Abstract
We have identified a cDNA from a human phytohemagglutinin-activated lymphoblast library encoding a protein that binds 125I-labeled human interleukin 12 (125I-huIL-12) with a Kd of about 5 nM when expressed in COS-7 cells. When coexpressed in COS-7 cells with the previously identified IL-12 β receptor (IL-12Rβ) protein, two classes of 125I-huIL-12 binding sites were measured with Kds of about 55 pM and 8 nM, corresponding to the high- and low-affinity binding sites seen on phytohemagglutinin-activated lymphoblasts. This newly identified huIL-12R subunit is a member of the cytokine receptor superfamily, with closest resemblance to the β-type cytokine receptor gp130 and the receptors for leukemia inhibitory factor and granulocyte colony-stimulating factor. Consequently, we have reclassified the previously identified IL-12Rβ subunit as huIL-12Rβ1 and designated the newly identified subunit as huIL-12Rβ2. huIL-12Rβ2 is an 862-amino acid type I transmembrane protein with a 595-amino-acid-long extracellular domain and a cytoplasmic tail of 216 amino acids that contains three tyrosine residues. A cDNA encoding the mouse homolog of the huIL12Rβ2 protein has also been isolated. Human and mouse IL-12Rβ2 proteins show a 68% amino acid sequence identity. When expressed in COS-7 cells, huIL-12Rβ2 exists as a disulfide-linked oligomer with an apparent monomeric molecular weight of 130 kDa. Coexpression of the two identified IL-12R subunits in Ba/F3 cells conferred IL-12 responsiveness, and clones of these cotransfected Ba/F3 cells that grew continuously in the presence of IL-12 were isolated and designated LJM-1 cells. LJM-1 cells exhibited dose-dependent proliferation in response to huIL-12, with an ED50 of about 1 pM huIL-12. Interestingly, Ba/F3 cells transfected with IL-12Rβ2 alone proliferated in response to huIL-12 with an ED50 of about 50 pM, although a role for endogenous mouse IL-12Rβ1 in IL-12 signal transduction in these transfectants cannot be ruled out. These results demonstrate that the functional high-affinity IL-12R is composed of at least two β-type cytokine receptor subunits, each independently exhibiting a low affinity for IL-12.
Interleukin 12 (IL-12) is a heterodimeric cytokine produced primarily by antigen-presenting cells that has important activities in the regulation of various aspects of the immune response. IL-12 stimulates proliferation of activated T and natural killer cells (1, 2, 3, 4), enhances lymphokine-activated killer/natural killer and cytotoxic T lymphocyte lytic activity (2, 5), and induces IFN-γ production by T and natural killer cells (6, 7, 8). IL-12 plays a central role in promoting Th1-type immune responses and thus cell-mediated immunity (9, 10, 11, 12, 13, 14). The biological activities of IL-12 are mediated via binding to specific cell surface receptors. Phytohemagglutinin (PHA)-activated lymphoblasts and IL-2-activated natural killer cells exhibit at least two classes of 125I-labeled human IL-12 (125I-huIL-12) binding sites with Kd’s of 5–20 pM and 2–6 nM (15, 16).
We have previously described the identification of a cDNA encoding an IL-12 receptor (R) protein, designated IL-12Rβ. When expressed in COS-7 cells, IL-12Rβ mediates binding of 125I-huIL-12 with a Kd of about 2–5 nM, corresponding to the low affinity binding sites seen on PHA-activated lymphoblasts (17). This IL-12Rβ chain is a member of the cytokine receptor superfamily, and is related most closely over its entire length to the “β type” (18) cytokine receptor glycoprotein (gp)130 and the receptors for leukemia inhibitory factor and granulocyte colony-stimulating factor. Although this IL-12Rβ protein has a short cytoplasmic tail, the box 1 and box 2 cytokine receptor motifs are present. In light of the important role of tyrosine phosphorylation in signal transduction pathways, it is interesting to note that there are no tyrosine residues present in the cytoplasmic tail of this IL-12Rβ protein. Nevertheless, antibody inhibition experiments demonstrated that anti-IL-12Rβ monoclonal antibodies were able to specifically inhibit IL-12-induced proliferation of PHA-activated lymphoblasts, production of interferon-γ from resting peripheral blood mononuclear cells, and development of lymphokine-activated killer activity (19), demonstrating that this IL-12Rβ protein is necessary for IL-12 signaling.
The inability to confer high affinity (Kd of 5–20 pM) 125I-huIL-12 binding to COS-7 cells transfected with the previously identified IL-12Rβ protein, as well as the lack of cytoplasmic tyrosine residues, suggested that at least one additional IL-12R subunit exists. We now report the identification, using expression cloning techniques, of an additional β type IL-12 receptor protein. This receptor is designated IL-12Rβ2, and the previously identified IL-12Rβ is therefore reclassified as IL-12Rβ1. IL-12Rβ2 by itself binds 125I-huIL-12 with low affinity. When coexpressed with IL-12Rβ1, it confers high-affinity 125I-huIL-12 binding and IL-12 responsiveness.
MATERIALS AND METHODS
Expression Cloning and Identification of IL-12R cDNA.
A plasmid expression library in the vector pEF-BOS (20) was established using mRNA isolated from lymphoblasts activated for 3 days with PHA as previously described (17). From this library, plasmid pools consisting of ≈100 clones were coexpressed with the IL-12Rβ1 cDNA (17) and screened for high-affinity IL-12 binding by a modification of the previously described method (15, 21) incorporating low concentrations of 125I-huIL-12 (about 30 pM) and a 3-hr wash at room temperature to allow for the selective measurement of high-affinity binding. By screening 440 pools and subsequent sib-selection, one positive cDNA was isolated and sequenced using an automated Applied Biosystems DNA sequencer. The corresponding mouse cDNA was isolated by crosshybridization-screening of a cDNA library generated from concanavalin A-stimulated splenocyte mRNA. Standard recombinant DNA methods were used for additional library screening, probe labeling, and RNA blotting (22).
Preparation of Cell Lines.
COS-7 cells were transfected by electroporation with expression constructs containing the indicated protein coding sequence in the pEF-BOS expression vector and harvested after 48–72 hr as described previously (17). Ba/F3-expressing huIL-12Rβ1 were produced by electroporation with linearized pRC-Rous sarcoma virus-IL-12R DNA as previously described for production of cytotoxic T lymphocyte lytic cells expressing huIL-12Rβ1 (17). G-418-resistant clones were subcloned by limiting dilution, screened for huIL-12Rβ1 expression by flow cytometry using the anti-huIL-12Rβ1 antibody 2–4E6 (19, 23), and the resultant cell line designated Ba/F3.huIL-12Rβ1 cells. Linearized expression constructs encoding huIL-12Rβ2 in the pEF-BOS expression vector and a puromycin resistance gene expression vector, pBSpacΔp (24), were introduced by electroporation into Ba/F3 or Ba/F3.huIL-12Rβ1 cells, and stable transfectants were selected by growth in medium containing 10 ng/ml IL-12 (a gift of Alvin Stern, Hoffmann–La Roche) or in IL-3-containing medium supplemented with 3 μg/ml puromycin (Sigma). The surviving cells were cloned by limiting dilution in IL-12- or puromycin-containing medium, as appropriate.
Characterization of huIL-12Rβ2 Protein.
Expression constructs encoding huIL-12Rβ2 with the addition of a C-terminal FLAG peptide tag (25) were prepared in the pEF-BOS expression vector. COS-7 cells were transfected by electroporation with huIL-12Rβ1 and/or huIL-12Rβ2-FLAG expression constructs as described above and metabolically labeled with [35S]cysteine for 6 hr at 37°C. The cells were then solubilized in 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfate lysis buffer (CHAPS; 10 mM CHAPS/50 mM Tris/0.3 M NaCl/500 mM iodoacetamide/100 mM phenylmethylsulfonyl fluoride) at a density of 1 × 106 cells per ml, and the cell lysates were immunoprecipitated by the addition of 5 μg of the anti-FLAG M2 antibody bound to 50 μl of GammaBind Plus Sepharose (Pharmacia). The immunoprecipitated proteins were analyzed by SDS/PAGE 4–12% gradient gels, and the labeled proteins visualized by autoradiography.
IL-12 Binding and Proliferation Assays.
125I-huIL-12 was prepared by the Iodogen method, and specific binding to cells was measured as described previously (15, 26). Specific binding was analyzed using the ligand nonlinear regression computer program (27) and plotted by the method of Scatchard (28). For measurement of IL-12-stimulated proliferation, Ba/F3 cells expressing the indicated proteins were washed and cultured in RPMI medium 1640 supplemented with 10% fetal bovine serum, 100 units/ml penicillin G, 100 μg/ml streptomycin, and 2 mM glutamine at 1 × 104 cells per well in flat-bottom 96-well microculture plates at 37°C for 24 hr. Various concentrations of IL-12 were added, the cells were cultured for an additional 42 hr at 37°C, and the wells were then pulsed with 0.5 μCi (1 Ci = 37 GBq) per well 3H-thymidine (New England Nuclear) for 6 hr at 37°C. Incorporation of 3H-thymidine into DNA was determined by harvesting the contents of each well onto glass fiber filters. All samples were assayed in triplicate.
RESULTS
Characterization of IL-12Rβ2 cDNA.
Using an expression cloning strategy that was specifically directed toward identifying high-affinity IL-12R, we have isolated a novel IL-12R cDNA. This cDNA, when coexpressed with the previously described IL-12R, reconstitutes a high-affinity IL-12R complex (see below). Due to its homology to other β-type cytokine receptor proteins (see below), we are proposing that this novel IL-12R be termed IL-12Rβ2, and that the previously isolated IL-12Rβ (17) be renamed IL-12Rβ1. The first IL-12Rβ2 cDNA that was isolated contained a very long (640 bp) and G+C-rich (73–78%) 5′ noncoding region (data not shown). Analysis of four additional independent clones confirmed the presence of this unusual 5′ end structure. Sequencing of the cDNAs showed the presence of one long open reading frame encoding an 862-amino acid (aa) protein (Fig. 1). This IL-12Rβ2 is a type I transmembrane protein, with a predicted molecular weight of 97,140 Da (including the signal peptide) and 94,059 Da (predicted mature protein). An extracellular domain of 595 aa is separated by a 24-aa transmembrane region from a 216-aa cytoplasmic tail that contains three tyrosine residues. Fig. 1 also shows that the corresponding mouse IL-12Rβ2 protein is very similar to the human receptor subunit, with an overall aa sequence identity of 68%.
Figure 1.

Comparison of aa sequences of human (Upper) and mouse (Lower) IL-12Rβ2 subunits. The gap program (Genetics Computer Group, Madison, WI) was used to construct the alignment. Signal peptides are underlined, potential N-linked glycosylation sites appear in bold italics and underlined, the hallmark sequence/structural motifs characteristic of the cytokine receptor superfamily are shaded, the putative transmembrane regions are doubly underlined, the cytoplasmic box I and II motifs are shaded, and the conserved cytoplasmic tyrosine residues are shown in bold and doubly underlined.
IL-12Rβ2 is a member of the cytokine receptor superfamily. Similar to IL-12Rβ1, IL-12Rβ2 is also most closely related to the members of the gp130-type subgroup (gp130, granulocyte colony-stimulating factor-R, leukemia inhibitory factor-R). Comparison of the huIL-12R extracellular domains using the align program (29) yields scores of 20 (IL-12Rβ2/gp130), 8 (IL-12Rβ1/gp130), and 3 (IL-12Rβ1/IL-12Rβ2). Scores greater than 3 are considered to demonstrate significant evolutionary relationships. A score of 3 indicates that the similarity would have occurred by chance with a probability of only 10−3, with increasing scores corresponding to decreasing probabilities. In contrast to IL-12Rβ1, IL-12Rβ2 contains an N-terminal Ig motif, and is thus more similar to gp130. The cytoplasmic region of IL-12Rβ2 also contains the characteristic box 1 motif and a possible box 2 motif: thus, both cloned IL-12 receptor chains have the general makeup of a β-type subunit (18).
RNA blot analysis of PHA-activated human lymphoblasts indicated the presence of two major transcripts of 4.5 and 6 kb (Fig. 2). Both mRNAs are up-regulated from undetectable or very low levels when peripheral blood mononuclear cells are treated with PHA. By analyzing uncloned cDNA from PHA-activated lymphoblasts using reverse transcription–PCR, we have not been able to detect any evidence for a soluble form of huIL-12Rβ2, since variant transcripts in which the transmembrane encoding region is absent have not been found (data not shown). However, production of soluble forms of huIL-12Rβ2 could result from proteolytic cleavage of the extracellular portion of this receptor subunit from the cell surface.
Figure 2.
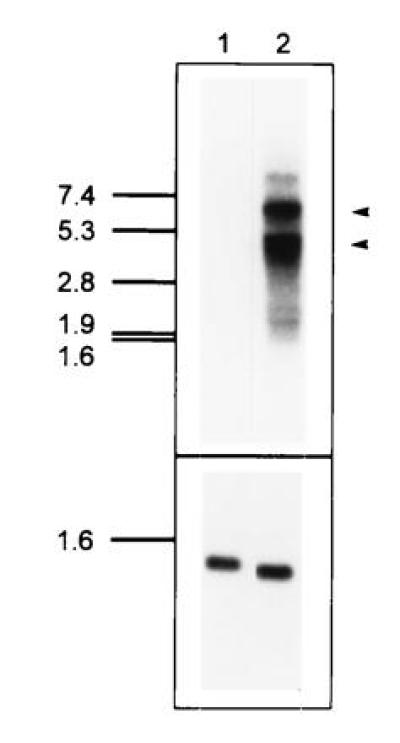
RNA blot analysis of huIL-12Rβ2 expression. (Upper) A blot containing 5 μg of poly(A)+ RNA from uninduced peripheral blood mononuclear cells (lane 1) or PHA-activated (3 days) peripheral blood mononuclear cells (lane 2) was probed with an IL-12Rβ2 cDNA insert. Two major RNAs at 4.5 kb and 6 kb are apparent in lane 2. Exposure time was 16 hr using an intensifying screen at −80°C. (Lower) As a loading control, the blot was rehybridized with a probe for a cell-cycle independent transcript, pHE7 (37). Exposure time was 20 min using an intensifying screen at −80°C.
Characterization of the IL-12R Subunits in COS-7 Cells.
COS-7 cells were transfected by electroporation with cDNA expression constructs for huIL-12Rβ1 and huIL-12Rβ2, alone and in combination, to investigate the functional properties of these proteins. As previously reported (17, 30), huIL-12Rβ1 mediates low affinity binding of 125I-huIL-12, with a Kd of about 10 nM and about 100,000 sites per cell (Fig. 3 Top). Expression of huIL-12Rβ2 in COS-7 cells yields similar results with a single class of 125I-huIL-12 binding sites with a Kd of about 5 nM (Fig. 3 Middle). When both proteins are coexpressed, two classes of binding sites can be detected (Fig. 3 Bottom). We have measured two classes of 125I-huIL-12 binding sites in cotransfected COS-7 cells with Kds of 55 ± 10 pM (1,860 ± 380 sites per cell) and 7.5 ± 0.6 nM (68,700 ± 16,500 sites per cell) (n = 8). The low number of high affinity sites detected on the cotransfected cells may be due to either competition for expression between huIL-12Rβ1 and huIL-12Rβ2 at the level of mRNA or protein, or may be due to competition between homo- and hetero-association of the β1 and β2 chains. Nevertheless, coexpression of the two IL-12R subunits creates 125I-huIL-12 binding sites that correspond to the high- and low-affinity binding sites measured on activated T cells (23).
Figure 3.
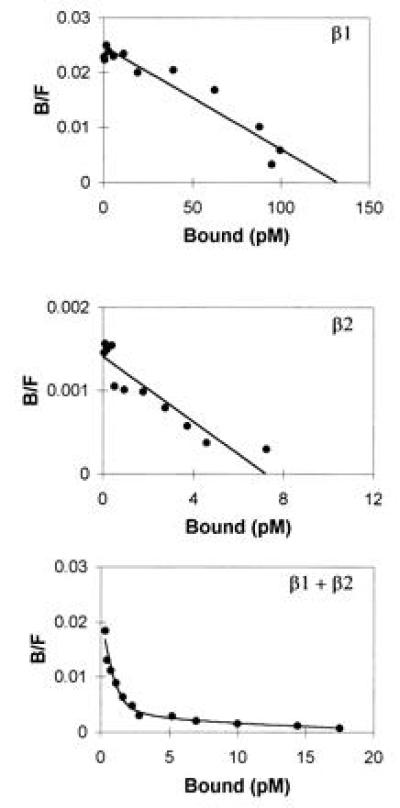
Coexpression of huIL-12Rβ1 and huIL-12Rβ2 in COS-7 cells yields high affinity binding of 125I-huIL-12. Specific binding of 125I-huIL-12 to COS-7 cells transfected with the indicated expression vectors was determined and analyzed by the method of Scatchard. (Top) huIL-12Rβ1, 6 nM Kd, 123,000 sites per cell; (Middle) huIL-12Rβ2, 5 nM Kd, 6500 sites per cell; (Bottom) both huIL-12Rβ1 and huIL-12Rβ2 expression vectors, 65 pM Kd, 1100 sites per cell and 10 nM Kd, 20,400 sites per cell.
Immunoprecipitation experiments were conducted to determine the biochemical properties of huIL-12Rβ2. C-terminally FLAG-tagged huIL-12Rβ2 protein was determined to have an apparent molecular weight of 130 kDa when expressed in COS-7 cells and analyzed by SDS/PAGE under reducing conditions (Fig. 4). Under nonreducing conditions, huIL-12Rβ2 protein migrates as a protein with an apparent molecular weight >220 kDa, suggesting the IL-12-independent formation of disulfide-bonded dimers of huIL-12Rβ2 in the transfected COS-7 cells. In addition, the existence of higher order multimers of huIL-12Rβ2 is suggested by the appearance of high molecular weight material at the top of the electrophoresis gels. However, this material may be an artifact of solubilization. It is interesting that this dimer/oligomer is still present when the samples are prepared in the presence of 700 mM 2-mercaptoethanol, demonstrating resistance of dimer/oligomer breakdown upon disulfide bond reduction. The protein migrating with an apparent molecular weight of 90 kDa is unrelated to the IL-12 receptor subunits as it appears in mock transfected COS-7 cells. Coimmunoprecipitation of the two IL-12 receptor subunits was not observed, as evidenced by the absence of the 100-kDa huIL-12Rβ1 protein from the pattern of proteins immunoprecipitated by anti-FLAG antibody from cell lysates of COS-7 cells cotransfected with huIL-12Rβ1 and huIL-12Rβ2-FLAG (Fig. 4). Additional experiments conducted in the presence of huIL-12 and chemical crosslinking agents also did not detect coimmunoprecipitation of heterodimeric β1/β2 protein complexes (data not shown).
Figure 4.
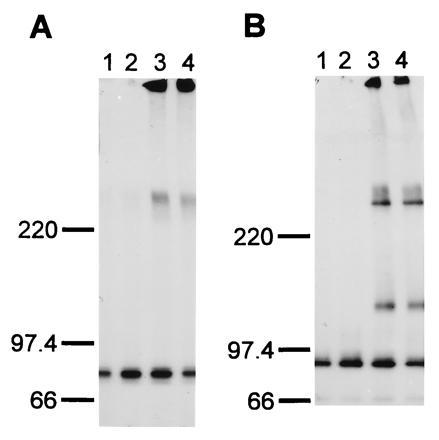
Analysis of huIL-12Rβ2 protein expressed on the surface of transfected COS-7 cells. COS-7 cells were mock-transfected (lane 1) or transfected with a huIL-12Rβ1 expression construct (lane 2), a huIL-12Rβ2-FLAG expression construct (lane 3), or cotransfected with huIL-12Rβ1 and huIL-12Rβ2-FLAG expression constructs (lane 4) as described in Materials and Methods. The cells were metabolically labeled with [35S]cysteine, cell lysates were prepared, and proteins were immunoprecipitated using the M2 anti-FLAG antibody. The precipitated proteins were analyzed on SDS/4-12% polyacrylamide gels under nonreducing (A) and reducing (B; 700 mM 2-mercaptoethanol) conditions. The migration of protein standards electrophoresed in parallel lanes is shown. Exposure time was 6 hr using an intensifying screen at −80°C.
Analysis of the Signaling Properties of the IL-12R Subunits.
To investigate the role the various IL-12R subunits play in mediating IL-12 signal transduction, the Ba/F3 IL-3-dependent mouse pro B cell line (31) was stably transfected with huIL-12Rβ1 and huIL-12Rβ2 alone and in combination. Stable Ba/F3 clones expressing huIL-12Rβ1, Ba/F3.huIL-12Rβ1 cells, were isolated on the basis of G-418 resistance. These cells express about 30,000 huIL-12Rβ1 molecules on the cell surface (data not shown), as determined using 125I-labeled Fab fragments of the anti-huIL-12Rβ1 monoclonal antibody 2–4E6 (23). As previously reported for the CTLL mouse IL-2-dependent T cell line (17), 125I-huIL-12 binds very inefficiently to the Ba/F3.huIL-12Rβ1 cells, with a Kd > 50 nM (data not shown). In contrast, Ba/F3 cells expressing huIL-12Rβ2 and selected in puromycin-containing medium exhibited 125I-huIL-12 binding with a Kd of about 5 nM (Fig. 5 Top). Similar results with Ba/F3.huIL-12Rβ2 cells selected for growth in IL-12-containing medium were seen. However, these IL-12-selected cells expressed about 5-fold fewer 125I-huIL-12 binding sites than Ba/F3.huIL-12Rβ2 cells selected for growth in puromycin-containing medium, and the combination of low affinity and small site number made Scatchard analysis of binding to the IL-12-selected cells difficult (data not shown). Cotransfected Ba/F3 cells, designated LJM-1 cells, exhibited two classes of 125I-huIL-12 binding sites with Kds of about 50 pM and 10 nM (Fig. 5 Middle), similar to the results seen with cotransfected COS-7 cells.
Figure 5.
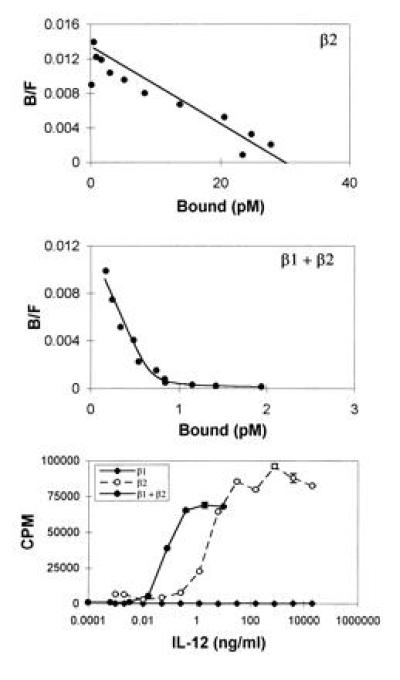
Coexpression of huIL-12Rβ1 and huIL-12Rβ2 in Ba/F3 cells yields high affinity 125I-huIL-12 binding and confers IL-12 responsiveness. Specific binding of 125I-huIL-12 to Ba/F3 cells expressing the indicated proteins was determined and analyzed by the method of Scatchard. (Top) huIL-12Rβ2, 3 nM Kd, 5600 sites per cell; (Middle) huIL-12Rβ1 and huIL-12Rβ2, 60 pM Kd, 120 sites per cell and 17 nM Kd, 500 sites per cell. (Bottom) IL-12-stimulated proliferation of Ba/F3 transfectants expressing huIL-12Rβ1 (♦), huIL-12Rβ2 (○), or both huIL-12Rβ1 and huIL-12Rβ2 (•) was determined as described.
We next examined the ability of huIL-12 to stimulate proliferation of the transfected Ba/F3 cells. In keeping with the ability to select these cells by long-term growth in huIL-12-containing medium, LJM-1 (cotransfected Ba/F3) cells proliferated in response to huIL-12 in a dose-dependent fashion (Fig. 5 Bottom). Ba/F3.huIL-12Rβ2 cells that grew slowly in huIL-12-containing medium were also identified, and subsequent cloning of these huIL-12Rβ2 transfectants in the presence of huIL-12 yielded clones that exhibited high levels of huIL-12-stimulated proliferation (Fig. 5 Bottom). In contrast, we were unable to isolate any Ba/F3.huIL-12Rβ1 cells by long-term growth in huIL-12-containing medium, and Ba/F3.huIL-12Rβ1 cells selected on the basis of G-418 resistance did not proliferate in response to huIL-12 (Fig. 5 Bottom). In parallel with the observed lack of high affinity 125I-huIL-12 binding to the Ba/F3.huIL-12Rβ2 cells, these cells exhibited an ED50 for IL-12-induced proliferation (50 pM) that was 50-fold greater than that observed with the LJM-1 cells (1 pM).
DISCUSSION
The identification of an IL-12 binding protein that is a β-type cytokine receptor subunit and that mediates low-affinity 125I-huIL-12 binding when expressed in COS-7 cells has been described (17). This IL-12 receptor protein was originally designated IL-12Rβ and has now been renamed IL-12Rβ1. By analogy to other cytokine receptor systems, the inability to confer high-affinity 125I-huIL-12 binding through expression of IL-12Rβ1 suggested that additional receptor protein(s) remained to be identified. An expression cloning strategy specifically directed toward the isolation of cDNAs that, upon coexpression with IL-12Rβ1 protein, would yield high-affinity IL-12 binding sites was therefore employed. The key features of this strategy were the use of picomolar concentrations of 125I-huIL-12 and a 3-hr wash period during which 125I-huIL-12 bound with low affinity was allowed to preferentially dissociate (21). A cDNA encoding a novel IL-12 receptor protein, designated IL-12Rβ2, was thus isolated. This IL-12Rβ2 protein, similar to the previously isolated IL-12Rβ1 subunit, is a member of the gp130-type subgroup of the cytokine receptor superfamily. Alignment scores for a sequence comparison between the β1 and β2 subunits of the IL-12R are significant. However, each of the two IL-12R subunits itself is more closely related to gp130 than to each other. In contrast to IL-12Rβ1, which does not contain any cytoplasmic tyrosine residues, the cytoplasmic region of IL-12Rβ2 contains three tyrosine residues, suggesting an important role for the β2 subunit in IL-12 signal transduction. Comparison of the human and mouse IL-12Rβ2 proteins reveals a high level of conservation, with a 68% aa sequence identity. This is in contrast to the situation for the IL-12Rβ1 subunit, which exhibits only a 54% sequence identity between the human and mouse proteins.
Expression of human IL-12Rβ2 in COS-7 cells yielded one class of 125I-huIL-12 binding sites with a Kd of about 5 nM. Although the binding affinity for 125I-huIL-12 of IL-12Rβ1 and IL-12Rβ2 expressed in COS-7 cells are similar, we have consistently observed the number of 125I-huIL-12 binding sites seen with IL-12Rβ2-transfected COS-7 cells to be about 10-fold lower than that seen with IL-12Rβ1-transfected cells. The reasons for this difference in expression efficiency are not known. In keeping with the fact that IL-12Rβ2 was identified using a screen that employed coexpression with IL-12Rβ1 and that was optimized for detecting high-affinity 125I-huIL-12 binding, coexpression of the β1 and β2 proteins in COS-7 cells yielded high-affinity 125I-huIL-12 binding. Two classes of binding sites for 125I-huIL-12, with Kds of about 50 pM and 10 nM, were measured in cotransfected COS-7 cells. These binding affinities are similar to the high- and low-affinity binding sites, with Kds of about 5–20 pM and 2–6 nM, detected on PHA-activated lymphoblasts and the IL-12 responsive Kit225/K6 T cell line (17, 23). The observed affinity difference between high-affinity binding in the cotransfected COS-7 cells vs. PHA-activated lymphoblasts is small but very reproducible. Besides this quantitative difference there are also qualitative differences. Certain classes of inhibitors, such as the anti-huIL-12Rβ1 mAb 2B10, are able to inhibit IL-12 bioactivity in huIL-12Rβ1/β2 cotransfected Ba/F3 cells but not in PHA-activated lymphoblasts (data not shown). However, it is clear that coexpression of both the β1 and β2 IL-12R chains in COS-7 cells is both necessary and sufficient for high-affinity 125I-huIL-12 binding. It is unclear whether the observed differences between cotransfected COS-7 or Ba/F3 cells and PHA-activated lymphoblasts reflect the involvement of additional IL-12R subunits in PHA-activated lymphoblasts or simply differences in posttranslational processing of the two known IL-12R subunits.
The low number of high-affinity binding sites measured on cotransfected cells may be due to the large excess of huIL-12Rβ1 expressed in transfected cells. Combined with the propensity for huIL-12Rβ2 to form homomultimers (as seen in Fig. 4), a small number of β1/β2-heterodimers may actually form in the cotransfected cells. Recent experiments have demonstrated that multiple interactions between huIL-12 and huIL-12Rβ1/β2 heterodimers are important for high-affinity 125I-huIL-12 binding. Experiments with homodimers of the mouse IL-12 p40 subunit, a known IL-12 antagonist, have shown that the p40 chain of IL-12 interacts primarily with the huIL-12Rβ1 protein (32), whereas other epitopes exist on the IL-12 heterodimer that primarily interact with huIL-12Rβ2 (D.H.P., L.J.M., S. Gillessen, U.G., R. Chizzonite, A. S. Stern, and M.K.G., unpublished work).
The apparent molecular weight of the IL-12Rβ2 protein monomer when expressed in COS-7 cells was 130 kDa, suggesting that about 36 kDa of carbohydrate are present on the predicted 94-kDa protein backbone. Similar to huIL-12Rβ1, huIL-12Rβ2 appears to exist at the cell surface as a disulfide-bonded dimer/oligomer. Formation of this huIL-12Rβ2 oligomer is not dependent on IL-12 binding, since it was observed in cells that were never exposed to IL-12. Coimmunoprecipitation experiments designed to examine the formation of β1/β2-containing heterodimers, conducted both in the absence or presence of huIL-12 and chemical crosslinking agents, have not yet been successful. This could be due to disruption of the β1/β2 heterodimers by the solubilization procedures, or simply due to insufficient sensitivity of the methods employed since only about 2000 high-affinity 125I-huIL-12 binding sites per cell are reconstituted on cotransfected COS-7 cells, in the presence of a large excess of β1 and β2 homomultimers that will interfere with the immunoprecipitation of the heterodimeric complexes.
To address the question of signaling through the IL-12R complex, we used the IL-3-dependent murine pro B cell line Ba/F3 as the host cell for expression of the various IL-12R proteins. The Ba/F3 cell line has previously been used to investigate signaling mediated by a number of different cytokine receptors, including erythropoietin (33), granulocyte/macrophage colony-stimulating factor (34), and IL-10 (35). Cotransfected Ba/F3 cells (designated LJM-1 cells) bound 125I-huIL-12 with two classes of binding sites with Kds of about 50 pM and 10 nM, similar to the binding sites observed in cotransfected COS-7 cells. However, the number of binding sites measured on LJM-1 cells (about 150 high-affinity sites per cell and 1,000 low-affinity sites per cell) was significantly lower than that detected in cotransfected COS-7 cells. It is possible that there may be negative selective effects against high level expression of high-affinity IL-12R complexes on the Ba/F3 cell surface, i.e., supranormal levels of IL-12R signaling may inhibit cell growth. Such selective pressure would influence the number of high-affinity sites on the stably transfected Ba/F3 cells but not the transiently transfected COS-7 cells. In support of this hypothesis, huIL-12Rβ2-transfected Ba/F3 cells isolated by growth in puromycin exhibited about 5-fold more 125I-huIL-12 binding sites per cell than similar cells isolated by growth in high concentrations of huIL-12.
LJM-1 cells exhibited dose-dependent proliferation in response to IL-12, with an ED50 of about 1 pM. This ED50 is ≈50-fold lower than the observed Kd of about 50 pM for high-affinity 125I-huIL-12 binding to these cells, and suggests that a very low level of receptor occupancy is sufficient for stimulation of proliferation by the IL-12R complex.
Ba/F3 cells transfected with huIL-12Rβ1 expressed about 30,000 huIL-12Rβ1 molecules per cell but bound 125I-huIL-12 very inefficiently (Kd > 50 nM) and did not respond to IL-12. In contrast, IL-12Rβ2-transfected Ba/F3 cells bound 125I-huIL-12 with a Kd of about 5 nM. Surprisingly, although the IL-12Rβ2-transfected cells did not exhibit high-affinity 125I-huIL-12 binding, transfectants exhibiting long-term growth in IL-12-containing medium were isolated. Therefore, expression of IL-12Rβ2 alone was sufficient to confer IL-12-responsiveness to Ba/F3 cells. It should be noted that reverse transcription–PCR analysis of Ba/F3 cells has demonstrated the presence of detectable levels of mouse IL-12Rβ1 mRNA (U.G., unpublished data), and this mouse homolog of IL-12Rβ1 may be necessary for signal transduction in the IL-12Rβ2-transfected Ba/F3 cells. Although the requirement for endogenous mouse IL-12Rβ1 in huIL-12Rβ2-mediated signaling in the transfected Ba/F3 cells has not been formally ruled out, additional evidence suggests that the homomultimers are mediating the IL-12 signaling. Ba/F3 transfectants expressing chimeric receptor proteins consisting of the epidermal growth factor-R extracellular domain fused to the huIL-12Rβ2 intracellular domain proliferate in response to epidermal growth factor treatment (J. Zou, D. H. Presky, C.-Y. Wu, and U. Gubler, unpublished work). In addition, Ba/F3 cells transfected with mouse IL-12Rβ2 alone did not exhibit long-term growth in medium containing mouse IL-12, whereas mouse IL-12Rβ1/mouse IL-12Rβ2 cotransfected Ba/F3 cells do (V. L. Wilkinson, D. Carvajal, L.J.M., A.O.C., U.G., M.K.G., and D.H.P., unpublished work). If mouse IL-12Rβ1 expression was significant in the host Ba/F3 cell line, the mouse IL-12Rβ2-transfected Ba/F3 cells should have been able to grow in mouse IL-12. These observations suggest that endogenous mouse IL-12Rβ1 is not playing a role in the observed IL-12 signaling in huIL-12Rβ2 Ba/F3 transfectants. It is intriguing that the IL-12Rβ2 protein, in contrast to the IL-12Rβ1 protein, contains cytoplasmic tyrosine residues. By analogy to many other cytokine receptors, these IL-12Rβ2 cytoplasmic tyrosine residues are likely to play an important role in IL-12 signal transduction. Within the heterodimeric IL-12R complex, IL-12Rβ2 could therefore be considered the signal specificity determining “driver” subunit based on the concept outlined by Lai et al. (36). By bringing Janus kin ase kinases to the receptor complex, the β1 subunit would then serve as the more generic “trigger” chain. Experiments are underway to determine if this is indeed the case, or if the β1 subunit primarily acts as an affinity converter.
In conclusion, we have identified IL-12Rβ2, an additional protein subunit of the functional, high-affinity IL-12R complex. IL-12Rβ2 appears to have an important function in the IL-12 signal transduction process, and in concert with the previously identified IL-12Rβ1 subunit, can confer high-affinity binding and IL-12 responsiveness to cells expressing these proteins. We have produced a cotransfected Ba/F3 cell line, LJM-1 cells, that binds 125I-huIL-12 with high affinity, and that proliferates in response to IL-12 with an ED50 of 1 pM. This IL-12-responsive cell line should be useful for further investigation into IL-12 signal transduction mechanisms.
Acknowledgments
We thank Lucy Foppiani, John Duker, and Doug Larigan (Hoffmann–La Roche, Bioinformatics Department) for oligonucleotide synthesis and DNA sequencing, and Frank Podlaski and Alvin Stern (Hoffmann–La Roche, Department of Inflammation/Autoimmune Diseases) for providing purified huIL-12.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
References
- 1.Kobayashi M, Fitz L, Ryan M, Hewick R M, Clark S C, Chan S, Loudon R, Sherman F, Perussia B, Trinchieri G. J Exp Med. 1989;170:827–845. doi: 10.1084/jem.170.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stern A S, Podlaski F J, Hulmes J D, Pan Y C, Quinn P M, Wolitzky A G, Familletti P C, Stremlo D L, Truitt T, Chizzonite R. Proc Natl Acad Sci USA. 1990;87:6808–6812. doi: 10.1073/pnas.87.17.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf S F, Temple P A, Kobayashi M, Young D, Dicig M, Lowe L, Dzialo R, Fitz L, Ferenz C, Hewick R M. J Immunol. 1991;146:3074–3081. [PubMed] [Google Scholar]
- 4.Gately M K, Desai B B, Wolitzky A G, Quinn P M, Dwyer C M, Podlaski F J, Familletti P C, Sinigaglia F, Chizzonite R, Gubler U. J Immunol. 1991;147:874–882. [PubMed] [Google Scholar]
- 5.Chouaib S, Chehimi J, Bani L, Genetet N, Tursz T, Gay F, Trinchieri G, Mami-Chouaib F. Proc Natl Acad Sci USA. 1994;91:12659–12663. doi: 10.1073/pnas.91.26.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan S H, Perussia B, Gupta J W, Kobayashi M, Pospisil M, Young H A, Wolf S F, Young D, Clark S C, Trinchieri G. J Exp Med. 1991;173:869–879. doi: 10.1084/jem.173.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy E E, Terres G, Macatonia S E, Hsieh C S, Mattson J, Lanier L, Wysocka M, Trinchieri G, Murphy K, O’Garra A. J Exp Med. 1994;180:223–231. doi: 10.1084/jem.180.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gately M K, Warrier R R, Honasoge S, Carvajal D M, Faherty D A, Connaughton S E, Anderson T D, Sarmiento U, Hubbard B R, Murphy M. Int Immunol. 1994;6:157–167. doi: 10.1093/intimm/6.1.157. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh C S, Macatonia S E, Tripp C S, Wolf S F, O’Garra A, Murphy K M. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 10.Manetti R, Parronchi P, Giudizi M G, Piccinni M P, Maggi E, Trinchieri G, Romagnani S. J Exp Med. 1993;177:1199–1204. doi: 10.1084/jem.177.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trinchieri G. Blood. 1994;84:4008–4027. [PubMed] [Google Scholar]
- 12.Gately M K, Brunda M J. In: Cytokines: Interleukins and Their Receptors. Kurzrock R, Talpaz M, editors. Norwell, MA: Kluwer; 1995. pp. 341–366. [Google Scholar]
- 13.Stern A S, Magram J, Presky D H. Life Sci. 1996;58:639–654. doi: 10.1016/s0024-3205(96)80003-8. [DOI] [PubMed] [Google Scholar]
- 14.Magram J, Connaughton S E, Warrier R R, Carvajal D M, Wu C-Y, Ferrante J, Stewart C, Sarmiento U, Faherty D A, Gately M K. Immunity. 1996;4:471–481. doi: 10.1016/s1074-7613(00)80413-6. [DOI] [PubMed] [Google Scholar]
- 15.Chizzonite R, Truitt T, Desai B B, Nunes P, Podlaski F J, Stern A S, Gately M K. J Immunol. 1992;148:3117–3124. [PubMed] [Google Scholar]
- 16.Desai B B, Quinn P M, Wolitzky A G, Mongini P K, Chizzonite R, Gately M K. J Immunol. 1992;148:3125–3132. [PubMed] [Google Scholar]
- 17.Chua A O, Chizzonite R, Desai B B, Truitt T P, Nunes P, Minetti L J, Warrier R R, Presky D H, Levine J F, Gately M K. J Immunol. 1994;153:128–136. [PubMed] [Google Scholar]
- 18.Stahl N, Yancopoulos G D. Cell. 1993;74:587–590. doi: 10.1016/0092-8674(93)90506-l. [DOI] [PubMed] [Google Scholar]
- 19.Wu C Y, Warrier R R, Carvajal D M, Chua A O, Minetti L J, Chizzonite R, Mongini P K A, Stern A S, Gubler U, Presky D H, Gately M K. Eur J Immunol. 1996;26:345–350. doi: 10.1002/eji.1830260212. [DOI] [PubMed] [Google Scholar]
- 20.Mizushima S, Nagata S. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara T, Miyajima A. EMBO J. 1992;11:1875–1884. doi: 10.1002/j.1460-2075.1992.tb05239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Chizzonite R, Truitt T, Nunes P, Desai B B, Chua A O, Gately M K, Gubler U. Cytokine. 1994;6:82. (abstr.). [Google Scholar]
- 24.de la Luna S, Soria I, Pulido D, Ortin J, Jimenez A. Gene. 1988;62:121–126. doi: 10.1016/0378-1119(88)90585-9. [DOI] [PubMed] [Google Scholar]
- 25.Hopp T P, Prickett K S, Price V, Libby R T, March C J, Cerretti P, Urdal D L, Conlon P J. Biotechnology. 1988;6:1205–1210. [Google Scholar]
- 26.Chizzonite R, Truitt T, Podlaski F J, Wolitzky A G, Quinn P M, Nunes P, Stern A S, Gately M K. J Immunol. 1991;147:1548–1556. [PubMed] [Google Scholar]
- 27.McPherson G A. J Pharmacol Methods. 1985;14:213–228. doi: 10.1016/0160-5402(85)90034-8. [DOI] [PubMed] [Google Scholar]
- 28.Scatchard G. Ann NY Acad Sci. 1949;51:660–672. [Google Scholar]
- 29.Dayhoff M O, Barker W C, Hunt L T. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- 30.Presky D H, Gubler U, Chizzonite R A, Gately M K. Res Immunol. 1995;146:439–445. doi: 10.1016/0923-2494(96)83013-6. [DOI] [PubMed] [Google Scholar]
- 31.Palacios R, Steinmetz M. Cell. 1985;41:727–734. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 32.Gillessen S, Carvajal D, Ling P, Podlaski F J, Stremlo D L, Familletti P C, Gubler U, Presky D H, Stern A S, Gately M K. Eur J Immunol. 1995;25:200–206. doi: 10.1002/eji.1830250133. [DOI] [PubMed] [Google Scholar]
- 33.Jones S S, D’Andrea A D, Haines L L, Wong G G. Blood. 1990;76:31–35. [PubMed] [Google Scholar]
- 34.Kitamura T, Hayashida K, Sakamaki K, Yokota T, Arai K I, Miyajima A. Proc Natl Acad Sci USA. 1991;88:5082–5086. doi: 10.1073/pnas.88.12.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho A S Y, Liu Y, Khan T A, Hsu D H, Bazan J F, Moore K W. Proc Natl Acad Sci USA. 1993;90:11267–11271. doi: 10.1073/pnas.90.23.11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai S Y, Xu W, Gaffen S L, Liu K D, Longmore G D, Greene W C, Goldsmith M A. Proc Natl Acad Sci USA. 1996;93:231–235. doi: 10.1073/pnas.93.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao X, Kozak C A, Liu Y J, Noguchi M, O’Connell E, Leonard W J. Proc Natl Acad Sci USA. 1993;90:8464–8468. doi: 10.1073/pnas.90.18.8464. [DOI] [PMC free article] [PubMed] [Google Scholar]


