Abstract
Human filamins are large actin-crosslinking proteins composed of an N-terminal actin-binding domain followed by 24 Ig-like domains (IgFLNs), which interact with numerous transmembrane receptors and cytosolic signaling proteins. Here we report the 2.5 Å resolution structure of a three-domain fragment of human filamin A (IgFLNa19–21). The structure reveals an unexpected domain arrangement, with IgFLNa20 partially unfolded bringing IgFLNa21 into close proximity to IgFLNa19. Notably the N-terminus of IgFLNa20 forms a β-strand that associates with the CD face of IgFLNa21 and occupies the binding site for integrin adhesion receptors. Disruption of this IgFLNa20–IgFLNa21 interaction enhances filamin binding to integrin β-tails. Structural and functional analysis of other IgFLN domains suggests that auto-inhibition by adjacent IgFLN domains may be a general mechanism controlling filamin–ligand interactions. This can explain the increased integrin binding of filamin splice variants and provides a mechanism by which ligand binding might impact filamin structure.
Keywords: cell adhesion, cytoskeleton, filamin, integrin, X-ray crystallography
Introduction
By crosslinking actin filaments and interacting with transmembrane receptors and cytosolic signaling proteins, filamins play important roles in regulating the dynamics of the actin cytoskeleton and integrating cellular mechanics and signaling (Stossel et al, 2001).
Vertebrate filamins are non-covalent dimers of 240–280 kDa subunits composed of an N-terminal actin-binding domain formed from two calponin homology domains followed by a rod region composed of 24 tandem immunoglobulin-like domains (IgFLN1–24) (Gorlin et al, 1990; Stossel et al, 2001; van der Flier and Sonnenberg, 2001). Dimerization is mediated via the C-terminal IgFLN24 (Pudas et al, 2005). Flexible hinges between IgFLN15 and 16 and IgFLN23 and 24 result in a V-shaped flexible actin crosslinker capable of stabilizing orthogonal networks with high-angle F-actin branching (Hartwig et al, 1980). In addition to crosslinking F-actin, filamins act as scaffolds for a growing list of transmembrane receptors, signaling and adapter proteins (Stossel et al, 2001; Feng and Walsh, 2004; Popowicz et al, 2006). In general, these interactions are mediated by the C-terminal domains, IgFLN 16–24, enabling filamin to complex multiple partners in close proximity to one another, potentially enhancing signal transduction (Ohta et al, 2006).
Humans and mice each have three homologous filamin genes encoding the proteins filamin A, B and C; of these, filamin A (FLNa) is the most abundant and widely expressed (Stossel et al, 2001; van der Flier and Sonnenberg, 2001). In mice, FLNa expression is essential for proper cardiac and vascular development (Feng et al, 2006; Hart et al, 2006), FLNb is required for skeletal and microvascular development (Zhou et al, 2007) and FLNc is necessary for normal myogenesis (Dalkilic et al, 2006). In humans, heterozygous null FLNa alleles result in defective neuronal migration causing periventricular heterotopia (Fox et al, 1998), while certain FLNa missense mutations cause familial cardiac valvular dystrophy (Kyndt et al, 2007) and putative gain-of-function mutations result in a spectrum of congenital malformations generally characterized by skeletal dysplasias (Robertson et al, 2003). Mutations in FLNb cause a class of diseases with abnormal vertebral segmentation, joint formation and skeletogenesis (Krakow et al, 2004) and an FLNc mutation causes an autosomal dominant myofibrillar myopathy (Vorgerd et al, 2005). The diversity in phenotypes associated with different filamin mutations reveals that filamins perform a variety of essential functions and the current evidence suggests that in many cases specific disease phenotypes will result from disruption of specific interactions between IgFLN domains and their binding partners.
Despite identification of more than 39 vertebrate filamin-binding proteins (Stossel et al, 2001; Popowicz et al, 2006), relatively little is known about how binding is regulated, how the IgFLN domains are arranged with respect to one another or how the arrangement of IgFLN domains modulates the ligand-binding activity of adjacent domains. We previously identified IgFLNa21 as the major binding site in FLNa for integrin adhesion receptors (Kiema et al, 2006). Integrins, αβ-heterodimers that span the plasma membrane, connect the extracellular environment to the actin cytoskeleton (Hynes, 2002). Thus, filamin–integrin complexes could provide a mechanical and biochemical link through which the dynamic actin cytoskeleton could respond to external cues. Indeed, modulation of integrin–filamin binding through both gain-of-function and loss-of-function mutations in integrin β-tails modulates cell migration (Calderwood et al, 2001) and alternative splicing of filamin genes, which results in deletions of portions of the rod domain, enhances integrin binding and affects myogenesis (Xie et al, 1998; Xu et al, 1998; van der Flier et al, 2002).
The structure of IgFLNa21 in complex with a β7 integrin peptide confirmed that IgFLNa21 is a β-sandwich composed of two β-sheets, similar to other human and Dictyostelium discoideum IgFLNs (Fucini et al, 1997; McCoy et al, 1999; Popowicz et al, 2004; Kiema et al, 2006; Nakamura et al, 2006). The integrin β7 peptide binds to the CD face of the IgFLNa21 β-sandwich and this may represent a general mechanism for IgFLN domain–ligand interactions as other IgFLNs also bind their respective ligands at the CD face (Kiema et al, 2006). Integrin binding to IgFLNa21 can be inhibited by phosphorylation of the integrin tail or by other integrin tail binding proteins that compete with filamin (Kiema et al, 2006); however, whether filamin's ligand-binding activity is itself regulated remained unclear. To date, the only reported multi-domain structures of filamin are of two- and three-domain fragments of D. discoideum filamin (ddFLN), where the domains form an elongated zigzag chain (McCoy et al, 1999; Popowicz et al, 2004). ddFLN is different from vertebrate filamins in that it contains only six IgFLN domains and dimerizes in an end-on antiparallel fashion rather than the proposed parallel or v-shaped arrangement for human filamins. To determine how adjacent human filamin domains are oriented, we investigated an integrin-binding three-domain fragment of FLNa, IgFLNa19–21. X-ray crystallography shows that within this three-domain protein, IgFLNa21 and IgFLNa19 are very similar to one another and to other IgFLNs, whereas IgFLNa20 is partially unfolded and its first strand binds the integrin-binding CD face of IgFLNa21. NMR and biochemical analyses indicate that the IgFLNa20–21 domain pair inhibits integrin β-tail binding and mutations perturbing the IgFLNa20–21 interaction enhance integrin binding. Analysis of other domain pairs suggests that this may be a general feature of filamin–ligand interactions.
Results
Crystal structure of IgFLNa19–21 reveals unexpected domain arrangement
To study the domain arrangement of the major integrin-binding site within filamin, we crystallized a fragment containing human FLNa domains 19–21 (IgFLNa19–21). Diffraction data to 2.5 Å resolution were used for the crystallographic calculations (Table I). The asymmetric unit of the crystal contained two molecules; accordingly, two copies of partial poly-Ala models for IgFLNa21 and IgFLNa19 were initially positioned in the asymmetric unit by the molecular replacement program Phaser (Storoni et al, 2004). In the final model, both copies of IgFLNa19 and IgFLNa21 could be completely built, but loops BC and DG of IgFLNa20 in chain A and 56 residues in chain B could not be included because of missing electron density (Figure 1A and B). The final R-factor values for the model remained moderate (R=25.3%, Rfree=29.8%) apparently because of disorder in the crystals resulting in missing electron density and high B-factor values, especially in IgFLNa20 (Figure 1B). Despite the disorder in IgFLNa20, its partial model could be validated by locating anomalous selenium signals in their appropriate positions of IgFLNa20 in chain A in crystals grown from SeMet-labeled protein (Supplementary Figure S1). As chain A is better resolved, and since non-crystallographic symmetry restraints were used in the refinement (Supplementary data), the structure of chain A was used in all further analyses.
Table 1.
Crystallographic statistics of IgFLNa19–21 (2J3S)
| Data collection | |
| Space group | C2221 |
| Cell dimensions | |
| a, b, c (Å) | 72.3, 78.4, 229 |
| α, β, γ (deg) | 90 |
| Resolution (Å) | 43.4–2.50 (2.56–2.50)a |
| Rsym | 7.3 (26.7) |
| I/σI | 17.3 (3.95) |
| Completeness (%) | 98.7 (89.9) |
| Redundancy | 6.59 (3.30) |
| Refinement | |
| Resolution (Å) | 43.4–2.50 |
| No. of reflections | 20 422 |
| Rwork/Rfree (%) | 25.3/29.8 |
| No. of atoms | |
| Protein | 3724 |
| Ligand/ion | 32 |
| Water | 23 |
| Average B-factors (Å2) | |
| Protein | 33.6 [42.8]b |
| Ligand/ion | 55.9 |
| Water | 24.2 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.016 |
| Bond angles (deg) |
1.45 |
| aHighest resolution shell is shown in parentheses. | |
| bThe values in brackets are calculated including the TLS tensor contribution with the program tlsanl. | |
Figure 1.
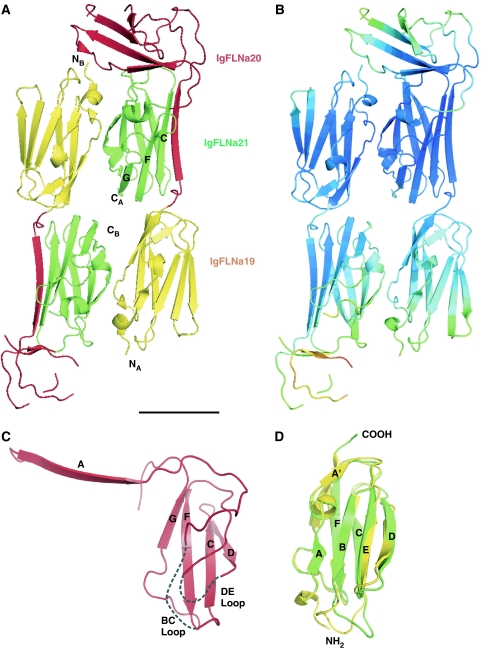
Structure of IgFLNa19–21. (A) Ribbon diagram of the asymmetric unit of the crystals containing two molecules of IgFLNa19–21, IgFLNa19 (yellow), IgFLNa20 (red) and IgFLNa21 (green). The N- and C-termini of molecules A and B are marked as well as β-strands G, F and C of IgFLNa21 in chain A. Scale bar in the bottom right, 25 Å. (B) The same structure as in panel A, now colored according to the calculated total atomic temperature factor (B-factor) values of Cα atoms. The range of B-factor values is from 11 to 143 A2; blue color indicates the lowest values (11–49 Å2), green and yellow higher values (50–79 Å2) and orange and red the highest values (80–143 A2). (C) Ribbon diagram of the isolated IgFLNa20 in the context of IgFLNa19–21. The three-stranded CFG β-sheet that is well ordered in the structure is labeled. Note that β-strand D partially interacts with this sheet. The two disordered loops (BC and DE) absent from the model are arbitrarily indicated with dashed lines. (D) Superimposition of IgFLNa19 (yellow) and IgFLNa21 (green) in the same orientation as panel C.
The domain arrangement of the IgFLNa19–21 fragment is unexpected and different from other immunoglobulin-like domain structures determined thus far (Figure 1A). The three domains form an elongated shape but the domain order along the long axis of the fragment is not sequential. Instead, IgFLNa19 is followed by IgFLNa21 and then IgFLNa20. The β-strands of IgFLNa19 and IgFLNa21 are arranged roughly along the long axis of the fragment, whereas the main part of IgFLNa20 is located across the loops of IgFLNa21 roughly perpendicular to the long axis. While the N-terminus of the fragment is at one end, the C-terminus is in the middle. This arrangement is only possible because IgFLNa20 does not have a complete immunoglobulin-like fold and interacts with IgFLNa21 in an unusual way. The first part of IgFLNa20 is separated from the rest of IgFLNa20 and, as discussed in more detail later, forms an additional β-strand next to the CFG β-sheet of IgFLNa21 (Figures 1A and 2A). The remainder of IgFLNa20 lies on top of IgFLNa21, interacting mainly with the BC loop of IgFLNa21. As a consequence of β-strand A being separated, the half of the IgFLNa20 immunoglobulin sandwich that should include β-strands ABED, as seen in IgFLNa19 and IgFLNa21 (Figure 1D), is rather distorted in IgFLNa20 (Figure 1C).
Figure 2.
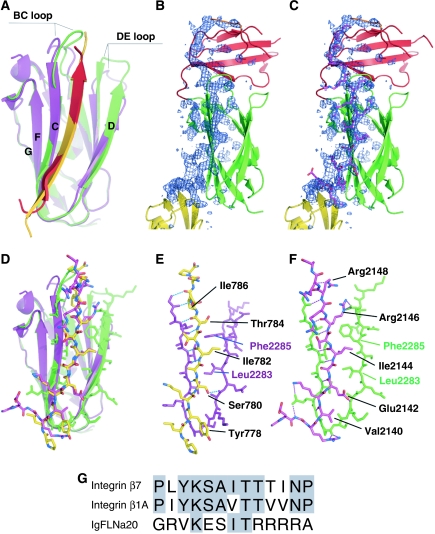
Comparison of interaction of IgFLNa21 with IgFLNa20 and integrin β7. (A) Superimposition of the ribbon diagram of the interaction between the CD face of IgFLNa21 (green) and the first part of IgFLNa20 (red) with the published complex (2BRQ) between IgFLNa21 (purple) and the integrin β7 cytoplasmic domain (gold). The BC and DE loops that differ between the two IgFLNa21 structures are indicated. (B, C) Electron density corresponding to the first strand of IgFLNa20, shown as difference (omit) map calculated without the residues 2136–2159 (shown in purple in panel C). The map is shown as a blue mesh at 2.5σ. The side chains pointing toward β-strand D of IgFLNa21 are well visible in the electron density. (D) Similar superimposition as in panel A with the peptides and IgFLNa21 strand C and D shown as stick models. Colors as in panel A. (E, F) Details of interaction between IgFLNa21 (purple in panel E, green in panel F) and the integrin β7 peptide (E: yellow) and IgFLNa20 (F: red). In both cases, hydrogen bonds are shown as dashed lines. (G) Sequence alignment shows that the IgFLNa20 sequence has little similarity to integrin sequences. Notably the most conserved residues Lys (2141 in IgFLNa20) and Thr (2145) point out from the interaction surface.
While IgFLNa20 has an unusual fold, IgFLNa19 and IgFLNa21 are very similar to one another (Figure 1C) and to previously published IgFLN structures (Fucini et al, 1997; McCoy et al, 1999; Popowicz et al, 2004; Pudas et al, 2005; Kiema et al, 2006; Nakamura et al, 2006). IgFLNa19 and IgFLNa21 can be superimposed with an r.m.s.d. of 1.50 Å for 89 Cα atoms, with the biggest differences observed in the BC and DE loops (Figure 1D). We previously reported the structure of IgFLNa21 bound to a peptide from the integrin β7 cytoplasmic tail (PDB code 2BRQ) (Kiema et al, 2006), and comparison of the two IgFLNa21 structures reveals good alignment of the β-strands (r.m.s.d., 0.60 Å for 40 Cα atoms) but BC and DE loops differ (overall r.m.s.d. 1.67 Å for 91 Cα atoms) (Figure 2A). These differences can be attributed to the contacts with IgFLNa20 (loop BC) in the three-domain structure and the presence of a covalently bound glutathione molecule in 2BRQ (loop DE).
The crystallographic results showed an unexpected domain arrangement of IgFLNa19–21. To test if the whole arrangement of IgFLNa19–21 is stable without the various interactions provided by crystal contacts, we performed a molecular dynamics simulation of a single IgFLNa19–21. As domain–domain movements usually occur on a nanosecond scale (Haran et al, 1992; Wang et al, 2006), the extension of the simulation to 10 ns should be long enough to detect at least the beginning of substantial domain movements. However, during the simulation, further domain–domain packing is observed that stabilize the IgFLNa19–21 structure (Supplementary Figure S2). When compared to the position of IgFLNa21, IgFLNa19 shows only slight movement, while the removal of crystal contacts releases IgFLNa20 to move closer to the head of IgFLNa21 (Supplementary Figure S2). Overall, the simulation results suggest that the domain arrangement of IgFLNa19–21 is rather stable as seen in the crystal.
IgFLNa20 binds the integrin-binding surface of IgFLNa21
We have previously shown that integrin β-tails bind to the CD face of IgFLNs, with the integrin tail forming a β-strand that extends the β-sheet formed by the CFG strands (Kiema et al, 2006). In the current structure, the first strand of IgFLNa20 extends the IgFLNa21 CFG β-sheet in a manner analogous to the integrin peptide and completely covers the integrin-binding site (Figure 2). Despite little primary sequence similarity between the IgFLNa21-binding portions of the integrin β7 tail and IgFLNa20 (Figure 2G), when bound to IgFLNa21 they adopt very similar structures (Figure 2D–F) (r.m.s.d. 0.74 Å for nine Cα atoms) and bury comparable areas (727 A2 for IgFLNa20 and 667 A2 for β7) of accessible surface on the CD face of IgFLNa21. As was observed for the β7 integrin–IgFLNa21 complex, the first strand of IgFLNa20 forms hydrogen bonds to strand C of IgFLNa21 (Figure 2D and F). We have predicted that the specificity of this kind of IgFLN–ligand interaction is mainly determined by the hydrophobic interactions between the binding partner and the side chains of IgFLN β-strand D side chains (Kiema et al, 2006). The common hydrophopic interaction shared between integrin β7 tail and IgFLNa20 is Ile (IgFLNa residue 2144 and β7 residue 782) that is sandwiched between Leu2283 and Phe2285 of IgFLNa21 β-strand D (Figure 2D–F). Other interactions of IgFLNa20 seem to be less optimal than those of the integrin tail. In particular, the four Arg residues (2146–2149) of IgFLNa20 appear to form suboptimal interactions and are quite uncommon for a β-strand (Figure 2F).
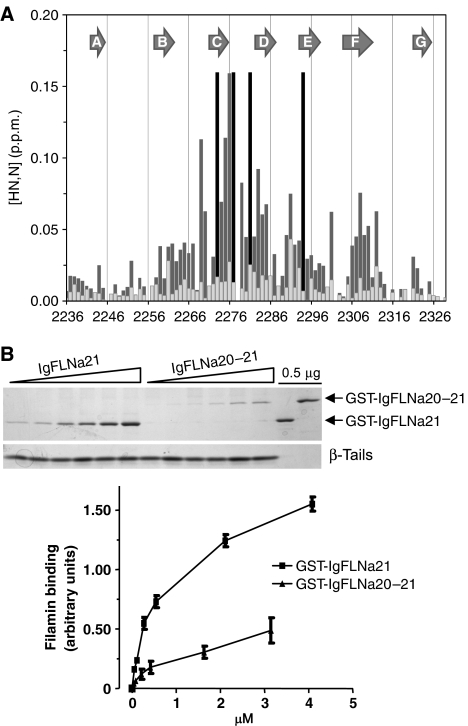
As described above, molecular dynamic simulations predict that the interaction between the first part of IgFLNa20 and the CD face of IgFLNa21 is stable, but to test this in solution NMR experiments were performed. The interaction between IgFLNa21 and IgFLNa20 was validated by solution-state NMR in two ways: (i) Addition of IgFLNa20 to 15N-labeled IgFLNa21 gave selective shifts concentrated on the CD face of IgFLNa21 (Figure 3A). This pattern is very similar to that observed with the β7 integrin (Kiema et al, 2006) and confirms that the CD face of IgFLNa21 is involved in the binding of IgFLNa20. Furthermore, the shifts induced by a truncated IgFLNa20 protein (residues 2167–2235), which lacks the residues forming the first β-strand, were very much reduced (Figure 3A), demonstrating that this region of IgFLNa20 is required for binding IgFLNa21. (ii) Since there is only one Ala–Leu pair in the IgFLNa19–21 sequence (Ala2272 and Leu2271), selective labeling (1-13C-Leu, 15N-Ala) of IgFLNa19–21 allowed Ala2272 in strand C to be uniquely identified in the complicated spectrum of the triple domain; this resonance was observed to be considerably shifted in IgFLNa19–21 at 37°C (δHN=8.080 p.p.m., δN=124.918 p.p.m.) compared to that in IgFLNa21 alone (δHN=8.921 p.p.m. and δN=124.970 p.p.m.). Taken together, these NMR experiments give strong support to the interaction of the N-terminus of IgFLNa20 with the CD face of IgFLNa21 as seen in the crystal structure.
Figure 3.
IgFLNa20–21 domain pair inhibits integrin binding. (A) Chemical shift perturbations of resonances in the U-15N-IgFLNa21 domain induced by a 30-fold excess of IgFLNa20 (dark gray) or IgFLNa20 var-1 (light gray). The combined chemical shifts of amide proton and nitrogen resonances in [1H,15N]-HSQC spectra were calculated using the formula ΔδHN,N=[(ΔδHN)2+(0.154 × ΔδN)2]½. The black lines correspond to residues whose resonances were severely broadened on addition of IgFLNa20. (B) Binding of purified GST-IgFLNa21 or GST-IgFLNa20–21 to β7 integrin tails was assessed by protein staining of pull-down assays. Protein binding was quantified by densitometry and filamin bound was calculated as the ratio of filamin bound to filamin in the loading control in each experiment (mean±s.e.; n⩾3).
IgFLNa20 inhibits integrin binding to IgFLNa21
IgFLNa20 and integrin β-tails bind to the same site on IgFLNa21 (Figure 2), suggesting that they may compete for binding to IgFLNa21. NMR analysis indicates that when free in solution integrin β7 tails bind IgFLNa21 with a higher affinity than IgFLNa20 does, and that β7 tails can displace free IgFLNa20 from IgFLNa21—evidenced by induction of a new pattern of shifts following addition of β7 tails (Supplementary Figure S3). However, in intact filamin, IgFLNa20 is tethered to IgFLNa21, thus increasing the effective local concentration and the occupancy of the interaction. To assess the impact of IgFLNa20 on integrin binding to IgFLNa21, we compared the binding of GST-IgFLNa21 and the two domain fragment GST-IgFLNa20–21. GST-IgFLNa21 bound to recombinant β7 integrin tails in a dose-dependent manner (Figure 3B). When quantified by scanning densitometry, curve fitting analysis indicated an apparent Kd of 0.7±0.1 μM, in good agreement with our previous data (Kiema et al, 2006). Notably, binding of the two-domain fragment to β7 was much lower (Figure 3B). A reliable calculation of the binding affinity for this interaction was not possible because saturation of GST-IgFLNa20–21 binding to β7 could not be achieved.
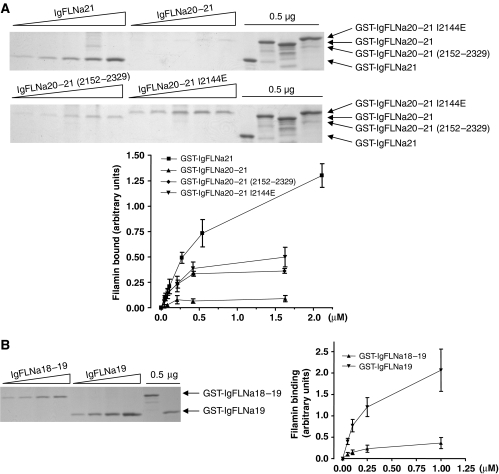
Structural analyses show that the first strand of IgFLNa20 is responsible for the interaction with IgFLNa21. We therefore generated a truncated IgFLNa20–21 protein (residues 2152–2329) lacking the first 13 amino acids of IgFLNa20, which normally form the interacting strand. This protein displayed a significant increase in β7 binding compared to wild-type IgFLNa20–21 (Figure 4A). To specifically disrupt the first strand interaction with IgFLNa21 without deleting a large stretch of amino acids, we substituted Ile2144 with Glu. Structurally Ile2144 corresponds to Ile782 in the integrin β7 tail (Figure 2E–G), which is important for β7 integrin binding to filamin (Calderwood et al, 2001), and occupies a hydrophobic pocket on IgFLNa21 that is important for integrin binding (Kiema et al, 2006). We predicted that the introduction of a large charged residue at this site should destabilize the interaction, and observed that IgFLNa20–21 (I2144E), like the N-terminal truncation, displayed enhanced β7 integrin binding in comparison to wild-type IgFLNa20–21 (Figure 4A). Thus, disruption of the IgFLNa20–IgFLNa21 interaction can enhance integrin binding, presumably through exposure of the integrin binding CD face on IgFLNa21.
Figure 4.
Mutations in the first strand of IgFLNa20 relieve inhibition of integrin binding. Binding of purified GST-IgFLNa21, -IgFLNa20–21, -IgFLNa20–21(2152–2329) or -IgFLNa20–21 I2144E (A) or GST-IgFLNa19 or -IgFLNa18–19 (B) to β7 integrin tails was assessed by protein staining of pull-down assays. Protein binding was quantified by densitometry and calculated as described in Figure 3 (mean±s.e.; n⩾3).
IgFLNa20 inhibits integrin binding to intact filamin
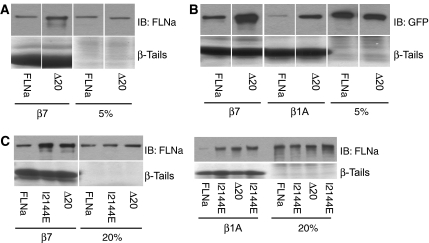
The experiments described above were performed using short, bacterially expressed recombinant fragments of filamin. To verify the results in the context of filamin expressed in cultured cells, we compared the ability of integrin β-tails to pull down wild-type and mutated filamin from cell lysates. FLNa lacking IgFLNa20 (FLNaΔ20) exhibited enhanced binding to β7 integrin tails (Figure 5A), consistent with an inhibitory role for IgFLNa20. Similar results were obtained using both untagged and GFP-tagged FLNa (Figure 5B); this effect was not limited to β7 integrins, as removal of IgFLNa20 also enhanced binding to β1A tails (Figure 5B), consistent with the general ability of β-integrin tails to bind to the CD face of IgFLNa21 (Kiema et al, 2006).
Figure 5.
IgFLNa20 inhibits integrin binding to intact filamin. Pull-down assays were performed with integrin tails using lysates from CHO cells transfected with untagged (A) or GFP-tagged (B) human FLNa and FLNaΔ20; bound proteins were detected by immunoblotting with anti-human FLNa or anti-GFP antibodies. (C) Binding of GFP-tagged human FLNa, FLNaΔ20 or FLNa(I2144E), expressed in CHO cells, to integrin tails was assessed in pull-down assays. 5 or 20% lanes represent corresponding percentage of the starting material in the binding assay.
The binding of FLNa (I2144E), containing the point mutation that destabilizes the IgFLNa20–IgFLNa21 interaction, was also assessed (Figure 5C). GFP-FLNa (I2144E) displayed enhanced binding to β7 and β1A integrin tails in pull-down assays from cell lysates (Figure 5C). Thus in the context of full-length filamin, IgFLNa20 masks the major integrin-binding site in IgFLNa21 and a single point mutation is sufficient to expose the integrin-binding site and enhance integrin–filamin interactions.
Filamin splice variants lack the inhibitory IgFLNa20 sequence
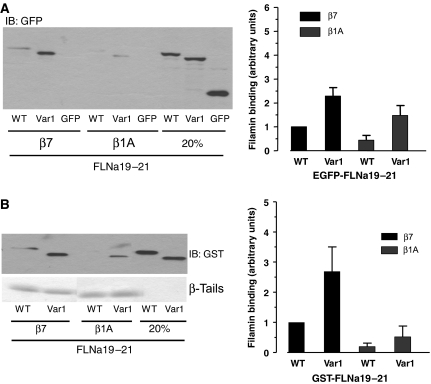
The observation that an intramolecular interaction between two adjacent IgFLNa domains reduces integrin binding to filamin suggests that this interaction may form part of a regulatory mechanism controlling filamin association with ligands. van der Flier et al (2002) have shown that naturally occurring FLNa and FLNb splice variants (var-1) exhibit enhanced binding to a variety of integrin β-tails. These splice variants lack a 41-amino-acid sequence encompassing the C-terminal part of IgFLNa19 and the N-terminal part of IgFLNa20, including the first strand of IgFLNa20. We have confirmed the previously reported increase in β1A integrin-binding activity of FLNa var-1 proteins using EGFP-tagged FLNa19–21 var-1 expressed in cultured cells and shown that binding to β7 tails is also increased (Figure 6A). We have also shown increased binding of purified bacterially expressed GST-FLNa19–21 var-1 protein to both β7 and β1A integrin tails (Figure 6B). Alternative splicing may therefore be one mechanism by which the inhibitory intramolecular IgFLNa20–IgFLNa21 interaction is regulated to control filamin's ligand-binding activities.
Figure 6.
Filamin splicing enhances integrin binding. (A) Pull-down assays were performed with integrin tails using lysates from CHO cells transfected with EGFP-tagged IgFLNa19–21 or IgFLNa19–21 var-1. Filamin bound was assessed by immunoblotting, quantified by densitometry and expressed as the ratio of filamin bound to filamin in the loading control in each experiment and normalized to 1 for IgFLNa19–21 binding to β7 tails (mean±s.e.; n⩾3). (B) Binding of purified GST-IgFLNa19–21 or IgFLNa19–21 var-1 (1 μg) to β7 integrin tails was assessed by immunoblotting of pull-down assays. Protein binding was quantified as previously described (mean±s.e.; n⩾3).
Auto-inhibition of ligand binding by other even-numbered domains
The preceding data indicate that IgFLNa20 negatively regulates ligand binding to IgFLNa21. Structural analysis of IgFLNa21 and IgFLNa19 reveals that both domains are very similar (Figure 1D), and mutagenesis and NMR analysis suggest that IgFLNa19 and IgFLNa21 both bind integrin β-tails in a very similar fashion (Kiema et al, 2006). We therefore tested whether IgFLNa18 could negatively regulate integrin binding to IgFLNa19. GST-IgFLNa19 bound β-integrins in a dose-dependent manner similar to IgFLNa21, but in comparison the two-domain construct GST-IgFLNa18–19 displayed severely reduced binding (Figure 4B). Thus, the auto-inhibition of ligand binding to IgFLN domains by the preceding even-numbered IgFLNa domains may be a more general phenomenon.
Discussion
We have described the molecular structure of a three-domain fragment of human FLNa. This reveals an unexpected arrangement with domains in a non-sequential order contrary to previous proposed models, and this arrangement is possible because IgFLNa20 adopts an unusual structure. The structure also reveals a mechanism of auto-inhibition, limiting accessibility to the integrin-binding site in filamin. This auto-inhibitory mechanism may be extended to other ligand-binding sites in filamin. Finally, loss of auto-inhibition in filamin splice variants provides a molecular explanation for their enhanced integrin-binding activity.
Structure of IgFLNa19–21
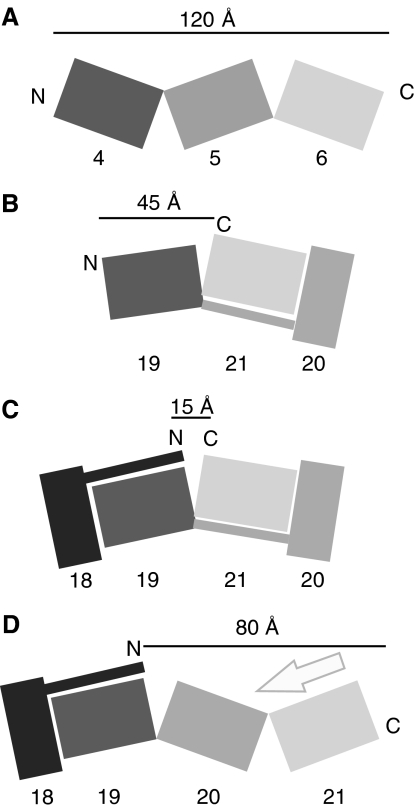
Vertebrate filamins contain 24 tandem immunoglobulin-like domains. Until now, our understanding of how adjacent IgFLN domains interact has been based on the structures of two- and three-domain fragments of ddFLN, where the domains form an elongated zigzag chain (Figure 7A) (McCoy et al, 1999; Popowicz et al, 2004). The structure of IgFLNa19–21 described here introduces another model for IgFLN packing (Figure 7B). In this structure, IgFLNa20 and IgFLNa21 fold together because IgFLNa20 is divided into two parts: the first part forms a β-strand next to strand C of IgFLNa21, and the rest lies on top of the BC loop of IgFLNa21. IgFLNa20 therefore forms an incomplete Ig-like fold. The usual arrangement of multiple Ig-like domains is an extended linear arrangement, for example that found in ddFLN (Popowicz et al, 2004), fibronectin (Leahy et al, 1996) or titin (Marino et al, 2005). Domain swapping (Rousseau et al, 2003) and interactions between non-contiguous domains (Pickford et al, 2001) have been observed previously but this is the first interdomain interaction of this type we are aware of.
Figure 7.
Illustration of filamin domain arrangements. (A) ddFLN Ig domain 4–6. (B) Human IgFLNa19–21. (C) A hypothetical model of two filamin domain pairs showing that in this kind of arrangement the N- and C-termini of the four-domain fragment would be close to each other. Additional IgFLN domains or domain pairs can be accommodated in this model by allowing flexibility in the interdomain linker regions. (D) A hypothetical model showing how binding of an integrin β-tail (arrow) to IgFLNa21 might affect the overall arrangement of IgFLNa18–21. Our data do not indicate whether ligand binding to one domain pair impacts adjacent domain pairs.
Using NMR techniques we tested whether the pairing between IgFLNa20 and IgFLNa21 also takes place in solution and showed that the interaction of the first part of IgFLNa20 and the CD face of IgFLNa21 took place between isolated domains, and also in the context of the three-domain IgFLNa19–21 fragment. Thus, we believe that this arrangement represents a genuine property of this filamin fragment.
Gorlin et al (1990) first reported the predicted amino-acid sequence of human filamin, and, noting that the N-terminal parts of some of the more C-terminal even-numbered domains (IgFLNa16, 18, 20 and 22) are different from other filamin domains (Supplementary Figure S4), proposed that these differences may change the way in which neighboring domains interact (Gorlin et al, 1990). We have now shown that the N-terminal portion of IgFLNa20 is involved in a domain–domain interaction that results in inhibition of ligand binding to IgFLNa21. Whether other similar structural and functional domain pairs exist in filamins remains to be determined; however, we have observed that IgFLNa18 inhibits integrin binding to IgFLNa19. Furthermore, recent NMR structures of individual IgFLNc16 (PDB code 2D7N), IgFLNb18 (2DMC) and IgFLNb20 (2DLG) domains are very similar to the structure of IgFLNa20 in our crystal; they all lack the first β-strand and the remaining BEG sheet is characteristically tilted when compared to complete IgFLNs. This may suggest that IgFLN16 and IgFLN17, and IgFLN18 and IgFLN19 also form domain pairs, possibly resulting in inhibition of ligand binding to the odd-numbered domains. Interestingly, the NMR structure of IgFLNb22 (2D7P) has all the β-strands in appropriate positions. Currently, we have no evidence for domain pairs in IgFLN1–15.
Considering that the chain of IgFLNs continues in both directions from the IgFLNa20–21 domain pair, it is interesting that the N- and C-termini of the domain pair are only about 20 Å apart, whereas in an arrangement as in the ddFLN, this distance would be 80 Å (Figure 7A and B). Thus, formation of domain pairs should reduce the overall length of the filamin rod region. If additional IgFLN domain pairs can form simultaneously, more significant effects on rod length are expected (Figure 7C). Because published EM images of filamin lack sufficient resolution to clearly identify individual domains and show considerable conformational heterogeneity in the rod (Tyler et al, 1980; Castellani et al, 1981; Hartwig and Stossel, 1981), it is difficult to reliably relate our findings to EM images. However, Gorlin et al (1990) report that while IgFLNa1–15 has a contour length of ∼55 nm, yielding an average span of ∼3.7 nm per IgFLNa, which is in good agreement with that seen in ddFLN Ig domains (Popowicz et al, 2004) (Figure 7A), IgFLNa16–23 only stretches over ∼15 nm (an average of ∼1.9 nm per IgFLNa). This reveals a difference in domain packing in the C-terminal portion of the filamin rod, consistent with the localization of domain pairs to this region.
Auto-inhibition of integrin binding by IgFLNa20
The N-terminal part of IgFLNa20 covered the integrin-binding surface of IgFLNa21. In fact, IgFLNa20 interacted with IgFLNa21 in a very similar way to the integrin β7 subunit cytoplasmic tail (Kiema et al, 2006). Structural analysis suggests that the intramolecular domain–domain interaction is less optimal than the integrin binding: the integrin peptide makes better hydrophopic interactions with IgFLNa21 than IgFLNa20 does. In particular, Arg2146 and Arg2148 that are located toward the end of the interacting β-strand of IgFLNa20 appear to have rather unfavorable interactions with IgFLNa21, although their side chains are well ordered in our structure (Figure 2B and C). In accordance with this, the presence of IgFLNa20 inhibited integrin binding to IgFLNa21, but this inhibition was only partial. The auto-inhibitory effect of IgFLNa20 was verified using deletion and point mutants in the context of both filamin fragments and full-length filamin.
Regulation of auto-inhibition of integrin binding
The discovery of intramolecular auto-inhibition of integrin binding raises the possibility that this provides a mechanism to regulate filamin–integrin interaction. If this is the case, the cell must be able to release the auto-inhibition in certain circumstances and so a mechanism for changing the conformation of the IgFLNa20–21 pair is required. Alternative splicing of filamin mRNA provides one such mechanism. Alternative splicing of FLNa has been shown to remove exon 40, generating FLNa var-1 lacking 41 amino acids that lie within IgFLNa19 and IgFLNa20 (van der Flier et al, 2002). This results in loss of the auto-inhibitory first β-strand of IgFLNa20 and increased integrin binding. The corresponding FLNb var-1 protein also exhibits enhanced integrin binding (van der Flier et al, 2002), suggesting that IgFLNb20 plays a similar auto-inhibitory role in modulating FLNb integrin interactions. While the mRNA for these splice variants has a weak but widespread distribution, factors regulating the splicing remain unknown, so it is unclear under what conditions splicing is used to release auto-inhibition of integrin binding.
Other possible ways to regulate the auto-inhibition of filamin's integrin-binding site include phosphorylation and mechanical force. Ser2152, a phosphorylation target for PKA, PAK1 and RSK (Jay et al, 2000; Vadlamudi et al, 2002; Woo et al, 2004), is located just after the first strand of IgFLNa20. In our structure, Ser2152 is exposed and susceptible to phosphorylation. It has been suggested that phosphorylation at this Ser may modulate integrin binding, but based on its location, phosphorylation is unlikely to affect auto-inhibition of integrin binding. Consistent with this, neither phospho-mimicking nor phospho-blocking mutations at this site impact integrin binding to an FLNa19–24 fragment (Travis et al, 2004). However, the effect that the negative charge introduced by phosphorylation may have on other neighboring domains remains to be tested.
In response to mechanical force transmitted through integrin adhesion receptors, FLNa is recruited to integrin-mediated adhesion contacts where it has a mechanoprotective effect (Glogauer et al, 1998; D'Addario et al, 2002). It is possible that mechanical forces acting on filamin alter the conformation of the partially unfolded IgFLNa20, modulating its auto-inhibitory effect and so controlling filamin integrin–integrin interactions. Single molecule force spectroscopy has shown that ddFLN domain 4 unfolds at lower forces than other domains via a stable unfolded intermediate (Schwaiger et al, 2004); similar studies on human filamin have shown that human IgFLN domains have a broad range of unfolding forces (Furuike et al, 2001). Stretching of filamin molecules would potentially pull the first strand of IgFLNa20 away from IgFLNa21 exposing the CD face and enhancing binding of ligands, including integrins, which could trigger signaling cascades to respond to increased mechanical stress.
Effects of ligand binding on the overall arrangement of filamin rod region
We have shown that IgFLNa20–21 is folded as a domain pair and that integrin binding can outcompete the main interaction between IgFLNa20 and IgFLNa21. As a consequence of integrin binding, we would expect release of the first part of IgFLNa20 from its interaction with FLNa21. It will be important to determine the effect this has on the overall conformation of the IgFLNa20–21 after integrin binding. If IgFLNa20 can form a complete immunoglobulin-like fold after its first strand is displaced from IgFLNa21, the overall topology of the two-domain pair might change to resemble the arrangement found in ddFLN (Figure 7D). This, in turn, might change the overall length and orientation of the filamin rod region. As noted above, the conformational heterogeneity and limited resolution of EM images of filamin (van der Flier and Sonnenberg, 2001) make it difficult to relate them to our atomic models. However, based on the domain packing seen in ddFLN (Popowicz et al, 2004), the predicted length of ‘activated' filamin monomers lacking any domain pairs would be ∼100 nm, in good agreement with 98±10 nm measurements by Castellani et al (1981) but longer than the ∼80 nm reported by Tyler et al (1980) and Hartwig and Stossel (1981), which conceivably represents a form containing domain pairs. Thus, we envision that by inducing changes in interdomain arrangements, integrin binding might have long-range effect in filamin. Likewise, similar effects are expected to be caused by ligand binding to other filamin domain pairs.
Although further studies are required for a complete molecular understanding of filamin topology and interactions, our current structural and biochemical studies have revealed new and interesting domain–domain interactions that provide mechanisms for regulating ligand binding to filamin and may allow filamin ligands to influence the length and architecture of the filamin rod region.
Materials and methods
Protein production
Recombinant His-tagged integrin cytoplasmic tail model proteins were produced and purified as previously described (Pfaff et al, 1998). IgFLNa19 (amino acids 2046–2141), IgFLNa21 (amino acids 2236–2329), IgFLNa20–21 (2142–2329), IgFLNa18–19 (amino acids 1955–2141), IgFLNa19–21 (amino acids 2046–2329) and IgFLNa19–21 var-1 (amino acids 2046–2125 then 2168–2329) were generated by polymerase chain reaction and subcloned into pGEX (Amersham) or EGFP (BD Biosciences) vectors for expression of GST or EGFP fusion proteins. Point mutations and deletions were introduced by QuikChange™ site-directed mutagenesis (Stratagene). All inserts were verified by DNA sequencing. GST fusion proteins were produced in Escherichia coli BL21 cells and purified on Glutathione Sepharose 4 Fast Flow medium (Amersham Biosciences) according to the manufacturer's instructions. The IgFLNa19–21 fragment used for crystallization was cloned in modified pET24d vector containing a His6 tag followed by tobacco etch virus (TEV) cleavage site (Pudas et al, 2005). Protein was produced in E. coli BL21(DE3) cells and purified on Ni-NTA Agarose (Qiagen) according to the manufacturer's instructions. The His6 tag was cleaved with TEV protease (Invitrogen) during overnight dialysis at 4°C to 50 mM NaCl and 50 mM Tris–HCl pH 8.0. Additional purification was achieved by anion-exchange chromatography on Acell QMA matrix (Waters) and gel filtration on HiLoad 16/60 Superdex 75 column (Amersham Biosciences).
Crystallography
The IgFLNa19–21 protein was crystallized using the hanging drop vapor diffusion method at 22°C by mixing 1 ml of 30 mg/ml protein solution in 100 mM NaCl, 50 mM Tris and 1 mM DTT pH 8.0 with an equal volume of 1.6 M (NH4)2SO4, 0.1 M citric acid pH 6.1 and 10% dioxane. The crystals were transferred to 0.25 M KBr, 20% glycerol, 1.6 M (NH4)2SO4 and 0.1 M citric acid pH 6.1 before freezing under liquid nitrogen. The data for final structure solution were collected at 100 K at European Synchrotron Radiation Facility (Grenoble, France) beam line ID23-1 by using MarMosaic 225 CCD detector (Marresearch GmbH). The data were processed with the XDS program package (Kabsch, 1993). Partial poly-Ala models for IgFLNa19 and IgFLNa21 were derived from IgFLNc24 structure (PDB code 1VO5) by replacing non-identical amino acids with Ala. Two copies of each of the models were initially positioned by molecular replacement program Phaser (Storoni et al, 2004) and the final model was generated by iterating between manual model building with programs O (Jones et al, 1991) or Coot (Emsley and Cowtan, 2004) and TLS+ restrained refinement with Refmac 5.2 (Murshudov et al, 1997). Tight non-crystallographic symmetry restrains between IgFLNa19 and IgFLNa21 of chains A and B were used in the final refinement. Crystallographic images were generated with PYMOL (DeLano Scientific, San Carlos, CA, USA; http://www.pymol.org). Further details of the crystallographic data, structure validation and molecular dynamics are given in Supplementary data. Atomic coordinates and structure factors have been deposited in the Protein Data Bank, accession code 2J3S.
NMR
IgFLNa20 (2141–2235) and IgFLNa20 var-1 (2167–2235) were expressed from a pGEX-6P-2 vector with a Precission protease cleavage site using BL21 codon plus cells (Stratagene) and uniform labeling was achieved by growing in M9 minimal media with 15N-NH4Cl and 13C-glucose. Selective labeling of IgFLNa19–21(1–13C-L2271,15N-A2272) was produced using a mixture of labeled (1-13C-leucine and 15N-alanine) and non-labeled amino acids (0.1 g for each per liter M9) (Peterson et al, 2001). The identity and purity of the products were confirmed by mass spectrometry and SDS–PAGE. The β7-derived peptide 776PLYKSAITTTINP788 (N-terminally acetylated and C-terminally amidated) was purchased from EZBiolab (USA).
All NMR samples were buffered with 50 mM sodium phosphate (pH 6.10) containing 100 mM NaCl and 5 mM DTT in 90% H2O and 10% D2O. NMR data were collected at 1H frequencies of 500, 600 and 750 MHz. The backbone amide 15N and 1H chemical shifts for wild-type IgFLNa21 were assigned using a 1 mM U-13C,15N-labeled protein sample and standard triple resonance experiments (Astrof et al, 1998). Gradient enhanced [1H,15N]-HSQC (Schleucher et al, 1994) experiments were used to carry out titrations for 100 μM IgFLNa21 with varying amounts of β7/IgFLNa20/IgFLNa20 var-1 at 25°C. The [1HN,15N] chemical shifts for (1-13C-L2271,15N-A2272)-IgFLNa19–21 were recorded with a 500 μM sample at 37°C on a Bruker cryoprobe-equipped Avance 500 MHz machine using a 3D HNCO pulse sequence (Kay et al, 1994). NMR data processing was carried out with NMRPipe (Delaglio et al, 1995) and SPARKY (www.cgl.ucsf.edu/home/sparky). Spectra were referenced to the water proton shift (4.766 p.p.m. at 25°C, 4.623 p.p.m. at 37°C) (Wishart et al, 1995) with indirect referencing in the nitrogen dimension using a 15N-1H frequency ratio of 0.101329118 (IUPAC).
Binding assays and analysis
Binding assays using recombinant integrin tail model proteins were performed as previously described (Pfaff et al, 1998; Calderwood et al, 2001). For GFP fusion proteins, or full-length human FLNa or FLNa mutants, Chinese hamster ovary (CHO) cells were transiently transfected with 3 μg of expression vector using Lipofectamine™ (Invitrogen), cells were harvested 24–48 h later, lysed as described previously (Calderwood et al, 2001) and binding assays were performed. Anti-filamin mAb1680 (Chemicon) and anti-GFP (Rockland) antibodies were purchased.
Supplementary Material
Supplementary Methods
Acknowledgments
This work was supported by a grant from the National Institutes of Health (ROI GM068600-01) to DAC. We acknowledge support from the Wellcome trust (CHC and IDC), the Clarendon Fund (PJ), the Academy of Finland (JY: 207021, 105211, 114713) and CSC-the Finnish IT Centre for Science for computational grant jyy2516. We thank the European Synchrotron Radiation Facility for beam time, Dr Didier Nurizzio for assistance using beam line ID23-1 and Dr Sosuke Yoshinaga, who prepared the IgFLNa20 and IgFLNa20 var-1 constructs for the NMR experiments. We thank Dr Titus Boggon for helpful discussions.
References
- Astrof N, Bracken C, Cavanagh J, Palmer AG III (1998) CO_H(N)CACB experiments for assigning backbone resonances in 13C/15N-labeled proteins. J Biomol NMR 11: 451–456 [DOI] [PubMed] [Google Scholar]
- Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, Schwartz MA, Ginsberg MH (2001) Increased filamin binding to β-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol 3: 1060–1068 [DOI] [PubMed] [Google Scholar]
- Castellani L, Offer G, Elliott A, O'Brien EJ (1981) Structure of filamin and the F-actin-heavy merofilamin complex. J Muscle Res Cell Motil 2: 193–202 [DOI] [PubMed] [Google Scholar]
- D'Addario M, Arora PD, Ellen RP, McCulloch CA (2002) Interaction of p38 and Sp1 in a mechanical force-induced, β1 integrin-mediated transcriptional circuit that regulates the actin-binding protein filamin-A. J Biol Chem 277: 47541–47550 [DOI] [PubMed] [Google Scholar]
- Dalkilic I, Schienda J, Thompson TG, Kunkel LM (2006) Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol 26: 6522–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, Nakamura F, Kwiatkowski DJ, Walsh CA (2006) Filamin A (FLNA) is required for cell–cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci USA 103: 19836–19841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Walsh CA (2004) The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol 6: 1034–1038 [DOI] [PubMed] [Google Scholar]
- Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, Graham DA, Scheffer IE, Dobyns WB, Hirsch BA, Radtke RA, Berkovic SF, Huttenlocher PR, Walsh CA (1998) Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron 21: 1315–1325 [DOI] [PubMed] [Google Scholar]
- Fucini P, Renner C, Herberhold C, Noegel AA, Holak TA (1997) The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold. Nat Struct Biol 4: 223–230 [DOI] [PubMed] [Google Scholar]
- Furuike S, Ito T, Yamazaki M (2001) Mechanical unfolding of single filamin A (ABP-280) molecules detected by atomic force microscopy. FEBS Lett 498: 72–75 [DOI] [PubMed] [Google Scholar]
- Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, McCulloch CA (1998) The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem 273: 1689–1698 [DOI] [PubMed] [Google Scholar]
- Gorlin JB, Yamin R, Egan S, Stewart M, Stossel TP, Kwiatkowski DJ, Hartwig JH (1990) Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J Cell Biol 111: 1089–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran G, Haas E, Szpikowska BK, Mas MT (1992) Domain motions in phosphoglycerate kinase: determination of interdomain distance distributions by site-specific labeling and time-resolved fluorescence energy transfer. Proc Natl Acad Sci USA 89: 11764–11768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AW, Morgan JE, Schneider J, West K, McKie L, Bhattacharya S, Jackson IJ, Cross SH (2006) Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet 15: 2457–2467 [DOI] [PubMed] [Google Scholar]
- Hartwig JH, Stossel TP (1981) Structure of macrophage actin-binding protein molecules in solution and interacting with actin filaments. J Mol Biol 145: 563–581 [DOI] [PubMed] [Google Scholar]
- Hartwig JH, Tyler J, Stossel TP (1980) Actin-binding protein promotes the bipolar and perpendicular branching of actin filaments. J Cell Biol 87: 841–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110: 673–687 [DOI] [PubMed] [Google Scholar]
- Jay D, Garcia EJ, Lara JE, Medina MA, de la Luz Ibarra M (2000) Determination of a cAMP-dependent protein kinase phosphorylation site in the C-terminal region of human endothelial actin-binding protein. Arch Biochem Biophys 377: 80–84 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallog 26: 795–800 [Google Scholar]
- Kay LE, Xu GY, Yamazaki T (1994) Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J Magn Reson A109: 129–133 [Google Scholar]
- Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, Wegener KL, Campbell ID, Ylanne J, Calderwood DA (2006) The molecular basis of filamin binding to integrins and competition with talin. Mol Cell 21: 337–347 [DOI] [PubMed] [Google Scholar]
- Krakow D, Robertson SP, King LM, Morgan T, Sebald ET, Bertolotto C, Wachsmann-Hogiu S, Acuna D, Shapiro SS, Takafuta T, Aftimos S, Kim CA, Firth H, Steiner CE, Cormier-Daire V, Superti-Furga A, Bonafe L, Graham JM Jr, Grix A, Bacino CA et al. (2004) Mutations in the gene encoding filamin B distrupt vertebral segmentation, joint formation and skeletogenesis. Nat Genet 36: 405–410 [DOI] [PubMed] [Google Scholar]
- Kyndt F, Gueffet JP, Probst V, Jaafar P, Legendre A, Le Bouffant F, Toquet C, Roy E, McGregor L, Lynch SA, Newbury-Ecob R, Tran V, Young I, Trochu JN, Le Marec H, Schott JJ (2007) Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation 115: 40–49 [DOI] [PubMed] [Google Scholar]
- Leahy DJ, Aukhil I, Erickson HP (1996) 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 84: 155–164 [DOI] [PubMed] [Google Scholar]
- Marino M, Svergun DI, Kreplak L, Konarev PV, Maco B, Labeit D, Mayans O (2005) Poly-Ig tandems from I-band titin share extended domain arrangements irrespective of the distinct features of their modular constituents. J Muscle Res Cell Motil 26: 355–365 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Fucini P, Noegel AA, Stewart M (1999) Structural basis for dimerization of the Dictyostelium gelation factor (ABP120) rod. Nat Struct Biol 6: 836–841 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nakamura F, Pudas R, Heikkinen O, Permi P, Kilpelainen I, Munday AD, Hartwig JH, Stossel TP, Ylanne J (2006) The structure of the GPIb–filamin A complex. Blood 107: 1925–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Hartwig JH, Stossel TP (2006) FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat Cell Biol 8: 803–814 [DOI] [PubMed] [Google Scholar]
- Peterson FC, Gordon NC, Gettins PG (2001) High-level bacterial expression and 15N-alanine-labeling of bovine trypsin. Application to the study of trypsin-inhibitor complexes and trypsinogen activation by NMR spectroscopy. Biochemistry 40: 6275–6283 [DOI] [PubMed] [Google Scholar]
- Pfaff M, Liu S, Erle DJ, Ginsberg MH (1998) Integrin β cytoplasmic domains differentially bind to cytoskeletal proteins. J Biol Chem 273: 6104–6109 [DOI] [PubMed] [Google Scholar]
- Pickford AR, Smith SP, Staunton D, Boyd J, Campbell ID (2001) The hairpin structure of the (6)F1(1)F2(2)F2 fragment from human fibronectin enhances gelatin binding. EMBO J 20: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popowicz GM, Muller R, Noegel AA, Schleicher M, Huber R, Holak TA (2004) Molecular structure of the rod domain of Dictyostelium filamin. J Mol Biol 342: 1637–1646 [DOI] [PubMed] [Google Scholar]
- Popowicz GM, Schleicher M, Noegel AA, Holak TA (2006) Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci 31: 411–419 [DOI] [PubMed] [Google Scholar]
- Pudas R, Kiema TR, Butler PJ, Stewart M, Ylanne J (2005) Structural basis for vertebrate filamin dimerization. Structure 13: 111–119 [DOI] [PubMed] [Google Scholar]
- Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, Kim CA, Morava E, Newbury-Ecob R, Orstavik KH, Quarrell OW, Schwartz CE, Shears DJ, Suri M, Kendrick-Jones J, Wilkie AO (2003) Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet 33: 487–491 [DOI] [PubMed] [Google Scholar]
- Rousseau F, Schymkowitz JW, Itzhaki LS (2003) The unfolding story of three-dimensional domain swapping. Structure 11: 243–251 [DOI] [PubMed] [Google Scholar]
- Schleucher J, Schwendinger M, Sattler M, Schmidt P, Schedletzky O, Glaser SJ, Sorensen OW, Griesinger C (1994) A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J Biomol NMR 4: 301–306 [DOI] [PubMed] [Google Scholar]
- Schwaiger I, Kardinal A, Schleicher M, Noegel AA, Rief M (2004) A mechanical unfolding intermediate in an actin-crosslinking protein. Nat Struct Mol Biol 11: 81–85 [DOI] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr D 60: 432–438 [DOI] [PubMed] [Google Scholar]
- Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS (2001) Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol 2: 138–145 [DOI] [PubMed] [Google Scholar]
- Travis MA, van der FA, Kammerer RA, Mould AP, Sonnenberg A, Humphries MJ (2004) Interaction of filamin A with the integrin β7 cytoplasmic domain: role of alternative splicing and phosphorylation. FEBS Lett 569: 185–190 [DOI] [PubMed] [Google Scholar]
- Tyler JM, Anderson JM, Branton D (1980) Structural comparison of several actin-binding macromolecules. J Cell Biol 85: 489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi RK, Li F, Adam L, Nguyen D, Ohta Y, Stossel TP, Kumar R (2002) Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat Cell Biol 4: 681–690 [DOI] [PubMed] [Google Scholar]
- van der Flier A, Kuikman I, Kramer D, Geerts D, Kreft M, Takafuta T, Shapiro SS, Sonnenberg A (2002) Different splice variants of filamin-B affect myogenesis, subcellular distribution, and determine binding to integrin β subunits. J Cell Biol 156: 361–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier A, Sonnenberg A (2001) Structural and functional aspects of filamins. Biochim Biophys Acta Mol Cell Res 1538: 99–117 [DOI] [PubMed] [Google Scholar]
- Vorgerd M, van der Ven PF, Bruchertseifer V, Lowe T, Kley RA, Schroder R, Lochmuller H, Himmel M, Koehler K, Furst DO, Huebner A (2005) A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am J Hum Genet 77: 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Unruh JR, Johnson CK, Kuczera K, Schowen RL, Borchardt RT (2006) Effects of ligand binding and oxidation on hinge-bending motions in S-adenosyl-L-homocysteine hydrolase. Biochemistry 45: 7778–7786 [DOI] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD (1995) 1H, 13Cand 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6: 135–140 [DOI] [PubMed] [Google Scholar]
- Woo MS, Ohta Y, Rabinovitz I, Stossel TP, Blenis J (2004) Ribosomal S6 kinase (RSK) regulates phosphorylation of filamin A on an important regulatory site. Mol Cell Biol 24: 3025–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Xu W, Davie EW, Chung DW (1998) Molecular cloning of human ABPL, an actin-binding protein homologue. Biochem Biophys Res Commun 251: 914–919 [DOI] [PubMed] [Google Scholar]
- Xu W, Xie Z, Chung DW, Davie EW (1998) A novel human actin-binding protein homologue that binds to platelet glycoprotein Ibα. Blood 92: 1268–1276 [PubMed] [Google Scholar]
- Zhou X, Tian F, Sandzen J, Cao R, Flaberg E, Szekely L, Cao Y, Ohlsson C, Bergo MO, Boren J, Akyurek LM (2007) Filamin B deficiency in mice results in skeletal malformations and impaired microvascular development. Proc Natl Acad Sci USA 104: 3919–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods