar/inverse hexagonal phase transition examined by high resolution x-ray diffraction. Biophys. J. 84:3111–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kucerka, N., S. Tristram-Nagle, and J. F. Nagle. 2005. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J. Membr. Biol. 208:193–202. [DOI] [PubMed] [Google Scholar]
- 44.Zamyatnin, A. A. 1972. Protein volume in solution. Prog. Biophys. Mol. Biol. 24:107–123. [DOI] [PubMed] [Google Scholar]
- 45.Perkins, J. P. 1988. Modern Physical Methods in Biochemistry, Part B. A. Neuberger and L. L. M. van Deenen, editors. Elsevier, Amsterdam, The Netherlands.
- 46.Hirai, M., H. Iwase, T. Hayakawa, M. Koizumi, and H. Takahashi. 2003. Determination of asymmetric structure of ganglioside-DPPC mixed vesicle using SANS, SAXS, and DLS. Biophys. J. 85:1600–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balgavy, P., M. Dubnickova, N. Kucerka, M. A. Kiselev, S. P. Yaradaikin, and D. Uhrikova. 2001. Bilayer thickness and lipid interface area in unilamellar extruded 1,2-diacylphosphatidylcholine liposomes: a small-angle neutron scattering study. Biochim. Biophys. Acta. 1512:40–52. [DOI] [PubMed] [Google Scholar]
- 48.Nagle, J. F., and S. Tristram-Nagle. 2000. Structure of lipid bilayers. Biochim. Biophys. Acta. 1469:159–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cerbon, J., and V. Calderon. 1995. Generation, modulation and maintenance of the plasma membrane asymmetric phospholipid composition in yeast cells during growth: their relation to surface potential and membrane protein activity. Biochim. Biophys. Acta. 1235:100–106. [DOI] [PubMed] [Google Scholar]
- 50.Sleight, R. G., and R. E. Pagano. 1983. Rapid appearance of newly synthesized phosphatidylethanolamine at the plasma membrane. J. Biol. Chem. 258:9050–9058. [PubMed] [Google Scholar]
- 51.Kobayashi, T., and R. E. Pagano. 1989. Lipid transport during mitosis. Alternative pathways for delivery of newly synthesized lipids to the cell surface. J. Biol. Chem. 264:5966–5973. [PubMed] [Google Scholar]
- 52.Rappolt, M., K. Pressl, G. Pabst, and P. Laggner. 1998. Lα-phase separation in phosphatidylcholine-water systems induced by alkali chlorides. Biochim. Biophys. Acta. 1372:389–393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.