Abstract
Context
A major determinant of osteoporotic fractures is peak bone mineral density (BMD), which is a highly heritable trait. Recently, we identified significant linkage for hip BMD in premenopausal sister pairs at chromosome 14q (LOD score = 3.5), where the estrogen receptor β gene (ESR2) is located.
Objective
The objective of the study was to determine whether ESR2 polymorphisms are associated with normal BMD variation.
Design
This was a population‐based genetic association study, using 11 single nucleotide polymorphisms (SNPs) distributed across the ESR2 gene.
Setting
The study was conducted at an academic research laboratory and medical center.
Patients and Other Participants
A total of 411 healthy men (aged 18–61 yr) and 1291 healthy premenopausal women (aged 20–50 yr) living in Indiana participated in the study.
Intervention(s)
There were no interventions.
Main Outcome Measure(s)
The main outcome measures were SNP genotype distributions and their association with BMD at the femoral neck and lumbar spine.
Results
Significant association of spine BMD was found with three SNPs in men and one SNP in women (P ≤ 0.05). The conditional linkage analysis using the ESR2 haplotypes showed that the ESR2 gene accounts for, at most, 18% of the original linkage.
Conclusions
ESR2 polymorphisms are significantly associated with bone mass in both men and women. However, the ESR2 gene is not entirely responsible for our original linkage, and an additional gene(s) in chromosome 14q contributes to the determination of BMD.
Osteoporosis is the most common skeletal disease, characterized by a reduction in bone strength and microarchitectural deterioration of bone tissue, leading to an increased risk of fracture. A major determinant of bone strength in later life is bone mineral density (BMD) achieved during young adulthood. BMD is a complex quantitative trait, which consists of genetic and environmental components. Environmental factors such as nutrition and physical activity influence the attainment of BMD. However, a major proportion of BMD variation is due to genetic factors (1,2).
Estrogen is essential for the acquisition and maintenance of bone mass in women and probably also in men (1,3). In adult women, the decrease in serum estrogen level after menopause leads to an increase in net bone resorption and fracture risk. Mutations in the aromatase gene cause estrogen deficiency in men, leading to increased bone turnover, reduced BMD, and osteoporosis (4,5). Estrogen acts through estrogen receptors α and β, which are encoded by ESR1 and ESR2 genes, respectively. Both receptors are highly expressed in bone (6–9).
In light of estrogen's functional role, ESR2 is a strong candidate gene underlying BMD variation in normal subjects. Several studies have investigated the relationship between BMD and a dinucleotide [cytosine‐adenine (CA)] repeat polymorphism (D14S1026) located within the ESR2 gene (10–13). The length of this CA repeat was associated with spine and hip BMD in four different populations (10–13). Furthermore, a common single nucleotide polymorphism (SNP) within the gene was estimated to account for 4.0% of the difference in hip BMD in men (12). However, all of these previous association studies were limited by sample size and/or number of markers analyzed.
We previously performed a genome screen in premenopausal sister pairs and identified quantitative trait locus (QTL) for hip BMD to genetic markers on chromosome 14q (14), where the ESR2 gene is located. Linkage to the same region was also found for lumbar spine BMD in the Framingham cohort (15). In light of these previous linkage and association findings, we investigated whether genetic polymorphisms in the ESR2 gene contribute to BMD variation at the spine and hip in our large sample of healthy Caucasian men and women.
Subjects and Methods
Subjects
Healthy Caucasian brother and sister pairs were recruited as a part of a sibling pair study to identify genes underlying bone mass. A total of 411 men in 192 brother sibships and 1291 women in 586 sister sibships were studied in the current study (Table 1). Of the total sibships, 25 were from the same family, yielding a total of 753 unrelated families. A blood sample from each subject was collected for genomic DNA extraction. In addition, 283 parents of the sister sibships and 138 parents of the brother sibships provided a blood sample for DNA extraction but did not undertake phenotypic assessments. A detailed medical history of prospective subjects was obtained through administration of health and lifestyle questionnaires. Those who had conditions known to affect BMD or cause artifactual readings of BMD were excluded from the study. Women who had irregular menses or a history of pregnancy or lactation within 3 months before enrollment were also excluded. However, women taking oral contraceptives were not excluded. Informed written consent was obtained from all subjects before their participation in the study. The study was approved by the Institutional Review Board of Indiana University‐Purdue University Indianapolis. All studies were performed at the General Clinical Research Center of Indiana University School of Medicine.
TABLE 1.
Characteristics of study subjects
| Men (n = 411)a | Women (n = 1291)b | |||
|---|---|---|---|---|
| Measure | ||||
| Mean ± SD | Range | Mean ± SD | Range | |
| Age (yr) | 34.6 ± 10.8 | 18–61 | 33.2 ± 7.1 | 20–50 |
| Height (cm) | 178.2 ± 6.8 | 160.2–200.9 | 165.5 ± 6.1 | 146.8–192.3 |
| Weight (kg) | 87.6 ± 16.7 | 52.6–136.0 | 69.2 ± 15.6 | 41.2–145.8 |
| L2–L4 lumbar spine BMD (g/cm²) | 1.282 ± 0.165 | 0.902–1.856 | 1.280 ± 0.139 | 0.796–1.782 |
| Femoral neck BMD (g/cm²) | 1.093 ± 0.159 | 0.695–1.780 | 1.012 ± 0.133 | 0.632–1.603 |
From 192 families.
From 586 families.
BMD and other measurements
Areal BMD (g/cm²) at the lumbar spine L2–L4 and femoral neck was measured by dual‐energy x‐ray absorptiometry, using two DPX‐L and one Prodigy machines (GE Lunar Corp., Madison, WI). All three dual‐energy x‐ray absorptiometry instruments were cross‐calibrated weekly using a step‐wedge phantom. There was no detectable systematic difference among the three machines; the mean difference was less than 1.5% at any point. The coefficient of variation (the precision of the measurement) measured in 115 sister pairs was 1.0% for femoral neck and 0.52% for lumbar spine. Sisters and brothers were measured on the same instrument as their sibling(s), usually at the same visit. Height and weight were measured using a Harpenden stadiometer and a Scale‐Tronix weighing scale, respectively.
SNP genotyping
The ESR2 reference gene (accession no. NM_001437) consists of nine exons, spanning approximately 61 kb of chromosome 14q23.2 (Fig. 1). The longest known mRNA isoform [estrogen receptor‐βcx; accession no. AB006589 (16)] contains additional 5′ and 3′ exons and spans 112 kb of the genomic region (Fig. 1). To fully cover the ESR2 gene, SNPs distributed throughout the longest isoform were selected from the July 2003 assembly of the University of California, Santa Cruz Genome Browser on Human (http://genome.ucsc.edu/cgi-bin/hgGateway) and the Molecular Variation Database (dbSNP) linked through LocusLink (http://www.ncbi.nlm.nih.gov/LocusLink) for ESR2. Potential SNPs were initially screened by direct sequencing of three pools of 10 unrelated Caucasian DNA samples each and then validated by direct sequencing or PCR restriction fragment length polymorphism analysis of the 30 DNA samples that comprised the pools.
Fig. 1.
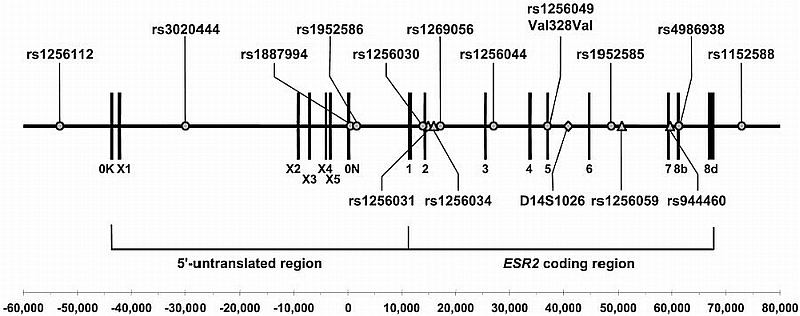
Gene structure of the ESR2 gene. Exon numbering is based on a literature review (32). The coding region comprises exons 1–7 and alternative exon 8s. The 5′ region contains seven known untranslated exons (0K, X1–X5, and 0N). Position and size of exons are indicated by vertical bars. SNPs (circles) tested in this study are shown above the gene. SNPs (triangle) and a microsatellite marker (diamond) tested in previous association studies (10–13) are shown below the gene. The scale in base pairs below the gene is based on the transcription start site of the ESR2 reference sequence (accession no. NM_001437). The negative numbers indicate the promoter region of the reference sequence, which starts from exon 0N.
A total of 11 validated SNPs (Table 2) were genotyped by matrix‐assisted laser desorption/ionization time‐of‐flight (MALDI‐TOF) mass spectrometry of allele‐specific primer extension products (MassARRAY System, Sequenom, Inc., San Diego, CA). The multiplex assays were designed using the SpectroDESIGNER 2.0 software (Sequenom). The PCR and extension reactions were run under conditions similar to SpectroPREP user's guide for homogeneous MassEXTEND (Sequenom). In brief, genomic DNA was first amplified by standard PCR methods. After removal of residual deoxynucleotide triphosphates, allele‐specific primer extension products were generated with a mixture of dideoxynucleotide triphosphates and deoxynucleotide triphosphates chosen such that one allele of each SNP would be extended by a single nucleotide and the other allele would be extended by at least two nucleotides. Aliquots of the extension products were then spotted onto SpectroCHIPs (Sequenom), and the alleles were determined by MALDI‐TOF mass spectrometry. Our prior work established that this technique has an error rate of less than 1 in 1000 genotypes. The missing rate for genotyped SNPs ranged from 3.4 to 7.8%, as shown in Table 2.
TABLE 2.
Properties of ESR2 SNPs tested in this study
| SNP | Allelea | Functionb | Minor allele frequencyc | HWE P valuec | Missing rate (%) | Validation method | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1256112 | A→G | Promoter | 0.44 | 0.81 | 5.0 | BsmAI | ||||||
| rs3020444 | T→C | Promoter | 0.32 | 0.67 | 4.0 | HphI | ||||||
| rs1887994 | G→T | 5′‐Untranslated region | 0.09 | 0.43 | 4.0 | ApoI | ||||||
| rs1952586 | A→G | 5′‐Untranslated region | 0.11 | 0.28 | 7.8 | TfiI | ||||||
| rs1256030 | C→T | Intron 1 | 0.46 | 0.33 | 4.5 | AluI | ||||||
| rs1269056 | G→A | Intron 2 | 0.43 | 0.52 | 4.1 | Sequencing | ||||||
| rs1256044 | T→C | Intron 3 | 0.43 | 0.43 | 4.6 | MspI | ||||||
| rs1256049 | G→A | Exon 5 | 0.03 | 0.20 | 3.9 | RsaI | ||||||
| rs1952585 | T→C | Intron 6 | 0.07 | 0.99 | 3.9 | DdeI | ||||||
| rs4986938 | G→A | 3′ Flanking region | 0.37 | 0.88 | 3.4 | AluI | ||||||
| rs1152588d | G→C | 3′ Flanking region | 0.45 | 0.43 | 5.3 | BsrI |
Given in the orientation of the ESR2 gene (i.e. minus/reverse strand).
Based on the ESR2 reference sequence (see Fig. 1).
Minor allele frequency and Hardy‐Weinberg equilibrium (HWE) P value were calculated from 753 unrelated samples.
Located in the spectrin repeat containing nuclear envelope 2 (SYNE2) gene.
Statistical analysis
Stepwise regression analysis was employed separately on both lumbar spine and femoral neck BMD using height, weight, oral contraceptive use, pack‐years of smoking, and age to identify significant covariates with BMD. P ≤ 0.10 was required for retention of a covariate in the regression model. Regression residuals, representing covariate‐adjusted BMD values, were computed and used in all analyses.
Hardy‐Weinberg equilibrium and minor allele frequency were calculated from a sample consisting of one sibling from each of the 753 unrelated families; these were the only calculations performed using the one‐per‐family sample. Hardy‐Weinberg equilibrium was tested for each genotyped SNP, using χ² statistics. Haplotypes were constructed from the SNP data of all siblings and parents, using the SimWalk2 computer package (17). Linkage disequilibrium statistics (Lewontin's D′) for each pair of SNPs were then calculated based on the observed haplotype and allele frequencies, using the HAPLOXT program (18).
To test for association due to linkage disequilibrium between ESR2 polymorphisms and BMD variation, we used a population‐based association method. Population‐based association tests have maximal power to detect even relatively small genetic effects in samples of moderate size but are susceptible to producing false‐positive results in admixed or stratified populations (19). However, it should be noted that our Caucasian sample was previously determined to be genetically homogeneous (20).
The hypothesis of population‐based association was tested using the total association model of Abecasis et al. (21), which uses a variance component framework to perform a test of association between a marker and a quantitative trait while properly accounting for related subjects in a family sample. The proportion of BMD variation accounted for by each SNP was estimated as a²/2Vp, where a is the mean allelic effect from the variance components model fitting (21) and Vp is the total variance of the BMD trait (22). The variance component method assumes additive genetic effects at the locus tested. To enable detection of nonadditive effects, we also performed a mixed‐model analysis for each genotyped SNP, using SAS (version 9.1, SAS Institute, Cary, NC). This model used family membership as a random effect, along with the fixed effect of SNP genotype, to correct for familial correlations among subjects.
The hypothesis that genetic variation at the ESR2 locus underlies the observed linkage finding on chromosome 14q was tested using a conditional linkage approach. LOD scores measuring linkage to markers in the 14q region (microsatellite markers D14S587, D14S592, D14S588, and D14S53) were computed using the software package SOLAR (23). These were then compared with LOD scores calculated when ESR2 SNP haplotypes computed using SimWalk2 (17) were included in the linkage model as covariates. The relative contribution of ESR2 to the linkage finding was measured by the decrease in the LOD score when the ESR2 data were included as covariates.
P ≤ 0.05 was considered significant for all the analyses. For SNPs and BMD phenotypes in which significance was observed, a multiple testing correction was obtained via a permutation framework (24). Five thousand permuted data sets were generated by uncoupling the BMD phenotypes from SNP genotype data but preserving the phenotypic correlation and linkage disequilibrium structure of the observed data. This was accomplished by retaining lumbar spine and femoral neck BMD from a particular subject together in each permutated replicate; likewise, the SNP genotypes from each subject were permuted as a unit. Empiric significance levels were then obtained from the observed distribution of test statistics from these permuted replicates.
Results
Sample demographics
Characteristics of the study subjects are shown in Table 1. Men ranged in age from 18 to 61 yr, and women ranged from 20 to 50 yr. The mean age of men was 34.6 yr, and that of women was 33.2 yr. BMD at lumbar spine was similar in men and women. However, mean femoral neck BMD was approximately 8% higher in men than women. Age and body weight were the only covariates tested that approached statistical significance (P < 0.10) in the regression model fitting of BMD. Regression residuals representing age‐ and weight‐adjusted BMD values were, therefore, used in all further analyses. In our female sample, body weight and age together explained 12.1 and 17.2% of the variation in lumbar spine and femoral neck BMD, respectively. In men, 19.4% of the spine and 29.1% of the neck BMD were explained by body weight and age combined.
SNP genotyping and linkage disequilibrium
Eleven SNPs distributed across the longest known ESR2 isoform were genotyped (Fig. 1). Detailed information about each SNP is summarized in Table 2. The genotype distribution of all SNPs was in Hardy‐Weinberg equilibrium (Table 2 2). Linkage disequilibrium coefficients (D′) between each pair of the genotyped markers were estimated from sibship genotype data (Table 3). Substantial linkage disequilibrium (D′ ≥ 0.60) exists among all informative markers with minor allele frequency of at least 0.3, indicating that our coverage of the ESR2 gene was adequate for association studies.
TABLE 3.
Linkage disequilibrium coefficient (D′) between SNPs spanning the ESR2 gene
| rs3020444 | rs1887994 | rs1952586 | rs1256030 | rs1269056 | rs1256044 | rs1256049 | rs1952585 | rs4986938 | rs1152588 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1256112 | 0.69 | 0.42 | 0.73 | 0.72 | 0.74 | 0.72 | 0.71 | 0.63 | 0.60 | 0.70 | ||||||||||
| rs3020444 | 0.71 | 0.75 | 0.67 | 0.68 | 0.67 | 0.85 | 0.41 | 0.67 | 0.64 | |||||||||||
| rs1887994 | 0.72 | 0.72 | 0.74 | 0.67 | 0.88 | 0.67 | 0.66 | 0.61 | ||||||||||||
| rs1952586 | 0.33 | 0.69 | 0.63 | 0.39 | 0.17 | 0.75 | 0.59 | |||||||||||||
| rs1256030 | 0.85 | 0.86 | 0.72 | 0.21 | 0.70 | 0.79 | ||||||||||||||
| rs1269056 | 0.82 | 0.70 | 0.69 | 0.69 | 0.82 | |||||||||||||||
| rs1256044 | 0.70 | 0.63 | 0.70 | 0.81 | ||||||||||||||||
| rs1256049 | 0.72 | 0.75 | 0.62 | |||||||||||||||||
| rs1952585 | 0.29 | 0.64 | ||||||||||||||||||
| rs4986938 | 0.69 |
SNP association analyses
Two population‐based association methods [variance component (vc) and mixed models] were used to test the evidence for association of the 11 SNPs with hip and spine BMD. We found evidence of significant association of lumbar spine BMD variation with three of the 11 SNPs in men and one of the 11 SNPs in women (Table 4). The most significant association (Pvc = 0.0072 in men and Pmixed = 0.0073 in women; empiric P < 0.01) was found with rs3020444, which is located between 5′‐untranslated exons, X1 and X2 (Table 4 and Fig. 1). Three SNPs with significant Pvc accounted for 1.1–2.3% of the spine BMD in men. None of the tested SNPs reached statistical significance for femoral neck BMD variation in men or women (Table 4). However, a marginal association was found between rs1256112 and femoral neck BMD in men (Pvc = 0.053). Mean spine BMD values are shown for each SNP genotype of rs3020444, the only SNP with P < 0.01 (Table 5).
TABLE 4.
Association of ESR2 SNPs with BMD
| Men (n = 411) | Women (n = 1291) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Lumbar spine | Femoral neck | Lumbar spine | Femoral neck | ||||||||||||
| Pvca | Pmixedb | Pvca | Pmixedb | Pvca | Pmixedb | Pvca | Pmixedb | |||||||||
| rs1256112 | 0.0098 | 0.038 | 0.053 | 0.11 | 0.87 | 0.97 | 0.84 | 0.95 | ||||||||
| rs3020444 | 0.0072 | 0.014 | 0.16 | 0.35 | 0.61 | 0.0073 | 0.85 | 0.21 | ||||||||
| rs1887994 | 0.44 | 0.72 | 0.97 | 0.85 | 0.93 | 0.70 | 0.61 | 0.28 | ||||||||
| rs1952586 | 0.85 | 0.98 | 0.50 | 0.71 | 0.49 | 0.65 | 0.21 | 0.13 | ||||||||
| rs1256030 | 0.057 | 0.098 | 0.48 | 0.72 | 0.75 | 0.67 | 0.70 | 0.73 | ||||||||
| rs1269056 | 0.074 | 0.20 | 0.45 | 0.33 | 0.78 | 0.93 | 0.44 | 0.74 | ||||||||
| rs1256044 | 0.085 | 0.20 | 0.53 | 0.31 | 0.89 | 0.99 | 0.37 | 0.66 | ||||||||
| rs1256049 | 0.15 | 0.36 | 0.91 | 0.66 | 0.77 | 0.72 | 0.49 | 0.57 | ||||||||
| rs1952585 | 0.41 | 0.68 | 0.37 | 0.30 | 0.064 | 0.11 | 0.22 | 0.25 | ||||||||
| rs4986938 | 0.12 | 0.28 | 0.18 | 0.36 | 0.48 | 0.80 | 0.99 | 0.98 | ||||||||
| rs1152588 | 0.040 | 0.12 | 0.37 | 0.38 | 0.90 | 0.95 | 0.69 | 0.79 | ||||||||
P value from variance component model.
P value from mixed model.
P values of ≤ 0.05 are bold.
TABLE 5.
Mean spine BMD values by rs3020444 SNP genotype
| Men | Women | |||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | ||||||||
| n | Mean ± SD (g/cm²) | n | Mean ± SD (g/cm²) | |||||
| TT | 180 | 1.265 ± 0.141 | 593 | 1.297 ± 0.132 | ||||
| TC | 170 | 1.305 ± 0.148 | 525 | 1.273 ± 0.127 | ||||
| CC | 46 | 1.336 ± 0.151 | 130 | 1.302 ± 0.136 | ||||
Conditional linkage analysis
Given the significant associations found with ESR2, we sought to determine whether the variation in the ESR2 gene accounted for our previously observed QTL on chromosome 14q in women (14). Conditional linkage analysis (23) was used to test whether inclusion of the ESR2 haplotypes as a covariate in linkage analysis significantly decreased the evidence of linkage to chromosome 14q. Haplotypes were inferred from the three SNPs that had significant associations with spine BMD in men: rs1256112, rs3020444, and rs1152588. The most frequent haplotype (G‐T‐C) among the eight possible haplotypes accounted for 29.7 and 30.4% of the alleles in men and women, respectively. The observed maximum LOD score in the linkage region of chromosome 14q was decreased by at most 18% when the ESR2 haplotypes were included as a covariate in the linkage analysis. This suggests that other gene(s) within the chromosome 14q region are primarily responsible for our original linkage evidence in women.
Discussion
In the present study, we investigated whether the ESR2 gene, located in the chromosome 14q region linked to hip BMD (14), is associated with variation in bone mass in healthy Caucasian men and women. The substantial linkage disequilibrium observed among the 11 SNPs demonstrated that the entire gene was adequately covered for association studies. Population‐based association tests found that the ESR2 SNPs were associated with lumbar spine BMD in both men and women and accounted for a small portion (<3%) of the BMD variation in men. These findings indicate that ESR2 may influence attainment of bone mass at the spine, particularly in men. The strong association between ESR2 SNPs and the spine BMD makes biological sense because the lumbar spine is particularly responsive to estrogen replacement therapy, compared with the femoral neck (25–27). Furthermore, trabecular bone, which is a major component of the spine, is known to have particularly high ESR2 gene expression (6). The discrepancy between the strength of the association between men and women is consistent with studies in mice (28) and humans (29) demonstrating sex‐specific determinants of bone mass. Alternatively, ESR2 polymorphisms may be more important at lower estrogen concentrations, as would occur in men and postmenopausal women, than at higher estrogen concentrations, as seen in premenopausal women.
Although mean age, sample size, or skeletal sites vary somewhat between published studies, our results are in general agreement with those of previous studies (summarized in Table 6). Four previous studies detected significant association between the CA repeat polymorphism (D14S1026) and BMD variation (10–13). One earlier study also demonstrated that ESR2 SNPs and haplotypes account for at least small BMD variation in both men and women (12). Similar to our study, the earlier study also estimated a 2–4% effect size of ESR2 variations on BMD in men (12). Although the level of association did not reach statistical significance, spine BMD was marginally associated with two SNPs (rs1256030 and rs1269056) located near rs1256031, which showed evidence of association with femoral neck BMD in the earlier study (12) (Fig. 1). Furthermore, the results in the present study are also consistent with the absence of association of BMD with the AluI (30) or RsaI polymorphism (31) (Table 6). Taken together, the findings from these studies indicate that, although the effect size may be small, genetic variations in the ESR2 gene are associated with BMD variation in normal subjects. Furthermore, associations found with BMD in elderly men and women as well as premenopausal women suggest that ESR2 polymorphisms may influence both acquisition of bone mass and rate of bone loss.
TABLE 6.
Summary of published association studies of BMD and ESR2
| Ref. | Study subject (mean age ± SDa) | Marker tested | Association found between marker and phenotypeb | |||
|---|---|---|---|---|---|---|
| 10 | 204 postmenopausal Japanese women (70.5 ±6.8 yr) | D14S1026 | D14S1026 and lumbar spine BMD | |||
| 11 | 120 premenopausal Chinese women (37.2 ± 7.8 yr) and 205 postmenopausal Chinese women (62.5 ± 8.6 yr) | D14S1026 | D14S1026 and lumbar spine and femoral neck BMD in premenopausal women but not in postmenopausal women | |||
| 12 | 723 Caucasian men (60.1 ± 9.5 yr) and 795 Caucasian women (59.5 ± 9.2 yr) | D14S1026 and four SNPs | D14S1026 and femoral neck BMD in men and women; rs1256031 and femoral neck BMD in men; haplotype and femoral neck BMD in men and women | |||
| 13 | 226 Caucasian women (60–98 yr) | D14S1026 | D14S1026 and lumbar spine and femoral neck BMD | |||
| 30 | 283 Caucasian men (22–90 yr) | AluI (rs4986938) | No association | |||
| 31 | 79 postmenopausal Slovenian women with osteoporosis (65.2 ± 6.7 yr) | RsaI (rs1256049) | No association | |||
| This study | 411 Caucasian men (34.6 ± 10.8 yr) and 1291 premenopausal Caucasian women (33.2 ± 7.1 yr) | 11 SNPs | Three SNPs (rs1256112, rs3020444, and rs1152588) and lumbar spine BMD in men; rs3020444 and lumbar spine in premenopausal women |
Age range is given when mean age ± SD is unavailable.
Includes association findings only with femoral neck or lumbar spine BMD.
Compared with previous association studies, our study had several strengths. First, we had a relatively large sample of 411 men and 1291 women who have undergone rigorous BMD phenotyping, yielding substantial statistical power to detect association between BMD variation and ESR2 genotypes. Second, we previously determined that our Caucasian sample is genetically homogeneous and thus is not susceptible to false‐positive association results due to population stratification (20). Third, rather than testing only a few SNPs or a single microsatellite marker in the ESR2 gene, we tested 11 SNPs distributed across the entire gene. Fourth, we selected seven SNPs that had high heterozygosity to maximize our statistical power to detect association with BMD. Fifth, we analyzed linkage disequilibrium between SNPs and used this information to help interpret the results of association analyses.
We tested 11 SNPs distributed throughout the ESR2 gene to capture effects of all functional elements. The only exonic SNP tested in this study was a synonymous SNP in exon 5 (rs1256049, Va1328Val). In agreement with a previous study on Slovenian patients with osteoporosis (31), we found no evidence of association (P > 0.1) with this SNP in our population. Because this polymorphism was present in 5.1% of the Slovenian sample (31) and 3.4% of our sample, it is likely a relatively uncommon exonic SNP with no apparent functional consequence. Other exonic SNPs were not tested in this study because of their low expected heterozygosity and concomitant limited power to test for association.
All other SNPs tested in this study were located in either introns or flanking regions of the gene (Fig. 1) and may not possess any direct functional significance. However, these SNPs are expected to be in linkage disequilibrium with functional sequence variations in regulatory regions of the ESR2 gene. In this regard, it is intriguing that the most significant association in both genders was found with the SNP (rs3020444) located in the 5′‐untranslated region, where multiple untranslated exons exist (Fig. 1). These untranslated exons are thought to act in tissue‐specific regulation of gene expression (32). Therefore, it is tempting to speculate that this SNP (or polymorphisms in linkage disequilibrium with this SNP) may influence the ESR2 gene expression in bone cells. Furthermore, some ESR2 isoforms transcribed from these 5′‐untranslated exons are known to lack ligand binding affinity but can preferentially bind to the ESR1 protein and thereby prevent it from binding to DNA (10). It should also be noted that we cannot rule out that the observed association may be due to association with adjacent genes in linkage disequilibrium with the SNPs tested in this study. There is evidence that substantial linkage disequilibrium extends beyond the ESR2 gene.
Our original linkage finding at chromosome 14q was for hip BMD in premenopausal women (14), whereas ESR2 polymorphisms were associated with spine BMD in men and, to a lesser extent, women. This discrepancy may result from the effect of sample size on linkage analysis and association study. Our sample size of 411 men and 1291 women has significant statistical power for population‐based association studies but does not have the power to identify all the possible QTLs underlying BMD at the hip or spine. Although significant association between ESR2 polymorphisms and spine BMD was identified, conditional linkage analysis of the ESR2 gene indicates that the gene accounts for at most 18% of our 14q QTL. Therefore, the ESR2 gene is not entirely responsible for our original linkage finding, and it is more likely that yet unidentified gene(s) contributes to the majority of the linkage evidence for hip BMD in our population. In this regard, the chromsome14q QTL contains other strong candidate genes such as estrogen‐related receptor β (ESRRB), which is structurally and functionally related to classic estrogen receptors (33), and bone morphogenetic protein 4.
In conclusion, we found evidence that the ESR2 gene is associated with normal BMD variation using SNP analyses. ESR2 polymorphisms account for at least a small portion of the spine BMD variation in men and have minimal effect in women. However, identification of multiple genes with small effects, such as ESR2, could lead to development of potential screening panel for osteoporosis risk in normal individuals and allow early intervention and treatment against future bone loss. The effect size of the polymorphisms, as well as the result of conditional linkage analysis, suggest that the ESR2 gene is not responsible for our original 14q QTL for hip BMD. Further analysis of chromosome 14q is necessary to identify a gene(s) contributing to normal BMD variation.
Acknowledgments
We thank siblings and their parents who participated in this study as well as the study coordinators, without whom this work would not have been possible. SNP genotyping by MALDI‐TOF mass spectrometry used the facilities of the Center for Medical Genomics at Indiana University School of Medicine, which is supported in part by a grant from the Indiana Genomics Initiative (INGEN). We are grateful for the assistance of Drs. Howard J. Edenberg and Xiaoling Xuei at the Center for Medical Genomics with SNP genotyping.
Abbreviations
- BMD
Bone mineral density
- CA
cytosine‐adenine
- ESR2
estrogen receptor β
- MALDI‐TOF
matrix‐assisted laser desorption/ionization time‐of‐flight
- QTL
quantitative trait locus
- SNP
single nucleotide polymorphism
- vc
variance component
Footnotes
This work was supported by National Institutes of Health Grants P01 AG‐18397, R01 AR‐43476, M01 RR‐00750, and K24 AR‐02095 and a grant from the Eli Lilly & Co. Centre for Women’s Health. INGEN is supported in part by the Lilly Endowment, Inc.
References
- 1.Rizzoli R, Bonjour J‐P, Ferrari SL. Osteoporosis, genetics and hormones. J Mol Endocrinol. 2001;26:79–94. doi: 10.1677/jme.0.0260079. [DOI] [PubMed] [Google Scholar]
- 2.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23:303–326. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 3.Riggs BL, Khosla S, Melton LJ., III Sex steroids and the construction and conservation of the adult skeleton. Endocr Rev. 2002;23:279–302. doi: 10.1210/edrv.23.3.0465. [DOI] [PubMed] [Google Scholar]
- 4.Morishima A, Grumbach MM, Simpson ER, Fisher C, Qin K. Aromatase deficiency in male and female siblings caused by a novel mutation and the physiological role of estrogens. J Clin Endocrinol Metab. 1995;80:3689–3698. doi: 10.1210/jcem.80.12.8530621. [DOI] [PubMed] [Google Scholar]
- 5.Carani C, Qin K, Simoni M, Faustini‐Fustini M, Serpente S, Boyd J, Korach KS, Simpson ER. Effect of testosterone and estradiol in a man with aromatase deficiency. N Engl J Med. 1997;337:91–95. doi: 10.1056/NEJM199707103370204. [DOI] [PubMed] [Google Scholar]
- 6.Bord S, Horner A, Beavan S, Compston J. Estrogen receptors α and β are differentially expressed in developing human bone. J Clin Endocrinol Metab. 2001;86:2309–2314. doi: 10.1210/jcem.86.5.7513. [DOI] [PubMed] [Google Scholar]
- 7.Braidman IP, Hainey L, Batra G, Selby PL, Saunders PTK, Hoyland JA. Localization of estrogen receptor β protein expression in adult human bone. J Bone Miner Res. 2001;16:214–220. doi: 10.1359/jbmr.2001.16.2.214. [DOI] [PubMed] [Google Scholar]
- 8.Batra GS, Hainey L, Freemont AJ, Andrew G, Saunders PTK, Hoyland JA, Braidman IP. Evidence for cell‐specific changes with age in expression of oestrogen receptor (ER) α and β in bone fractures from men and women. J Pathol. 2003;200:65–73. doi: 10.1002/path.1332. [DOI] [PubMed] [Google Scholar]
- 9.Bland R. Steroid hormone receptor expression and action in bone. Clin Sci (Lond) 2000;98:217–240. [PubMed] [Google Scholar]
- 10.Ogawa S, Hosoi T, Shiraki M, Orimo H, Emi M, Muramatsu M, Ouchi Y, Inoue S. Association of estrogen receptor β gene polymorphism with bone mineral density. Biochem Biophys Res Commun. 2000;269:537–541. doi: 10.1006/bbrc.2000.2285. [DOI] [PubMed] [Google Scholar]
- 11.Lau HHL, Ho AYY, Luk KDK, Kung AWC. Estrogen receptor β gene polymorphisms are associated with higher bone mineral density in premenopausal, but not postmenopausal southern Chinese women. Bone. 2002;31:276–281. doi: 10.1016/s8756-3282(02)00827-x. [DOI] [PubMed] [Google Scholar]
- 12.Shearman AM, Karasik D, Gruenthal KM, Demissie S, Cupples LA, Housman DE, Kiel DP. Estrogen receptor β polymorphisms are associated with bone mass in women and men: the Framingham Study. J Bone Miner Res. 2004;19:773–781. doi: 10.1359/JBMR.0301258. [DOI] [PubMed] [Google Scholar]
- 13.Scariano JK, Simplicio SG, Montoya GD, Garry PJ, Baumgartner RN. Estrogen receptor β dinucleotide (CA) repeat polymorphism is significantly associated with bone mineral density in postmenopausal women. Calcif Tissue Int. 2004;74:501–508. doi: 10.1007/s00223-003-0170-x. [DOI] [PubMed] [Google Scholar]
- 14.Peacock M, Koller DL, Hui S, Johnston CC, Foroud T, Econs MJ. Peak bone mineral density at the hip is linked to chromosomes 14q and 15q. Osteoporos Int. 2004;15:489–496. doi: 10.1007/s00198-003-1560-7. [DOI] [PubMed] [Google Scholar]
- 15.Karasik D, Myers RH, Cupples LA, Hannan MT, Gagnon DR, Herbert A, Kiel DP. Genome screen for quantitative trait loci contributing to normal variation in bone mineral density: the Framingham Study. J Bone Miner Res. 2002;17:1718–1727. doi: 10.1359/jbmr.2002.17.9.1718. [DOI] [PubMed] [Google Scholar]
- 16.Ogawa S, Inoue S, Watanabe T, Orimo A, Hosoi T, Ouchi Y, Muramatsu M. Molecular cloning and characterization of human estrogen receptor βcx: a potential inhibitor of estrogen action in human. Nucleic Acids Res. 1998;26:3505–3512. doi: 10.1093/nar/26.15.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker‐sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 18.Abecasis GR, Cookson WOC. GOLD—graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- 19.Schork NJ, Fallin D, Thiel B, Xu X, Broeckel U, Jacob HJ, Cohen D. The future of genetic case‐control studies. Adv Genet. 2001;42:191–212. doi: 10.1016/s0065-2660(01)42023-2. [DOI] [PubMed] [Google Scholar]
- 20.Koller DL, Peacock M, Lai D, Foroud T, Econs MJ. False positive rates in association studies as a function of degree of stratification. J Bone Miner Res. 2004;19:1291–1295. doi: 10.1359/JBMR.040409. [DOI] [PubMed] [Google Scholar]
- 21.Abecasis GR, Cardon LR, Cookson WOC. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falconer DS. Introduction to quantitative genetics. 3rd ed. Essex, UK: Longman Scientific and Technical; 1989. [Google Scholar]
- 23.Almasy L, Blangero J. Exploring positional candidate genes: linkage conditional on measured genotype. Behav Genet. 2004;34:173–177. doi: 10.1023/B:BEGE.0000013731.03827.69. [DOI] [PubMed] [Google Scholar]
- 24.Westfall PH, Zaykin DV, Young SS. Multiple tests for genetic effects in association studies. Methods Mol Biol. 2002;184:143–168. doi: 10.1385/1-59259-242-2:143. [DOI] [PubMed] [Google Scholar]
- 25.Moore M, Bracker M, Sartoris D, Saltman P, Strause L. Long‐term estrogen replacement therapy in postmenopausal women sustains vertebral bone mineral density. J Bone Miner Res. 1990;5:659–664. doi: 10.1002/jbmr.5650050616. [DOI] [PubMed] [Google Scholar]
- 26.Grey AB, Cundy TF, Reid IR. Continuous combined oestrogen/progestin therapy is well tolerated and increases bone density at the hip and spine in post‐menopausal osteoporosis. Clin Endocrinol (Oxf) 1994;40:671–677. doi: 10.1111/j.1365-2265.1994.tb03020.x. [DOI] [PubMed] [Google Scholar]
- 27.Duan Y, Tabensky A, DeLuca V, Seeman E. The benefit of hormone replacement therapy on bone mass is greater at the vertebral body than posterior processes or proximal femur. Bone. 1997;21:447–451. doi: 10.1016/s8756-3282(97)00177-4. [DOI] [PubMed] [Google Scholar]
- 28.Orwoll ES, Belknap JK, Klein RF. Gender specificity in the genetic determinants of peak bone mass. J Bone Miner Res. 2001;16:1962–1971. doi: 10.1359/jbmr.2001.16.11.1962. [DOI] [PubMed] [Google Scholar]
- 29.Duncan EL, Cardon LR, Sinsheimer JS, Wass JAH, Brown MA. Site and gender specificity of inheritance of bone mineral density. J Bone Miner Res. 2003;18:1531–1538. doi: 10.1359/jbmr.2003.18.8.1531. [DOI] [PubMed] [Google Scholar]
- 30.Khosla S, Riggs BL, Atkinson EJ, Oberg AL, Mavilia C, Del Monte F, Melton LJ, III, Brandi ML. Relationship of estrogen receptor genotypes to bone mineral density and to rates of bone loss in men. J Clin Endocrinol Metab. 2004;89:1808–1816. doi: 10.1210/jc.2003-031448. [DOI] [PubMed] [Google Scholar]
- 31.Arko B, Preželj J, Komel R, Kocijančič A, Marc J. No major effect of estrogen receptor β gene RsaI polymorphism on bone mineral density and response to alendronate therapy in postmenopausal osteoporosis. J Steroid Biochem Mol Biol. 2002;81:147–152. doi: 10.1016/s0960-0760(02)00061-4. [DOI] [PubMed] [Google Scholar]
- 32.Hirata S, Shoda T, Kato J, Hoshi K. Isoform/variant mRNAs for sex steroid hormone receptors in humans. Trends Endocrinol Metab. 2003;14:124–129. doi: 10.1016/s1043-2760(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 33.Giguère V. To ERR in the estrogen pathway. Trends Endocrinol Metab. 2002;13:220–225. doi: 10.1016/s1043-2760(02)00592-1. [DOI] [PubMed] [Google Scholar]


