Abstract
The steroid receptor RNA activator (SRA) is a unique modulator of steroid receptor transcriptional activity, as it is able to mediate its coregulatory effects as a RNA molecule. Recent findings, however, have painted a more complex picture of the SRA gene (SRA1) products. Indeed, even though SRA was initially thought to be noncoding, several RNA isoforms have now been found to encode an endogenous protein (SRAP), which is well conserved among Chordata. Although the function of SRAP remains largely unknown, it has been proposed that, much like its corresponding RNA, the protein itself might regulate estrogen and androgen receptor signaling pathways. As such, data suggest that both SRA and SRAP might participate in the mechanisms underlying breast, as well as prostate tumorigenesis. This review summarizes the published literature dealing with these two faces of the SRA gene products and underscores the relevance of this bifaceted system to breast cancer development.
Estrogen and breast cancer
Through its mitogenic action on breast epithelial cells, estrogen not only controls the biology and the development of the normal mammary gland, but also participates in breast tumor growth promotion and breast cancer progression (for a review see [Jensen and Jordan, 2003]). Estrogen action is mainly mediated through two estrogen receptors (ERs), α and β [Green et al., 1986; Mosselman et al., 1996; Ogawa et al., 1998], that belong to the steroid/thyroid/retinoic acid receptors superfamily and primarily act as ligand-dependent transcription factors [Evans, 1988]. These receptors share the same functional and structural organization: a variable N-terminal region containing a hormone-independent activation domain AF-1; a DNA binding domain (DBD) responsible for the specificity of DNA recognition; and a C-terminal extremity containing both the ligand binding domain (LBD) and a ligand-dependent activation domain AF-2. Once bound to their ligand, the receptors undergo conformational changes, dimerize and specifically recognize regulatory DNA sequences (hormone responsive elements, HRE) upstream of target genes. Activated receptors, through a dynamic interplay involving coregulators, chromatin remodeling, histone modification and proteosomal activity, direct the assembly and the stabilization of a pre-initiation complex that will ultimately lead to the transcription of these genes [Dennis and O'Malley, 2005; McKenna et al., 1999; Shibata et al., 1997; Xu, 2005] .
Acknowledgement of the importance of estrogen signaling pathways in the growth of a large number of breast cancers has led to the development of endocrine therapies. For example, Tamoxifen, through competitive binding to ERs, antagonizes the mitogenic action of estrogen. It has been successfully used as an endocrine therapy for more than 20 years and an estimated 400,000 women are alive today because of long-term adjuvant Tamoxifen therapy [Jordan and Morrow, 1999]. The demonstration that Tamoxifen could also prevent the occurrence of breast cancer in women at risk [Fisher et al., 1998; Fisher et al., 2000] raised the hope that many more lives will be spared through the better understanding and manipulation of ER signaling pathways.
Over the last few years, it has become apparent that the balance between coactivators and corepressors, which respectively enhance and repress receptor activity, has an important role in the control of steroid receptor action in a given tissue [Lonard and O'Malley, 2006]. A direct participation of this balance during breast tumorigenesis and cancer progression is now suspected, and a search for possible means to control it and develop new targets for preventive and therapeutic endocrine strategies has started worldwide [Hall and McDonnell, 2005; Perissi and Rosenfeld, 2005].
In this context, the discovery of the steroid receptor RNA activator, which not only differentially coactivates ER-α and ER-β as a RNA, but can also encode a protein likely involved in the regulation of steroid receptor activity, brings a new layer of complexity.
An atypical coregulator: the noncoding steroid receptor RNA activator (SRA)
Discovery
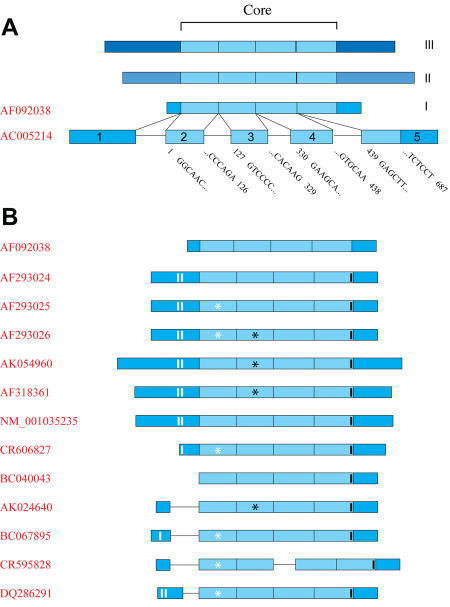
In an effort to discover new coregulators interacting with the AF-1 domain of the progesterone receptor (PR), Lanz et al. screened a human B-lymphocyte library using this domain as bait in a typical Yeast two-hybrid assay [Lanz et al., 1999]. They identified a new clone they called SRA, for steroid receptor RNA activator. Pursuing the analysis of the transcript corresponding to this clone, they subsequently identified three human SRA cDNAs (Figure 1A; SRA I, II and II) via conventional screening of skeletal muscle, heart and HeLa S3 cell line cDNA libraries. These sequences differed in their 5' and 3' extremities, but shared a central 687 bp core region (Figure 1A).
Figure 1. SRA1 genomic structure and transcripts.
A. Original SRA transcripts. Three SRA sequences (I, II and II) were originally described, differing in their 5' and 3' extremities, but sharing a central core sequence depicted in light blue [Lanz et al., 1999]. One sequence has been registered with the NCBI nucleotide database (AF092038). Alignment with chromosome 5q31.3 genomic sequence is provided. Introns and exons are represented by black lines and blue boxes, respectively. B. Currently identified SRA transcripts. Thirteen sequences, corresponding to all SRA transcripts identified to date, have been aligned with the genomic sequence of chromosome 5q31.3 (AC005214). White and black strips indicate the position of SRAP translation start and stop codons, respectively. White and black stars correspond to a point mutation in exon-2 (position 98 of the core: U to C) and a point mutation followed by a full codon (position 271 of the core: G to CGAC), respectively.
Only one of these sequences (AF092038) has been registered in the nucleotide database at the National Center for Biotechnology Information (NCBI). This sequence fully aligns with a portion of chromosome 5q31.3, defining the SRA1 gene overlapping 5 exonic and 4 intronic regions. The core sequence, identified as common among the 3 original cDNAs, encompasses exon-2 to exon-5 (Figure 1A). The SRA1 gene is flanked on the 5' terminus by the Fe64-LIKE2 gene (Fe64L2) and on the 3' reverse strand by the gene encoding the eukaryotic translation initiation factor 4E binding protein 3 (EIF4EBP3). Despite their close proximity, expression pattern analyses confirmed that SRA was an autonomous gene whose expression was independent of the concurrent expression of the flanking genes [Lanz et al., 2002].
In their original report, Lanz et al. presented solid functional evidence supporting the role of SRA as a steroid receptor coactivating molecule. Using cotransfection and reporter assays, they showed that SRA selectively enhanced the AF-1 activity of class I nuclear receptors (i.e., steroid receptors: androgen receptor "AR", ER-α, progesterone receptor "PR", and glucocorticoid receptor "GR"), while it did not affect, in their model, the activity of class II nuclear receptors (thyroid hormone "TR-β", all-trans retinoic acid receptor "RAR-γ", 9-cis retinoic acid "RXR-γ", and peroxisome proliferator-activated receptor "PPAR-γ).
SRA is a RNA coactivator
Surprisingly, although the Yeast two-hybrid screening system is based upon protein-protein interaction, Lanz et al. reported that their original Gal/SRA fusion clone contained a stop codon upstream of the SRA sequence. This construction, even though unable to generate a Gal/SRA fusion protein, was however required for the growth of the yeast colony. This led the authors to speculate that SRA, as a RNA, might have acted as a bridge between the PR-AF-1/Gal4 DNA binding domain and endogenous yeast transcriptional activators. All attempts by these authors to generate SRA protein products in vitro using the three original SRA cDNAs were unsuccessful, except when carboxyl-, but not N-terminal, fusions of SRA with GST or GAL4 were made [Lanz et al., 1999]. This suggested that none of the ATG codons contained in the three identified SRA transcript sequences could be used for the initiation of an efficient translation.
Because the concept of an RNA coactivating steroid receptor was entirely unprecedented, Lanz et al. performed a series of convincing experiments to prove an action at the RNA, rather than the protein level. They first established that SRA was able to coactivate the progesterone receptor in an open reading frame-independent manner by showing that all three alternate open reading frames fused to the translation initiation region of the HSV-thymidine kinase were able to activate transcription with similar efficiency. Furthermore, the introduction of point mutations changing any putative open reading frame or adding premature translation stop codons did not affect the ability of SRA to coactivate PR-mediated transcription. Finally, inhibition of de novo protein synthesis with cycloheximide had no effect on the coactivating properties of SRA on glucocorticoid receptor-mediated transcription, but efficiently reduced the activity of other known coactivator peptides, such as the steroid receptor coactivator 1 (SRC-1) and the CREB-binding protein (CBP). Altogether, these data confirmed that the observed coactivator role of SRA was mediated through a RNA transcript rather than any peptide product.
SRA functional core
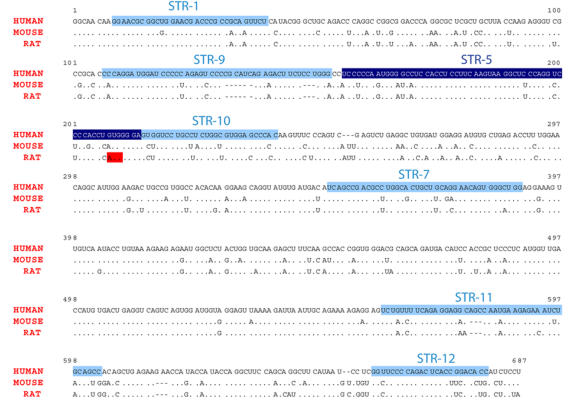
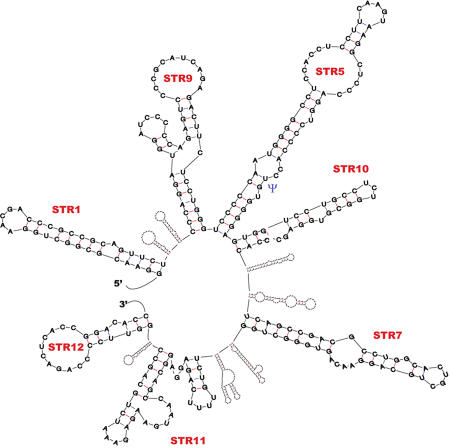
SRA core sequence, found to be necessary and sufficient for SRA to act as a coactivator [Lanz et al., 1999], is fairly well conserved between rodent and human (Figure 2). Serial removal of both ends of the core region reduced SRA coactivation. Removed sections however, were not, by themselves, sufficient for coactivation [Lanz et al., 1999; Lanz et al., 2002]. These results hinted that SRA functional regions were not limited to a single, discrete domain, but rather to several sections distributed throughout the whole core sequence. Low-resolution RNA modeling [Zuker, 2003] predicts several substructures in SRA secondary structure (Figure 3). Through mutation experiments, six secondary structural motifs (STR-1, -9, -10, -7, -11 and -12) individually participating in SRA’s coactivator role have been identified [Lanz et al., 2002]. These observations not only underlined the functional importance of SRA structural features, but also suggested their potential role(s) in modulating the ability of SRA to interact with other molecules.
Figure 2. Alignment of human, mouse and rat core SRA RNA sequences.
Nucleotide sequences corresponding to structures shown to be functionally relevant (STR1, 9, 5, 10, 7, 11 and 12) are boxed in blue [Lanz et al., 2002]. STR5 structure, containing an important pseudouridylation site at position 207 ([Zhao et al., 2007], and see the subsection "Effect of SRA on ER-α and ER-β-mediated transcription") is shaded in dark blue. AUG codon at position 208 in the rat sequence (and referred to in the subsection "SRAP function") is boxed in red.
Figure 3. Schematic profile of the predicted secondary structure of human core SRA RNA.
The secondary structure profile of SRA core sequence has been modeled using Mfold software [Zuker, 2003]. Detailed structure of STR1, 9, 5, 10, 7, 11 and 12 [Lanz et al., 2002] is provided. The position of Uridine residue 207 in STR-5, found to be a site of pseudouridylation (see the "Emerging mechanism of action subsection"), is shown by a blue Ψ.
Effect of SRA on ER-α- and ER-β-mediated transcription
SRA, in many different cell models, increases E2-induced activity of both full-length ER subtypes [Cavarretta et al., 2002; Coleman et al., 2004; Deblois and Giguere, 2003; Hatchell et al., 2006; Klinge et al., 2004; Lanz et al., 1999; Shi et al., 2001; Watanabe et al., 2001; Zhao et al., 2007]. Functional analyses performed with constructions lacking the AF-1 domains showed that SRA coactivates the AF-2 regions of ER-α and ER-β [Coleman et al., 2004; Deblois and Giguere, 2003]. This somewhat contradicts the earlier results, which showed that AF-1-deleted PR and GR mutants were not activated by SRA, and that the N-terminal domain was needed for this RNA to coactivate steroid receptors [Lanz et al., 1999]. This suggests that either estrogen receptors have differential SRA-mediated mechanisms and/or that the cell system used or HREs investigated have a critical effect on the observed action of SRA on a given receptor. The impact of the sequence of EREs used to drive the expression of reporter genes on measured SRA effects has indeed been reported [Klinge et al., 2004].
SRA can also enhance AF-1 activity of ER-α, but not that of ER-β [Coleman et al., 2004; Deblois and Giguere, 2003]. Conflicting reports have been published regarding the role played by E2 and the phosphorylation of the S118 residue in ER-α in this SRA effect [Coleman et al., 2004; Deblois and Giguere, 2003]. In the study by Deblois et al., a construction consisting of ER-α AF-1/DNA binding domain (hence not containing the ligand binding domain) was active only when SRA was present and E2 added. The phosphorylation of S118 residue, known to be critical for ER-α activity [Ali et al., 1993; Kato et al., 1995; Kato et al., 2000; Le Goff et al., 1994; Weigel and Moore, 2007a; Weigel and Moore, 2007b], was necessary to see this effect [Deblois and Giguere, 2003]. It was therefore proposed that E2, even though unable to bind the receptor itself, was activating the mitogen-activated protein kinase (MAPK) pathway and indirectly induced the S118 phosphorylation necessary for SRA to act as a coactivator of the AF-1 region. In the study by Coleman et al., a fusion protein consisting of ER-α AF-1 domain fused to the DBD domain of Gal 4 induced transcription in the presence of SRA [Coleman et al., 2004]. In this system, neither E2 treatment or phosphorylation of S118 was needed for SRA to coactivate ER-α AF-1 activity. These opposite findings may again result from the different systems used, but they also raised the possibility of a role played by ER-α sequences (mainly DBD) present in the first report, but absent in the second study. The DBD of nuclear receptors is indeed known to be the target of coregulatory molecules [Ko et al., 2002; Mathur et al., 2001; Tao et al., 2001]. Further studies are warranted to further address this issue.
The ability of SRA to enhance, in the presence of E2, AF-2 activity of both estrogen receptors, but only ER-α AF-1, suggests at least two different mechanisms of action of this SRA in participating in ligand-mediated transcription. The observation that it can also coactivate the response of ER-α, but not ER-β, to Tamoxifen [Coleman et al., 2004], raises the possibility that, in addition, it could participate in the events leading to the known differential response of these two receptors to antagonist molecules [Barkhem et al., 1998; Watanabe et al., 1997].
Emerging mechanism of action
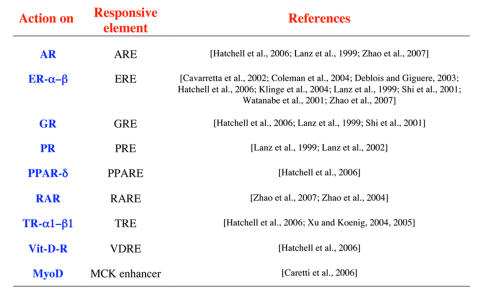
Several studies have now been published shedding light on SRA’s mechanism of action (Table 1). SRA action appears not to be solely limited to enhancing steroid receptor activity. Indeed, it was found to increase the activity of other nuclear receptors, as well. The discrepancy with the original findings in the ability to modulate the activity of nuclear receptors other than steroid receptors may, as shown for ERs, result from differences in cell-type and reporter systems used. It is also likely that SRA’s coactivator role on a given nuclear receptor will depend upon the presence or absence of other regulatory molecules (see below). The recent report that SRA modulates the activity of MyoD, a transcription factor participating in skeletal myogenesis [Caretti et al., 2006], suggests that the role of SRA might be broader than originally predicted.
Table 1. Transcription factors coactivated by SRA RNA.
For each protein, the responsive element involved and the references reporting the effect are listed.
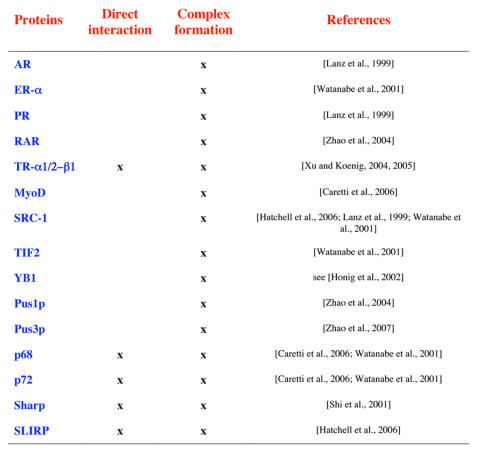
Several proteins participating in the formation of ribonucleoprotein complexes with SRA RNA have now been identified (Table 2). These include the transcription factors whose activity is increased by SRA, as well as accessory proteins acting as positive or negative regulators of nuclear receptor activity.
Table 2. Proteins forming complexes with or directly binding to SRA RNA.
For each protein, the formation of complexes or direct binding, together with reporting references are given.
Association with nuclear receptors
Using an in vitro system consisting of extracts from Xenopus oocytes, it was found that SRA could form complexes with full-length AR, but not with AF-1-deleted AR (ΔAF-1-AR) mutants [Lanz et al., 1999]. This suggests that sequences within this domain directly or indirectly participate in the association of SRA/NR. Interestingly, it has been shown that SRA RNA could directly bind to a 40 amino acid long segment immediately following the second Zinc finger of the DNA binding domain of TR-α1, TR-α2 and TR-β [Xu and Koenig, 2004; Xu and Koenig, 2005], emphasizing that this nuclear receptor might have different options/sites to recruit SRA.
Nuclear receptor coactivators
(i) SRC-1 and TIF2 - SRC-1 and TIF2 belong to the p160 family of nuclear receptor coactivators [Xu and Li, 2003]. These proteins, which directly bind to the AF-2 region of nuclear receptors upon agonist binding, are also able to interact with the AF-1 domain of ERs [Dutertre and Smith, 2003; Metivier et al., 2001; [Tremblay et al., 1999]. They therefore participate in the functional synergy existing between AF-1 and AF-2, as well as in the recruitment of other coregulators [McKenna et al., 1999; Smith and O'Malley, 2004; Xu, 2005]. SRA and SRC-1 can be associated in a large ribonucleoprotein complex of 600-700 kDa, which does not contain the other coactivators p300 or CBP [Lanz et al., 1999]. This led to the hypothesis that SRA might act by modulating the activity of very distinct coactivator complexes. Interestingly, SRA can form complexes with ΔAF-1-AR in the presence of SRC-1 [Lanz et al., 1999], confirming the ability of activated receptors to recruit SRA through different mechanisms.
(ii) p68 and p72 - p68 and p72 belong to the large DExD/H box family of RNA-helicases, which are involved in all aspects of RNA biology, from synthesis and splicing to transport and translation [Fuller-Pace, 2006]. p68/72 directly interact with all members of the SRC-1 family, with the AF-1 region of ER-α, but not other nuclear receptors (including ER-β) and with SRA [Watanabe et al., 2001]. As such, they are able to specifically coactivate not only the agonist-induced AF-2 activity, but the ligand-independent or antagonist-induced AF-1 activity of this receptor as well. It should be stressed that p68 interaction with ER-α is potentiated by the phosphorylation of ER-α [Watanabe et al., 2001]. Interestingly, the physical interaction of p68/72 with SRA is required for these helicases to act as ER-α-specific coactivators. This suggests a crucial role played by SRA in the proper folding/interplay of the different molecules needed to lead to an efficient transcription of target genes. More details on the possible roles of SRA/p68/p72 interactions can be found in a recently published review [Caretti et al., 2007].
(iii) Pus1p and Pus3p - Pus1p and Pus3p belong to the pseudouridine synthase (PUS) family of proteins, which isomerize uridine (U) to pseudouridine (Ψ) in noncoding RNAs such as tRNA, rRNA or snRNA [Charette and Gray, 2000; Ferre-D'Amare, 2003]. Such post-transcriptional modification was found to alter the structural and rigidity features of the target RNAs and to modulate RNA/RNA, as well as RNA/protein interactions. Pus1p and Pus3p pseudouridylate several common, as well as different residues within SRA. Both Pus proteins physically interact with the first Zinc finger of the DNA binding domain of the class II nuclear receptor RAR [Zhao et al., 2007; Zhao et al., 2004]. In the presence of either Pus protein, pseudouridylated SRA becomes able to coactivate this class of nuclear receptor. In contrast, only Pus1p associates with class I receptors and synergizes with SRA to act as a coactivator of these receptors. It was shown that the mutagenesis of a site of pseudouridylation common to both Pus1p and Pus3p, and located in the STR-5 substructure (change from U to A, position 207, see Figure 2 and Figure 3), leads to an overall hyper-pseudouridylation of SRA. Interestingly, in such a hyper-pseudouridylated state, SRA switches from being a coactivator of class I receptors to a dominant negative regulator. Pseudouridylation of SRA by Pus1p and Pus3p therefore appears as a major player in regulating the effect of this RNA on nuclear receptor activity.
Proteins antagonizing nuclear receptor activity
(i) SHARP (SMRT/HDAC1 Associated Repressor Protein) - SHARP was originally identified [Shi et al., 2001] as a protein directly interacting with the C-terminal extremity of the nuclear receptor corepressor SMRT (silencing mediator of retinoic acid and thyroid hormone receptor). SHARP contains a transcriptional repressor domain (RD) and an RNA-interacting domain (RRM). Through the former domain, SHARP is able to recruit HDAC1 and SMRT, whereas the latter domain is needed to interact with STR-7, one of the important functional/structural domains of SRA involved in SRA action [Hatchell et al., 2006; Lanz et al., 2002; Shi et al., 2001]. Full-length SHARP, but not RD only, repressed SRA coactivation of ER and GR. It has been suggested that this repressive action might result from the sequestration of SRA and associated coactivators or from the recruitment of corepressor on the site of target genes.
(ii) SLIRP (SRA stem-loop interacting RNA binding protein) - SLIRP was recently identified as a protein binding to STR7 [Hatchell et al., 2006]. This small (109 amino acids) protein mainly consists of a RNA recognition motif (RRM) and represses nuclear receptor activity through binding SRA. SLIRP, which is recruited on the promoter of target genes, also controls the amount of SRA associated with SRC-1. SLIRP siRNA experiments showed a reduced level of nuclear receptor corepressor (NCoR) associating with ER on the pS2 target gene promoter in the absence of ligand. This led the authors to hypothesize that SLIRP could participate in the recruitment of this corepressor to the promoter of target genes. Interestingly, the majority of the endogenous SLIRP is found in the mitochondria, raising the possibility that SRA may also have a role in this cell compartment.
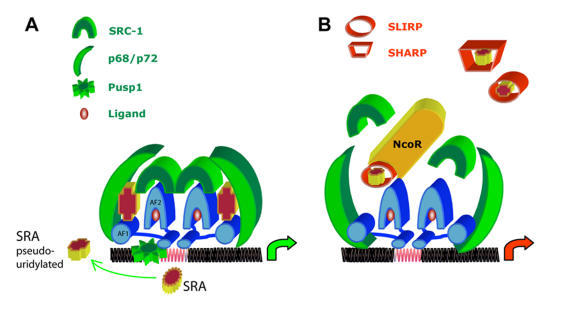
An emerging model of the mechanism of action of SRA on ER-α is presented in Figure 4. It should be stressed that the presence of molecules such as p68, SLIRP or Pus1/3p at the promoter of target genes has been demonstrated by Chromatin Immunoprecipitation (ChIP) assays [Caretti et al., 2006; Hatchell et al., 2006; Zhao et al., 2007; Zhao et al., 2004], though the recruitment of SRA RNA at these sites remains to be experimentally established. Similarly, further studies are needed to establish the exact kinetics of events involving these different partners, as well as the potential differential effects of receptor ligands. The active participation of SRA RNA in the different interactions between the receptor and its coregulators has however led to the proposition that SRA serves as a “gasket” or “molecular adapter”, facilitating the interactions between these molecules. It should be emphasized that the exact participation of the different SRA secondary substructures (detailed in Figure 3) in these physical interactions remains to be elucidated. Indeed, as of today, only STR-7 has been clearly shown, through its ability to be recognized by SLIRP and SHARP, to modulate the inhibitory effect of these molecules [Hatchell et al., 2006]. Further analyses are therefore needed to establish the involvement of other SRA substructures in establishing adequate physical interfaces between SRA RNA and its interacting proteins. The observation that pseudouridylation, known to stabilize RNA structures, affects SRA function [Zhao et al., 2007], strongly suggests that this posttranscriptional modification might also participate in the establishment of functional interfaces between the different partners.
Figure 4. Emerging putative model of SRA RNA action on ER-α signaling.
A. Activation of ER-α gene expression by SRA RNA. Pus1p (green wheel), which is able to bind the DNA binding domain of all nuclear receptors, pseudouridylates specific SRA RNA uridine residues, leading to an optimum configuration of this RNA. The resulting SRA-ψ (cross), could now form a stabilizing complex with p68 (green crescent able to bind SRC-1 and ER-α AF-1 region) and SRC-1 (green horseshoe). Transcription of target genes with suitable ERE (red elements on DNA) will occur. It should be stressed that the physical presence of SRA RNA at the level of promoter has not yet been established experimentally. Similarly, the kinetics of events involving these molecules at the promoter site, as well as the possible effect of specific ligands (red sphere), remain to determined. B. Inhibition of SRA RNA-mediated ER-α. SLIRP (hollow red cylinder) and SHARP (hollow trapeze) act as negative regulators. It has been proposed that they might act by sequestrating SRA, by destabilizing the complex SRA/SRC-1 or by recruiting the nuclear receptor corepressor N-CoR at the promoter region of silenced genes. ER-α ligand binding domain, DNA binding domain and AF-1 domain are shown in blue horseshoe, flat elliptic cylinder and blue cylinder, respectively.
SRA expression and relevance to breast cancer
Different SRA transcripts, detected by Northern blot with apparent migration size of 0.7, 0.85, and ~1.5 kb, have been observed in human tissues [Lanz et al., 1999]. SRA appears highly expressed in liver, skeletal muscle, adrenal gland and the pituitary gland, whereas intermediate expression levels are seen in the placenta, lung, kidney and pancreas. Interestingly, brain and other typical steroid-responsive tissues such as prostate, breast, uterus and ovary contained low levels of SRA RNA [Lanz et al., 2003; Lanz et al., 1999]. In all established cancer cell lines, the 0.85 kb SRA appears to be the main detectable transcript [Lanz et al., 1999]. It should be stressed that the additional 0.7 kb SRA transcript is also observed in cells from breast origin (MCF-7, T47-D). Altogether, SRA is expressed in most tissues. The presence of different transcripts, together with their relative levels of expression, suggest alternative mechanisms of regulation are likely to be tissue- and cell type-specific.
Using RT-PCR targeting core SRA sequences, several reports have shown that SRA expression was increased during breast, uterus and ovarian tumorigenesis [Hussein-Fikret and Fuller, 2005; Lanz et al., 2003; Leygue et al., 1999; Murphy et al., 2000]. Interestingly, SRA overexpression might characterize particular subtypes or subgroups of lesions. Indeed, serous ovarian tumors expressed higher levels of SRA than granulosa cell tumors or mucinous cystadenocarcinoma [Hussein-Fikret and Fuller, 2005]. Similarly, ER-α-positive/PR-negative breast tumors expressed more SRA than ER-α-positive/PR-positive breast tumors [Leygue et al., 1999], whereas Tamoxifen-sensitive and -resistant breast tumors express similar levels [Murphy et al., 2002].
A possible direct involvement of SRA in the mechanisms underlying breast tumorigenesis and tumor progression has been proposed [Lanz et al., 2003; Leygue et al., 1999]. The generation of transgenic mice has however demonstrated that overexpression of the core SRA sequence in the mammary gland was not sufficient per se to induce tumorigenesis [Lanz et al., 2003]. Indeed, even though elevated proliferation leading to the formation of preneoplastic lesions were found to take place in the mammary gland of transgenic mice, none of these lesions led to the development of malignant tumors. The inability of the core SRA to lead, by itself, to a full malignant phenotype may result from the lack of other necessary mechanisms involving additional factors. It may also, as underlined by Lanz et al., result from the high level of apoptosis observed in transgenic mice breast tissue [Lanz et al., 2003]. Apoptosis, through cell loss, may indeed slow down the growth of potential tumor cells. The interpretation of the phenotype observed in these transgenic mice is further complicated by the ability of SRA to regulate the activity of both ERs and PRs. Indeed, both receptors are known to play crucial roles in normal, as well as abnormal mammary gland biology [Anderson and Clarke, 2004; Conneely et al., 2003; Hennighausen and Robinson, 1998; Curtis Hewitt et al., 2000]. The increased expression of PR (known target gene of ER) in the mammary gland of transgenic virgins, together with the apparent phenotype similarities with previously generated PR-A transgenic mice [Shyamala et al., 1998] led the authors to propose that they were observing a SRA-enhanced ER transactivation of PR expression [Lanz et al., 2003] . Altogether, the phenotype observed likely results from a complex interplay involving steroid receptor activities and possibly other still to be discovered SRA-regulated factors.
Coding SRAs and SRA protein (SRAP)
Heterogeneity of SRA transcripts
Several additional human SRA sequences have now been deposited in the nucleotide database of the National Center for Biotechnology Information (Figure 1B). As expected, most of these sequences contain an intact core sequence (exon-2 to exon-5) and differ in their 5'-extremity. The presence of a full core sequence suggests that these isoforms could be fully functional as transcriptional coactivators. Compared to the original AF092038 SRA sequence, some of the sequences contain a point mutation in exon-2 (position 98 of the core: U to C) or a point mutation followed by a full codon (position 271 of the core: G to CGAC), which likely correspond to gene polymorphisms [Emberley et al., 2003].
Four sequences have a full or partial retention of intron-1. Interestingly, the 5' end of six sequences consists of an extended exon-1, which contains two methionine codons in frame with a large open reading frame defining a 236/237 amino acid long peptide. These cDNAs, as opposed to the original SRA, were translatable in vitro, as well as in vivo, leading to the production of a protein localized both in the cytoplasm and the nucleus [Emberley et al., 2003]. Altogether, this raised the possibility that these transcripts, even though still functional at the RNA level through their core sequence, might also encode a protein.
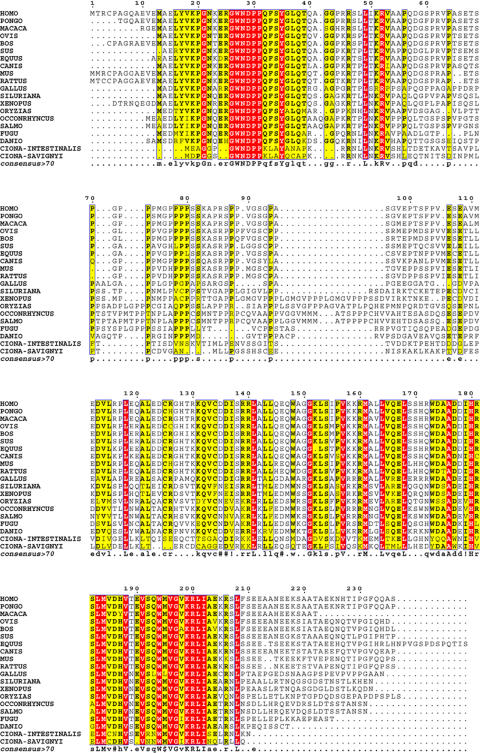
Sequence conservation of the protein encoded by coding SRAs
The sequence of the protein encoded by SRA, referred to as SRAP, is highly conserved in all Chordata (Figure 5). The most conserved amino acids define two distinct domains (N- and C-terminal) that represent the typical signature of this new family of proteins, and which are likely both participating in SRAP function. The presence of an endogenous SRA protein (SRAP) has now been confirmed in the muscle of several vertebrates including mouse, birds, cows and humans [Chooniedass-Kothari et al., 2004]. This protein is also ubiquitously found in human cancer cell lines from the breast [Emberley et al., 2003], the prostate [Kurisu et al., 2006], and other tissues as well (Leygue et al., personal observations), even though levels of expression appear to vary from one cell type to another.
Figure 5. Alignment of SRA protein-related sequences.
Putative SRA protein sequence homologues corresponding to 20 Chordata species are aligned. The numbers indicated on top of the alignment correspond to amino acid numbering of the human SRAP isoform 1. Amino acids conserved in all species are in red letters, whereas those observed in between 70% and 100% of all species are in yellow. Within the consensus sequence, #, ! and $ stand for D or E, I or V, M or L, respectively.
SRAP expression in breast tumors
In a small subset of patients with primary ER-α-positive tumors that was subsequently treated with Tamoxifen, it has been found that SRAP was detectable by Western blot in some patients, but not others [Chooniedass-Kothari et al., 2006]. Interestingly, the apparent overexpression of SRAP in some cases correlated with an overall better survival of the patients. This indicates that SRAP expression is differentially regulated in breast tumors. This also suggests that an increase in SRAP expression might characterize a less aggressive type of tumor, and possibly that this protein contributes to the overall improved outcome after Tamoxifen treatment. Further expression and functional studies are needed to clarify this issue.
SRAP function
To date, most studies have focused mainly on the coactivating function of the noncoding SRA. Exact functions of SRAP therefore remain generally underexplored. Nonetheless, SRAP was shown to directly interact with the AF-2 domain of AR in vitro [Kawashima et al., 2003]. In this study, it was proposed that SRAP, instead of SRA RNA, was coactivating the response to androgen. This proposition is based on the observation that an introduced shift in SRAP reading frame inhibited the translation of the SRAP protein studied, and led to the loss of coactivation function. This appears to be in direct contrast with the observations of Lanz et al. [Lanz et al., 2003]. Besides the already underlined differences, possibly resulting from the cell type and reporter system used, alternative hypotheses can be raised to explain these apparently contrasting results. Indeed, while suppression of SRAP protein production leads to a suppression of coactivating activity, which certainly suggests a functional role of the protein studied, it does not necessarily exclude the possibility that SRA RNA can be a coactivator. Indeed, the sequence used by Kawashima et al. in their experiments corresponds to the coding sequence of a short putative rat SRAP starting at the AUG codon at position 208 in Figure 2 [Kawashima et al., 2003]. This construct is therefore missing the 5' end of the SRA core and might generate a nonfunctional SRA RNA. It should also be stressed that the SRAP rat protein analyzed in these experiments was also missing the first domain of SRAP (as defined in Figure 5). This region is strongly conserved in Chordata. It is therefore reasonable to assume it might have an important functional role. The transient transfection of full-length SRA coding sequence also led to an activation of the response to androgen [Kurisu et al., 2006]. This contrasts with the decreased response to estrogen observed in breast cells stably transfected with coding SRA, which suggested that SRAP might repress the activity of ER in these cells [Chooniedass-Kothari et al., 2006].
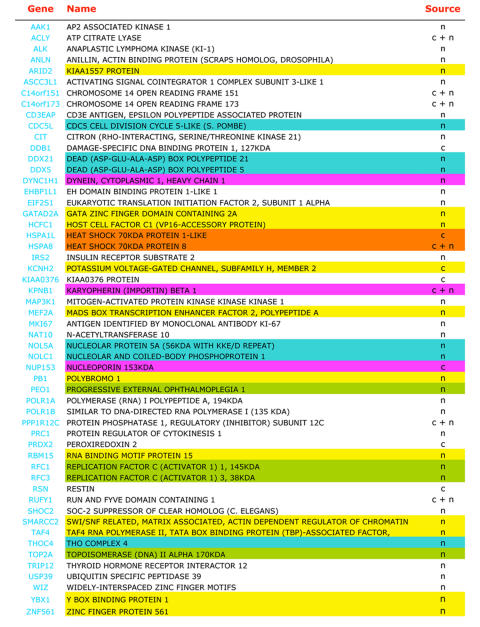
Interestingly, SRAP has recently been shown to interact with several transcription factors and transcription regulators ([Jung et al., 2005a] and Chooniedass-Kothari et al., personal observations). As a result of a concerted effort to generate a database of proteins interacting with nuclear receptor coregulators [Jung et al., 2005b; Jung et al., 2005a], a series of SRAP-interacting proteins has been recently listed. These proteins have been identified by coimmunoprecipitation of nuclear or cytoplasmic extracts from nuclear HeLa cell extracts using commercially available polyclonal rabbit anti-SRAP antibodies (Bethyl Laboratories, Inc.). As shown in Table 3, among the 54 proteins characterized as coimmunoprecipitating with SRAP, are protein chaperones and proteins involved in transport. Interactions with such molecules either reflect a "house-keeping" stage (folding, stabilization and transport) of the SRAP protein processing (common to many proteins), or indicate its possible functional involvement in these processes. The identification, however, of a large number of partners directly involved in RNA processing, regulation of transcription or DNA replication strongly suggests that SRAP may play a role in gene expression. The characterization of p68 (DDX5) as a SRAP-interacting protein is of particular interest, as it underlines the likelihood of crosstalk between SRA RNA and SRAP signaling. Altogether, emerging data suggest that SRAP, similar to its RNA counterpart, might also participate in the regulation of transcription. The exact roles of SRAP on nuclear receptor signaling pathways remain to be elucidated.
Table 3. Proteins forming complexes with SRAP.
Proteins identified by protein sequencing as coimmunoprecipitating with SRAP in nuclear (n) or cytoplasmic (c) extracts of HeLa cells [Jung et al., 2005a] are listed. Examples of proteins considered, using the Gene Ontology terms (biological processes: GOTERM-BP or molecular functions: GOTERM-MF, http://www.geneontology.org/), as molecular chaperones, involved in DNA replication, RNA processing, regulation of transcription or transport are highlighted in orange, green, blue, yellow and pink, respectively.
Coding/noncoding SRA RNAS: possible regulatory mechanisms?
Understanding how cells could regulate such a bifaceted system, involving both noncoding and coding RNAs, possibly sending contradictory and/or intertwined signals to the transcriptional machinery, represents an important question that currently remains unexplored. It has however been demonstrated that both coding and noncoding SRA RNAs coexist in breast cancer cells and that their relative proportions vary from one cell line to another [Hube et al., 2006]. The retention of intron-1, which introduces a shift in the SRAP reading frame and results in the production of noncoding SRA RNAs, has therefore been proposed as a potential mechanism used by cells to control the balance between these two RNA species.
The demonstrated nuclear function of noncoding SRA RNAs (i.e., coactivation of nuclear receptor), together with the existence of SRAP (resulting from the translation of coding RNAs), underlines the need for these transcripts to be present, at least temporarily, in a specific cell compartment. Such a need suggests that cells, through controlling the cellular localization of SRA transcripts, might have a simple way to regulate the functional effects of both partners. One might speculate that cis-elements, located within the transcripts or at their 5' or 3' extremities could, as described in other systems [Chabanon et al., 2004; Jambhekar and Derisi, 2007; Kindler et al., 2005; Li et al., 2006], participate in the targeting of a given RNA to a particular cellular compartment. In situ hybridization results have corroborated the likelihood of the existence of cellular mechanisms regulating the localization of SRA transcripts. Indeed, whereas SRA RNA is detected mainly in the nucleus of transgenic mice mammary epithelial cells overexpressing SRA [Lanz et al., 2003], this transcript is primarily found in the cytoplasm of transiently transfected MCF-7 breast cancer cells [Zhao et al., 2007]. Interestingly, these two groups used the same SRA construction (SRA II, see Figure 1A). Besides possible differences in the 5' and 3' extremities introduced by construction variations (such as different transcription start sites), which might contribute to the observed differential localization of SRA RNAs in the two systems, one might speculate that normal [Lanz et al., 2003] and cancer [Zhao et al., 2007] cells could differentially target the same SRA sequence. Studies are critically needed to fully characterize the mechanisms controlling the generation and the localization of noncoding and coding SRA RNAs, as well as their possible impact on breast tumorigenesis and breast tumor progression.
Summary
The overexpression of SRA core sequence during breast tumorigenesis, together with a higher expression of SRAP in patients more likely to survive under Tamoxifen treatment, make the study of these molecules highly relevant to breast cancer research. Indeed, one might foresee that the full understanding of this bifaceted system might provide new targets for curative or preventive strategies to fight this disease. Unfortunately, the complexity of the bifaceted SRA/SRAP system makes function difficult to address. For example, the interpretation of specific experiments performed using approaches decreasing SRA RNA [Caretti et al., 2006; Cavarretta et al., 2002; Hatchell et al., 2006; Kurisu et al., 2006] might be impaired by the fact that both RNA and protein are likely affected. This makes such approaches unsuitable for the further dissection of respective mechanisms of action of these two protagonists. It is likely that new techniques, specifically targeting the RNA or the protein production/function, will be needed to overcome this problem.
SRA can now definitively be seen as a functional coding RNA. It provides a fascinating link between two worlds, which, up to now, appeared to be quite delineated. Indeed, functional RNAs were thought to be inherently different from their messenger counterparts [Costa, 2005; Mattick, 2001]. mRNAs (coding RNAs) were only seen as a transitional step of genetic information, a passive link between DNA and a defined biological function filled by the corresponding protein. The duo SRA/SRAP is now forcing us to reconsider this concept.
Acknowledgments
Acknowledgements: I would like to thank Drs. D. Gietz and L.C. Murphy (University of Manitoba) for their constructive suggestions and comments during the preparation of this manuscript. This work was supported by the Canadian Institute for Health Research (CIHR), the Canadian Breast Cancer Research Alliance (CBCRA) and the Cancer Care Manitoba Foundation (CCMF).
Abbreviations
- AF-1
activation function 1
- AF-2
activation function 2
- AR
androgen receptor
- CBP
CREB-binding protein
- CHIP
chromatin immunoprecipitation
- DBD
DNA-binding domain
- ER
estrogen receptor
- ERE
estrogen receptor element
- GR
glucocorticoid receptor
- HRE
hormone-responsive element
- LBD
ligand binding domain
- MAPK
mitogen-activated protein kinase
- NCoR
nuclear corepressor
- PPAR
peroxisome proliferator-activated receptor
- PR
progesterone receptor
- Pus1p
pseudouridine synthase 1
- Pus3p
pseudouridine synthase 3
- RAR
all-trans retinoic acid receptor
- RD
transcriptional repressor domain
- RRM
RNA-interacting domain
- RXR
9-cis retinoic acid receptor
- SHARP
SMRT/HDAC1-associated repressor protein
- SLIRP
SRA stem-loop interacting RNA binding protein
- SRA
steroid receptor RNA activator
- SRA1
steroid receptor RNA activator gene
- SRAP
steroid receptor RNA activator protein
- SRC-1
steroid receptor coactivator 1
- STR
secondary structural motif
- TIF2
transcriptional intermediary factor 2
- TR
thyroid hormone
- YB-1
Y-box binding protein
References
- Ali S., Metzger D., Bornert J. M., Chambon P. Modulation of transcriptional activation by ligand-dependent phosphorylation of the human oestrogen receptor A/B region. Embo J. 1993;12:1153–60. doi: 10.1002/j.1460-2075.1993.tb05756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E., Clarke R. B. Steroid receptors and cell cycle in normal mammary epithelium. J Mammary Gland Biol Neoplasia. 2004;9:3–13. doi: 10.1023/B:JOMG.0000023584.01750.16. [DOI] [PubMed] [Google Scholar]
- Barkhem T., Carlsson B., Nilsson Y., Enmark E., Gustafsson J., Nilsson S. Differential response of estrogen receptor α and estrogen receptor β to partial estrogen agonists/antagonists. Mol Pharmacol. 1998;54:105–12. doi: 10.1124/mol.54.1.105. [DOI] [PubMed] [Google Scholar]
- Caretti G., Lei E. P., Sartorelli V. The DEAD-box p68/p72 proteins and the noncoding RNA steroid receptor activator SRA: eclectic regulators of disparate biological functions. Cell Cycle. 2007;6:1172–6. doi: 10.4161/cc.6.10.4228. [DOI] [PubMed] [Google Scholar]
- Caretti G., Schiltz R. L., Dilworth F. J., Di Padova M., Zhao P., Ogryzko V., Fuller-Pace F. V., Hoffman E. P., Tapscott S. J., Sartorelli V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell. 2006;11:547–60. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Cavarretta I. T., Mukopadhyay R., Lonard D. M., Cowsert L. M., Bennett C. F., O'Malley B. W., Smith C. L. Reduction of coactivator expression by antisense oligodeoxynucleotides inhibits ERalpha transcriptional activity and MCF-7 proliferation. Mol Endocrinol. 2002;16:253–70. doi: 10.1210/mend.16.2.0770. [DOI] [PubMed] [Google Scholar]
- Chabanon H., Mickleburgh I., Hesketh J. Zipcodes and postage stamps: mRNA localisation signals and their trans-acting binding proteins. Brief Funct Genomic Proteomic. 2004;3:240–56. doi: 10.1093/bfgp/3.3.240. [DOI] [PubMed] [Google Scholar]
- Charette M., Gray M. W. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–51. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- Chooniedass-Kothari S., Emberley E., Hamedani M. K., Troup S., Wang X., Czosnek A., Hube F., Mutawe M., Watson P. H., Leygue E. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004;566:43–7. doi: 10.1016/j.febslet.2004.03.104. [DOI] [PubMed] [Google Scholar]
- Chooniedass-Kothari S., Hamedani M. K., Troup S., Hube F., Leygue E. The steroid receptor RNA activator protein is expressed in breast tumor tissues. Int J Cancer. 2006;118:1054–9. doi: 10.1002/ijc.21425. [DOI] [PubMed] [Google Scholar]
- Coleman K. M., Lam V., Jaber B. M., Lanz R. B., Smith C. L. SRA coactivation of estrogen receptor-α is phosphorylation-independent, and enhances 4-hydroxytamoxifen agonist activity. Biochem Biophys Res Commun. 2004;323:332–8. doi: 10.1016/j.bbrc.2004.08.090. [DOI] [PubMed] [Google Scholar]
- Conneely O. M., Jericevic B. M., Lydon J. P. Progesterone receptors in mammary gland development and tumorigenesis. J Mammary Gland Biol Neoplasia. 2003;8:205–14. doi: 10.1023/a:1025952924864. [DOI] [PubMed] [Google Scholar]
- Costa F. F. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Curtis Hewitt S., Couse J. F., Korach K. S. Estrogen receptor transcription and transactivation: Estrogen receptor knockout mice: what their phenotypes reveal about mechanisms of estrogen action. Breast Cancer Res. 2000;2:345–52. doi: 10.1186/bcr79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblois G., Giguere V. Ligand-independent coactivation of ERalpha AF-1 by steroid receptor RNA activator (SRA) via MAPK activation. J Steroid Biochem Mol Biol. 2003;85:123–31. doi: 10.1016/s0960-0760(03)00225-5. [DOI] [PubMed] [Google Scholar]
- Dennis A. P., O'Malley B. W. Rush hour at the promoter: how the ubiquitin-proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J Steroid Biochem Mol Biol. 2005;93:139–51. doi: 10.1016/j.jsbmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Dutertre M., Smith C. L. Ligand-independent interactions of p160/steroid receptor coactivators and CREB-binding protein (CBP) with estrogen receptor-α: regulation by phosphorylation sites in the A/B region depends on other receptor domains. Mol Endocrinol. 2003;17:1296–314. doi: 10.1210/me.2001-0316. [DOI] [PubMed] [Google Scholar]
- Emberley E., Huang G. J., Hamedani M. K., Czosnek A., Ali D., Grolla A., Lu B., Watson P. H., Murphy L. C., Leygue E. Identification of new human coding steroid receptor RNA activator isoforms. Biochem Biophys Res Commun. 2003;301:509–15. doi: 10.1016/s0006-291x(02)03070-x. [DOI] [PubMed] [Google Scholar]
- Evans R. M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–95. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D'Amare A. R. RNA-modifying enzymes. Curr Opin Struct Biol. 2003;13:49–55. doi: 10.1016/s0959-440x(02)00002-7. [DOI] [PubMed] [Google Scholar]
- Fisher B., Costantino J. P., Wickerham D. L., Redmond C. K., Kavanah M., Cronin W. M., Vogel V., Robidoux A., Dimitrov N., Atkins J., Daly M., Wieand S., Tan-Chiu E., Ford L., Wolmark N. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- Fisher B., Powles T. J., Pritchard K. J. Tamoxifen for the prevention of breast cancer. Eur J Cancer. 2000;36:142–50. doi: 10.1016/s0959-8049(99)00269-5. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–15. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S., Walter P., Greene G., Krust A., Goffin C., Jensen E., Scrace G., Waterfield M., Chambon P. Cloning of the human oestrogen receptor cDNA. J Steroid Biochem. 1986;24:77–83. doi: 10.1016/0022-4731(86)90035-x. [DOI] [PubMed] [Google Scholar]
- Hall J. M., McDonnell D. P. Coregulators in nuclear estrogen receptor action: from concept to therapeutic targeting. Mol Interv. 2005;5:343–57. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- Hatchell E. C., Colley S. M., Beveridge D. J., Epis M. R., Stuart L. M., Giles K. M., Redfern A. D., Miles L. E., Barker A., MacDonald L. M., Arthur P. G., Lui J. C., Golding J. L., McCulloch R. K., Metcalf C. B., Wilce J. A., Wilce M. C., Lanz R. B., O'Malley B. W., Leedman P. J. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol Cell. 2006;22:657–68. doi: 10.1016/j.molcel.2006.05.024. [DOI] [PubMed] [Google Scholar]
- Hennighausen L., Robinson G. W. Think globally, act locally: the making of a mouse mammary gland. Genes Dev. 1998;12:449–55. doi: 10.1101/gad.12.4.449. [DOI] [PubMed] [Google Scholar]
- Hube F., Guo J., Chooniedass-Kothari S., Cooper C., Hamedani M. K., Dibrov A. A., Blanchard A. A., Wang X., Deng G., Myal Y., Leygue E. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25:418–28. doi: 10.1089/dna.2006.25.418. [DOI] [PubMed] [Google Scholar]
- Hussein-Fikret S., Fuller P. J. Expression of nuclear receptor coregulators in ovarian stromal and epithelial tumours. Mol Cell Endocrinol. 2005;229:149–60. doi: 10.1016/j.mce.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Jambhekar A., Derisi J. L. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. Rna. 2007;13:625–42. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen E. V., Jordan V. C. The estrogen receptor: a model for molecular medicine. Clin Cancer Res. 2003;9:1980–9. [PubMed] [Google Scholar]
- Jordan V. C., Morrow M. Tamoxifen, raloxifene, and the prevention of breast cancer. Endocr Rev. 1999;20:253–78. doi: 10.1210/edrv.20.3.0368. [DOI] [PubMed] [Google Scholar]
- Jung S. Y., Luo H., Malovannaya A., Kim T., Zhang J., Qin J., O'Malley B. W. Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes. 2005a. [DOI] [PubMed]
- Jung S. Y., Malovannaya A., Wei J., O'Malley B. W., Qin J. Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes. Mol Endocrinol. 2005b;19:2451–65. doi: 10.1210/me.2004-0476. [DOI] [PubMed] [Google Scholar]
- Kato S., Endoh H., Masuhiro Y., Kitamoto T., Uchiyama S., Sasaki H., Masushige S., Gotoh Y., Nishida E., Kawashima H., Metzger D., Chambon P. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–4. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- Kato S., Masuhiro Y., Watanabe M., Kobayashi Y., Takeyama K. I., Endoh H., Yanagisawa J. Molecular mechanism of a cross-talk between oestrogen and growth factor signalling pathways. Genes Cells. 2000;5:593–601. doi: 10.1046/j.1365-2443.2000.00354.x. [DOI] [PubMed] [Google Scholar]
- Kawashima H., Takano H., Sugita S., Takahara Y., Sugimura K., Nakatani T. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem J. 2003;369:163–71. doi: 10.1042/BJ20020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S., Wang H., Richter D., Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21:223–45. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge C. M., Jernigan S. C., Mattingly K. A., Risinger K. E., Zhang J. Estrogen response element-dependent regulation of transcriptional activation of estrogen receptors α and β by coactivators and corepressors. J Mol Endocrinol. 2004;33:387–410. doi: 10.1677/jme.1.01541. [DOI] [PubMed] [Google Scholar]
- Ko L., Cardona G. R., Henrion-Caude A., Chin W. W. Identification and characterization of a tissue-specific coactivator, GT198, that interacts with the DNA-binding domains of nuclear receptors. Mol Cell Biol. 2002;22:357–69. doi: 10.1128/MCB.22.1.357-369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu T., Tanaka T., Ishii J., Matsumura K., Sugimura K., Nakatani T., Kawashima H. Expression and function of human steroid receptor RNA activator in prostate cancer cells: role of endogenous hSRA protein in androgen receptor-mediated transcription. Prostate Cancer Prostatic Dis. 2006;9:173–8. doi: 10.1038/sj.pcan.4500867. [DOI] [PubMed] [Google Scholar]
- Lanz R. B., McKenna N. J., Onate S. A., Albrecht U., Wong J., Tsai S. Y., Tsai M. J., O'Malley B. W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- Lanz R. B., Razani B., Goldberg A. D., O'Malley B. W. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA) Proc Natl Acad Sci U S A. 2002;99:16081–6. doi: 10.1073/pnas.192571399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz R. B., Chua S. S., Barron N., Soder B. M., DeMayo F., O'Malley B. W. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol Cell Biol. 2003;23:7163–76. doi: 10.1128/MCB.23.20.7163-7176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff P., Montano M. M., Schodin D. J., Katzenellenbogen B. S. Phosphorylation of the human estrogen receptor. Identification of hormone-regulated sites and examination of their influence on transcriptional activity. J Biol Chem. 1994;269:4458–66. [PubMed] [Google Scholar]
- Leygue E., Dotzlaw H., Watson P. H., Murphy L. C. Expression of the steroid receptor RNA activator in human breast tumors. Cancer Res. 1999;59:4190–3. [PubMed] [Google Scholar]
- Li Y., Bor Y. C., Misawa Y., Xue Y., Rekosh D., Hammarskjold M. L. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. 2006;443:234–7. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- Lonard D. M., O'Malley B. W. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–4. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Mathur M., Tucker P. W., Samuels H. H. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol. 2001;21:2298–311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–91. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N. J., Lanz R. B., O'Malley B. W. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–44. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- Metivier R., Penot G., Flouriot G., Pakdel F. Synergism between ERalpha transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: requirement for the AF-1 α-helical core and for a direct interaction between the N- and C-terminal domains. Mol Endocrinol. 2001;15:1953–70. doi: 10.1210/mend.15.11.0727. [DOI] [PubMed] [Google Scholar]
- Mosselman S., Polman J., Dijkema R. ER β: identification and characterization of a novel human estrogen receptor. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- Murphy L. C., Simon S. L., Parkes A., Leygue E., Dotzlaw H., Snell L., Troup S., Adeyinka A., Watson P. H. Altered expression of estrogen receptor coregulators during human breast tumorigenesis. Cancer Res. 2000;60:6266–71. [PubMed] [Google Scholar]
- Murphy L. C., Leygue E., Niu Y., Snell L., Ho S. M., Watson P. H. Relationship of coregulator and oestrogen receptor isoform expression to de novo tamoxifen resistance in human breast cancer. Br J Cancer. 2002;87:1411–6. doi: 10.1038/sj.bjc.6600654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S., Inoue S., Watanabe T., Hiroi H., Orimo A., Hosoi T., Ouchi Y., Muramatsu M. The complete primary structure of human estrogen receptor β (hER β) and its heterodimerization with ER α in vivo and in vitro. Biochem Biophys Res Commun. 1998;243:122–6. doi: 10.1006/bbrc.1997.7893. [DOI] [PubMed] [Google Scholar]
- Perissi V., Rosenfeld M. G. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6:542–54. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- Shibata H., Spencer T. E., Onate S. A., Jenster G., Tsai S. Y., Tsai M. J., O'Malley B. W. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent Prog Horm Res. 1997;52:141–64. [PubMed] [Google Scholar]
- Shi Y., Downes M., Xie W., Kao H. Y., Ordentlich P., Tsai C. C., Hon M., Evans R. M. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15:1140–51. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamala G., Yang X., Silberstein G., Barcellos-Hoff M. H., Dale E. Transgenic mice carrying an imbalance in the native ratio of A to B forms of progesterone receptor exhibit developmental abnormalities in mammary glands. Proc Natl Acad Sci U S A. 1998;95:696–701. doi: 10.1073/pnas.95.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., O'Malley B. W. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- Tao Y., Williams-Skipp C., Scheinman R. I. Mapping of glucocorticoid receptor DNA binding domain surfaces contributing to transrepression of NF-κ B and induction of apoptosis. J Biol Chem. 2001;276:2329–32. doi: 10.1074/jbc.C000526200. [DOI] [PubMed] [Google Scholar]
- Tremblay A., Tremblay G. B., Labrie F., Giguere V. Ligand-independent recruitment of SRC-1 to estrogen receptor β through phosphorylation of activation function AF-1. Mol Cell. 1999;3:513–9. doi: 10.1016/s1097-2765(00)80479-7. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Yanagisawa J., Kitagawa H., Takeyama K., Ogawa S., Arao Y., Suzawa M., Kobayashi Y., Yano T., Yoshikawa H., Masuhiro Y., Kato S. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor α coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. Embo J. 2001;20:1341–52. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Watanabe T., Inoue S., Ogawa S., Ishii Y., Hiroi H., Ikeda K., Orimo A., Muramatsu M. Agonistic effect of tamoxifen is dependent on cell type, ERE-promoter context, and estrogen receptor subtype: functional difference between estrogen receptors α and β. Biochem Biophys Res Commun. 1997;236:140–5. doi: 10.1006/bbrc.1997.6915. [DOI] [PubMed] [Google Scholar]
- Weigel N. L., Moore N. L. Kinases and protein phosphorylation as regulators of steroid hormone action. Nucl Recept Signal. 2007a;5:e005. doi: 10.1621/nrs.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel N. L., Moore N. L. Steroid Receptor Phosphorylation: A Key Modulator of Multiple Receptor Functions. Mol Endocrinol. 2007b:In Press. doi: 10.1210/me.2007-0101. [DOI] [PubMed] [Google Scholar]
- Xu B., Koenig R. J. An RNA-binding domain in the thyroid hormone receptor enhances transcriptional activation. J Biol Chem. 2004;279:33051–6. doi: 10.1074/jbc.M404930200. [DOI] [PubMed] [Google Scholar]
- Xu W. Nuclear receptor coactivators: the key to unlock chromatin. Biochem Cell Biol. 2005a;83:418–28. doi: 10.1139/o05-057. [DOI] [PubMed] [Google Scholar]
- Xu B., Koenig R. J. Regulation of thyroid hormone receptor alpha2 RNA binding and subcellular localization by phosphorylation. Mol Cell Endocrinol. 2005b;245:147–57. doi: 10.1016/j.mce.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Xu J., Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17:1681–92. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- Zhao X., Patton J. R., Ghosh S. K., Fischel-Ghodsian N., Shen L., Spanjaard R. A. Pus3p- and Pus1p-dependent pseudouridylation of steroid receptor RNA activator controls a functional switch that regulates nuclear receptor signaling. Mol Endocrinol. 2007;21:686–99. doi: 10.1210/me.2006-0414. [DOI] [PubMed] [Google Scholar]
- Zhao X., Patton J. R., Davis S. L., Florence B., Ames S. J., Spanjaard R. A. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell. 2004;15:549–58. doi: 10.1016/j.molcel.2004.06.044. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]