Natural selection will tend in the long run to reduce any part of the organization, as soon as it becomes, through changed habits, superfluous, without by any means causing some other part to be largely developed in a corresponding degree.
—Charles Darwin, The Origin of Species
INTRODUCTION
Cancer cells are typically held to be rugged and hardy. They take over the body, resisting medical efforts to contain them, and ultimately kill the patient. A great deal of attention has been focused on the special abilities that allow malignant cells to grow, commandeer the body's resources, and acquire drug resistance. One of the exciting developments of the past two decades is the discovery that tumor cells profit from mutation and other types of genomic instability, enabling them to evolve readily.
Malignant cells are adapted to their pathological condition, and their genomes bear hallmarks of response to selective pressures of their environment. The most common genetic changes in tumors are activating point mutations in oncogenes like K-ras, inactivating lesions in tumor suppressor genes such as P53, and various forms of aneuploidy, including loss of heterozygosity (LOH) and gene amplification. All tumors display at least a subset of these features.
However, evolvability bears a cost. Unstable genomes may have a short-term evolutionary advantage. But nearly all mutagenic processes are random. Therefore, adaptive changes come from a large pool of stochastic alterations, most of which are neutral or deleterious to gene function. Gene mutations that individually produce negligible effects on tumor growth are degenerative, because they erode the information encoded in the cancer cell's genome. Relatively little effort has been expended to investigate, and possibly exploit, this flip side of evolutionary change: nonadaptive alterations.
Here I explore the nonadaptive consequences of genome instability for cancer. I present a perspective that tumor cells, though they possess distinct strengths and exceptional abilities, are in some respects weak compared with normal cells. This frailty is rooted in fundamental principles of genetics and evolution and may lead to some new strategies for cancer therapy.
Overlapping Gene Function and Genetic Streamlining
Early evolutionists, including Darwin and Lamarck, appreciated that useless organs and structures disappear over-time. Sightless crustaceans and flightless birds were of great interest to Darwin in particular. Modern biologists have extended these observations to biochemical pathways and genes. In the absence of selective pressures that maintain gene function, coding sequences degenerate and are ultimately lost entirely.
A good example are genomes of obligate parasites such as the intracellular bacterium Rickettsia. On average such parasites possess less than half the number of genes of free-living bacteria (see www.ncbi.nlm.nih.gov/COG). Parasites inhabit a relatively constant, nourishing environment compared with free-living species. Presumably, random mutation and deletion gradually obliterate the unnecessary genes that do not contribute to fitness. This evolutionary process has been called genetic streamlining. Perhaps the ultimate manifestation of streamlining is the mitochondrian, believed to be a distant relative of Rickettsia. Over the countless generations since its establishment as an endosymbiont, the mitochondrial genome has lost nearly all of its original coding capacity. Mitochondrial DNA encodes a handful of electron transport system components and little else. In this respect, it is a highly degenerate genome.
At first glance, the observation that a large subset of Escherichia coli and Saccharomyces cerevisiae genes are nonessential appears to fly in the face of genetic streamlining (Ross-Macdonald et al., 1999; www.shigen.nig.ac.jp/ecoli/PEC). Exhaustive analysis of loss-of-function mutations reveals that individual disruptions in ∼80% of genes produce viable organisms. Experiments in worms, fruit flies, zebra fish, and mice suggest about the same level of functional overlap (Driever et al., 1996; Adams et al., 2000), and humans are probably similar.
But most nonessential genes may prove helpful or vital under certain conditions that are atypical in the laboratory. For instance, one yeast gene, SAC1, is required for growth only below ∼17°C (Novick et al., 1989). It appears that SAC1 evolved to increase the temperature range over which yeast cells could grow, allowing the yeast to occupy more diverse ecological niches. In the wild, low temperatures might kill off yeast that lack SAC1 function, whereas SAC1 makes no difference to cells that grow in a normal laboratory setting.
Therefore, nonessential genes may, in general, provide a buffer to various types of stress, including temperature, malnourishment, poisons, pathogens, and lack of water. Under stressful circumstances, these genes may enhance survival. Thus, they are maintained through intermittent selective pressures. An intracellular parasite such as Rickettsia does not need the same level of redundancy as a free-living organism. But, having dispensed with the extra apparatus, it is confined to its habitat and cannot survive without the host. It is hemmed in.
Cancer cells are akin to parasites. They are linked less closely to human evolutionary history than the normal cells from which they originate. Normal cells, like free-living bacteria, must be prepared for the unpredictable assaults of the world. Malignant cells have a shorter-term evolutionary memory. Thus, we may expect them to accrue mutations in nonessential genes. What are the specific genetic origins of such degenerative changes and the possible consequences?
Genetic Load
The idea of natural mutation loads in populations was pointed out and treated quantitatively first by Haldane (1937) and in more depth (and with genetic interactions) by others (Kimura and Maruyama, 1966; Maynard Smith, 1978; Kimura and Crow, 1979; Kondrashov, 1982). In the absence of selection, genotypic variation increases. In support of this view, experiments that compare fitness levels of two yeast strains, one wild-type and one deficient in DNA repair, show that mutations accumulate under mild selection conditions that compromise growth and viability only under stress (Szafraniec et al., 2001). Under normal growth conditions, the wild-type and mutant strains display similar properties. Interestingly, the yeast strains used were diploid. Thus, heterozygous mutations may be sufficient to generate declines in fitness, manifested only when cells must endure difficult circumstances such as growth at high temperature. Presumably, hemi- and homozygous mutations would produce more severe effects on fitness. Thus, stress actualizes the cryptic mutation load in cells grown under mild conditions.
Most mutations are deleterious or neutral to gene function. Thus, we expect tumors to accumulate a genetic load in nonessential genes, commensurate with their mutation rates and cell division numbers. The load of mutations should increase with time until it impacts cell viability. Indeed, a large genetic load may partly explain the high apoptotic rates of tumor cells.
Estimates for mutation rates in tumor cells range widely. Some have suggested that mutation frequencies in cancers could increase as much as 10,000-fold, at least transiently (Loeb, 1991). Such high rates may arise from a combination of factors, including rapid cell division, mutations in genome stability functions such as mismatch repair genes, and high-stress conditions similar to those that induce the error-prone replication (SOS) system in bacteria. Others, however, argue that mutation rates in malignant cells need not be higher than normal somatic replication error frequencies (Tomlinson et al., 1996; Tomlinson and Bodmer, 1999; Wang et al., 2002). Regardless, tumors demonstrably accumulate alterations, genetic and epigenetic in nature.
As mentioned above, the total mutation load depends on mutation rate and cell division number. Thirty cell divisions can generate a 10 g tumor from a single cell, assuming no attrition. In reality, however, tumors typically display significant apoptosis and necrosis, and it is likely that far more cell divisions are required to form a macroscopic growth (Wang et al., 2002).
In general, one suspects that the mutation load engenders some potential cost to the malignant cell, though possibly only under specific types of stressful conditions to which the tumor is seldom or never exposed. Based on such reasoning, tumors may have reduced thresholds for resistance to specific stresses and, overall, a compromised ability to buffer certain environmental changes and affronts.
LOH and Heterosis
Genomic instability, involving wholesale chromosome losses and large deletions, may exacerbate the problem. Tumors are riddled with hemizygosity, a feature thought to be driven in part by selection for inactivation of tumor suppressor genes. Because these mutations are mainly recessive, loss of function requires two hits. Often, one event involves loss of an entire homologous chromosome or a portion thereof (Knudson, 1971). This increases the chance to uncover recessive mutations in tumor suppressor genes on the remaining chromosome, an obvious selective advantage. Some LOH may also exist in tumors due to random occurrence and the lack of strong negative effects on fitness. Such lesions become fixed in tumor cell populations because they arise early in the tumor lineage and/or through genetic drift.
Whether selected or unselected, a variety of studies estimate that 10–30% of all tumor loci fall in regions of LOH (Gupta et al., 1997). LOH not only uncovers tumor suppressor mutations, but also extant germline mutations. Based on sequence analysis of 331 human genes in 82 normal individuals, every person is expected to carry 50 radical changes (including 10 nonsense mutations) in his/her genome, excluding mutations that affect splicing and transcription (Stephens et al., 2001). If a tumor displays LOH at 20% of its loci, 10 of these would be exposed by allelic loss, assuming allele losses are random. Somatic mutations that arise during tumorigenesis are an added, and probably substantial, burden. The protection from mutations afforded by diploidy is seriously compromised in most tumors.
Reduction to monoallelism is the opposite of heterosis, a well-established genetic phenomenon where outbred heterozygotes are fitter than their inbred parents. Darwin himself puzzled over hybrid vigor, and today plant breeders often exploit heterotic crosses to generate more robust progeny (e.g., for commercial corn varieties). Heterosis also occurs at the single-cell level in budding yeast (Steinmetz et al., 2002). Thus, monoallelism on a genome-wide scale, a seminal feature of cancers, is a general fitness liability.
Gene Dosage Imbalance
Another consequence of LOH is gene dosage imbalance and the presumptive abnormal gene expression ratios that accompany it. Relative expression of genes can be critical, perhaps more so when large numbers of genes are involved (Baker et al., 1994). Consider Drosophila males, which, like humans, have one X chromosome, whereas females have two. Drosophila males that fail to upregulate, or dosage compensate, genes on the X-chromosome die. They produce insufficient levels of X-chromosome gene products. Females can carry large deletions of X-chromosome material, but with deletions beyond a certain size, the animals cannot cope. This deletion-size threshold may arise from a small number of haploinsufficient loci or from the cumulative effects of many genes.
Gene dosage imbalance also contributes to several human diseases. Down (trisomy 21), Klinefelter (XXY), and Turner (XO) syndromes are the most familiar examples. Several haploinsufficient regions have also been delineated (Fisher and Scambler, 1994). In addition, one may conclude from their absence among viable offspring that most chromosome imbalances in humans are lethal.
Cancer cells are marked by extensive aneuploidy, with monosomy, extra chromosomes, deletions, and amplified genetic material. High frequencies of LOH translate into half the amounts of many proteins compared with normal cells. Epigenetic changes (e.g., methylation) and mutation further perturb the normal gene expression pattern. In an experiment that compared RNA expression of 6831 genes among a sample of 60 tumor cell lines, no gene varied <2-fold in at least one cell line compared with others in the set (Ross et al., 2000). Thus, gene expression differences of this order are the rule in cancer cells, not the exception. As shown by elegant yeast experiments, 2-fold differences in the expression of specific genes can affect fitness levels in stressed cells (Giaever et al., 1999). Gene dosage imbalance due to LOH and other factors likely creates further hazards for cancer cells (see Table 1).
Table 1.
Sources of weakness in cancer cells
| Weakness | Mechanism |
|---|---|
| High genetic load | High mutation rate |
| Loss of redundant functions | Genetic streamlining |
| Loss of heterosis (monoallelism) | LOH |
| Gene expression imbalance | Monosomy, polysomy, LOH, methylation |
| Reduced thresholds | Genetic streamlining, LOH |
LOH, loss of heterozygosity
Evidence for Genetic Degeneration in Tumors
The extent of degeneration depends on the number of genetic lesions that arise in cancer cells. LOH and other types of aneuploidy are clearly high, as are certain neutral alterations, including microsatellite repeat variants (Perucho et al., 1994). Epigenetic differences between tumor and normal cells are also well documented (Jones and Laird, 1999).
For point mutation, it is difficult to know the somatic mutation burden without systematic examination of tumor cell genomes. Such analysis may prove problematic due to “contamination” from normal tissues infiltrated throughout the neoplasm and tumor heterogeneity. Cancer molecular geneticists have amassed gigabytes of DNA sequence data for primary tumors and cancer cell lines. The vast majority of sequence information applies to a small group of genes implicated in tumorigenesis. P53 may be the most-studied cancer gene of all at the DNA sequence level.
The cancer research community has focused little on changes that may have no direct relation to the progression of the disease. But such changes have been documented and may be rather frequent among DNA sequences from tumor samples. In one study of colorectal cancers, nonsynonymous somatic coding sequence mutations were detected at a frequency of ∼200 per cancer cell genome (Wang et al., 2002). But do these mutations increase vulnerability?
The inability to withstand generalized stresses from chemotherapy may be one manifestation of the expected degeneration process that accompanies build-up of mutations. The literature contains some support for the view that tumor cells may be less able to cope with chemotherapeutics than normal cells (Harrison and Lerner, 1991). Despite the spotty track record of chemotherapy, there is no doubt that most tumors are at least initially vulnerable to such agents.
There is also at least one specific instance of possible relevance to this discussion. The response to the specific chemotherapeutic asparaginase, a bacterial enzyme that catabolizes asparagine to aspartate and ammonia, may be a manifestation of genetic (or epigenetic) streamlining in tumors. Intravenous administration of this enzyme depletes asparagine from the circulation, forcing cells to upregulate asparagine synthase to compensate for the shortfall. Sensitive lymphoid tumors have low levels of asparagine synthase and are more likely to starve from asparagine deprivation than normal cells (Capizzi, 1993). The genetic or epigenetic basis for this difference is not known so far as I am aware.
Overlapping Function, Paralogy, and Cancer Therapy
The chance observation involving tumor-specific changes in asparagine metabolism could be the tip of the iceberg. We may be able to define other instances of functional loss in tumors that can be exploited by appropriate therapies. Techniques such as gene expression profiling are capable of supplying information to identify these weak points. Indeed, a study of drug sensitivity in 60 tumor cell lines, coupled with gene expression data from the same cells, reveals a correlation between asparaginase response and asparagine synthase levels (Scherf et al. 2000). This retrospective analysis supports the view that gene expression studies can delineate novel therapeutic targets, if they exist. We may find other biochemical pathways in which tumors have lost functions, creating therapeutic vulnerabilities.
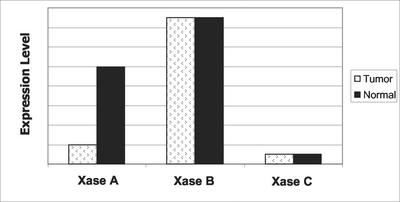
A related approach is to examine paralogs (Figure 1). It is well known that closely related genes often form synthetic lethal partners. For example, the yeast genetic interaction database contains 1423 synthetic lethal gene pairs. Of these, 186 (13%) involve paralogs (http://mips.gsf.de/proj/yeast/tables/interaction/genetic_interact.html). Individually, paralogs may be nonessential because one covers for the other's absence. However, if both genes are removed, the organism dies.
Figure 1.
Example of a relationship among expression patterns of a hypothetical paralog family, the Xases, in tumor and normal cells.
Using the public SAGE and UniGene databases (www.ncbi.nih.gov/SAGE or/UniGene) and standard BLAST sequence alignments, I searched for paralog pairs with the following pattern of expression in colon tumor/normal datasets: expression of both paralogs in normal tissue, and consistent expression of only one paralog in tumor tissue and cell lines. The paralog expressed in both tissue types (tumor and normal) is a candidate for an anticancer drug target. Specific inhibitors may lead to total loss of activity in tumor cells, perhaps resulting in cell death, but only partial diminution of function in normal cells. I recovered several paralog pairs, representing several protein classes, with the desired expression properties (see examples in Table 2). Some of these, including the 14-3-3 pair (σ and θ), are especially interesting. 14-3-3σ is known to be downregulated in several cancer types (Hermeking et al., 1997). Moreover, 14-3-3 proteins have been implicated as molecules whose inhibition leads to apoptosis (Masters and Fu, 2001). Finally, mutations in the only two yeast 14-3-3 orthologues (BMH1 and BMH2) are synthetic lethal.
Table 2.
Paralog pairs in colon tissue and cell lines
| HS No. | Description | Normal | Tumor | HCT116 | CaCo2 | SW837 | RKO |
|---|---|---|---|---|---|---|---|
| 79172 | Adenine translocator, 5 | 6 | 5 | 2 | 3 | 2 | 2 |
| 164280a | Adenine translocator, 6 | 13 | 36 | 17 | 21 | 37 | 27 |
| 321677 | STAT3 | 2 | 1 | 0 | 0 | 0 | 0 |
| 21486a | STAT1 | 2 | 2 | 3 | 1 | 4 | 2 |
| 858 | V-relB | 3 | 1 | 1 | 0 | 1 | 1 |
| 75569a | V-relA | 2 | 2 | 4 | 5 | 12 | 6 |
| 184510 | 14-3-3σ | 15 | 5 | 2 | 3 | 12 | 1 |
| 74405a | 14-3-3τ | 7 | 11 | 15 | 16 | 10 | 4 |
| 100090 | Tetraspan 3 | 30 | 15 | 3 | 5 | 2 | 4 |
| 121068a | Tetraspan 4 | 2 | 3 | 2 | 4 | 1 | 0 |
| 282975 | Carboxylesterase 2 | 38 | 8 | 2 | 4 | 5 | 2 |
| 76688a | Carboxylesterase 1 | 1 | 3 | 1 | 4 | 1 | 0 |
| 1244 | CD9 | 29 | 14 | 7 | 8 | 16 | 4 |
| 54457a | CD81 | 3 | 7 | 10 | 36 | 25 | 13 |
| 861 | MAPK3 | 29 | 8 | 3 | 4 | 7 | 4 |
| 79107a | MAPK14 | 2 | 2 | 0 | 2 | 1 | 4 |
| 80350 | Phosphatase 2 | 11 | 1 | 0 | 2 | 5 | 1 |
| 80324a | Phosphatase 6 | 1 | 1 | 0 | 4 | 2 | 1 |
| 167013 | Dynamin2 | 14 | 5 | 4 | 2 | 11 | 1 |
| 180628a | Dynamin1-like | 0 | 0 | 2 | 1 | 2 | 2 |
Normal, average of NC1+NC2 (primary colonic epithelium samples) SAGE counts; Tumor, average of Tu98+Tu102 (primary colon tumor samples) counts. Other categories are colon tumor cell lines. Numbers are SAGE counts (observations/∼50,000 tags). Paralog pairs are among the top 5 matches
The target candidate for each paralog pair
There are, of course, limitations to this strategy. They include 1) the observation that not all close paralog combinations display synthetic lethality in cells (e.g., P15 and P16); 2) heterogeneity among tumors and within a given tumor; 3) reliance on expression level as a surrogate for magnitude of protein function; 4) statistical fluctuations in the gene expression data; 5) possible adaptive responses of tumor cells to inhibition; 6) incompleteness (and errors) in genomic sequence and annotation; and 7) the necessity of devising drugs that discriminate among closely related molecules (paralogs). However, the pharmaceutical industry has confronted molecular selectivity issues vis-à-vis paralogs for years. And, as genome annotation improves, it may be simpler to predict the behavior of specific paralog double-mutant combinations. For example, further sequence analysis, extrapolation from genetic studies in model organisms such as yeast, protein interaction data and other types of genomic information may significantly improve the odds of forecasting synthetic-lethal paralog partners in human cells.
CONCLUSIONS
Tumors possess marvelous adaptations that facilitate their pathological activities. They forestall apoptosis, they invade tissues that exclude other nonnative cells, and they pump out drugs. But, given the extensive genome instability that they exhibit, it seems plausible that they also accumulate mutations in nonessential genes. In aggregate, these defects may increase susceptibility to therapies of nonspecific (i.e., traditional) types that induce stress, as well as to more targeted (i.e., synthetic lethal) forms (Hartwell et al., 1997).
Paralog pairs, potential synthetic-lethal partners with the desired expression properties, can be found in gene expression databases. Because of the unpredictable nature of biological systems, these genes encode only candidate targets that must be validated in cancer models. Practitioners of comparative gene expression technology in the cancer target discovery area mainly seek tumor suppressors, oncogenes, and tumor-associated antigens (Clark et al., 2000; Saha et al., 2001). But the target candidates derived from the paralog strategy outlined here may be expressed at equivalent levels in both the tumor and normal tissues. Approaches that search for targets with selective expression in tumors will miss such candidates.
References
- Adams, M.D. et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185-2195. [DOI] [PubMed] [Google Scholar]
- Baker, B.S., Gorman, M., and Marin, I. (1994). Dosage compensation in Drosophila. Annu. Rev. Genet. 28, 491-521. [DOI] [PubMed] [Google Scholar]
- Capizzi, R.L. (1993). Asparaginase revisited. Leuk. Lymphoma 10, 147-150. [DOI] [PubMed] [Google Scholar]
- Clark, E.A., Golub, T.R., Lander, E.S., and Hynes, R.O. (2000). Genomic analysis of metastasis reveals an essential role for RhoC. Nature 406, 532-535. [DOI] [PubMed] [Google Scholar]
- Driever, W. et al. (1996). A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123, 37-46. [DOI] [PubMed] [Google Scholar]
- Fisher, E., and Scambler, P. (1994). Human haploinsufficiency— one for sorrow, two for joy. Nat. Genet. 7, 5-7. [DOI] [PubMed] [Google Scholar]
- Giaever, G. et al. (1999). Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat. Genet. 21, 278-283. [DOI] [PubMed] [Google Scholar]
- Gupta, P.K. et al. (1997). High frequency in vivo loss of heterozygosity is primarily a consequence of mitotic recombination. Cancer Res 57, 1188-1193. [PubMed] [Google Scholar]
- Haldane, J.B.S. (1937). The effect of variation on fitness. Am. Nat. 71, 337-349. [Google Scholar]
- Harrison, D.E., and Lerner, C.P. (1991). Most primitive hematopoietic stem cells are stimulated to cycle rapidly after treatment with 5-fluorouracil. Blood 78, 1237-1240. [PubMed] [Google Scholar]
- Hartwell, L.H., Szankasi, P., Roberts, C.J., Murray, A.W., and Friend, S.H. (1997). Integrating genetic approaches into the discovery of anticancer drugs. Science 278, 1064-1068. [DOI] [PubMed] [Google Scholar]
- Hermeking, H. et al. (1997). 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol. Cell 1, 3-11. [DOI] [PubMed] [Google Scholar]
- Jones, P.A., and Laird, P.W. (1999). Cancer epigenetics comes of age. Nat. Genet. 21, 163-7. [DOI] [PubMed] [Google Scholar]
- Kimura, M., and Crow, J.F. (1979). Efficiency of truncation selection. Proc. Natl. Acad. Sci. USA 76, 396-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., and Maruyama, T. (1966). The mutational load with epistatic gene interactions in fitness. Genetics 54, 1337-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson, A.G., Jr. (1971). Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 68, 820-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A.S. (1982). Selection against harmful mutations in large sexual and asexual populations. Genet Res 40, 325-332. [DOI] [PubMed] [Google Scholar]
- Loeb, L.A. (1991). Mutator phenotype may be required for multi-stage carcinogenesis. Cancer Res 51, 3075-3079. [PubMed] [Google Scholar]
- Masters, S.C., and Fu, H. (2001). 14-3-3 proteins mediate an essential anti-apoptotic signal. J. Biol. Chem. 276, 45193-45200. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J. (1978). The Evolution of Sex. Cambridge: Cambridge University Press.
- Novick, P., Osmond, B.C., and Botstein, D. (1989). Suppressors of yeast actin mutations. Genetics 121, 659-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perucho, M., Peinado, M.A., Ionov, Y., Casares, S., Malkhosyan, S., and Stanbridge, E. (1994). Defects in replication fidelity of simple repeated sequences reveal a new mutator mechanism for oncogenesis. Cold Spring Harb. Symp. Quant. Biol. 59, 339-348. [DOI] [PubMed] [Google Scholar]
- Ross, D.T., Scherf, U., Eisen, M.B., Perou, C.M., Rees, C., et al. (2000). Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 24, 227-35.t. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald, P. et al. (1999). Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402, 413-418. [DOI] [PubMed] [Google Scholar]
- Saha, S. et al. (2001). A phosphatase associated with metastasis of colorectal cancer. Science 294, 1343-1346. [DOI] [PubMed] [Google Scholar]
- Scherf, U. et al. (2000). A gene expression database for the molecular pharmacology of cancer. Nat. Genet. 24, 236-244. [DOI] [PubMed] [Google Scholar]
- Steinmetz, L.M. et al. (2002). Dissecting the architecture of a quantitative trait locus in yeast. Nature 416, 326-330. [DOI] [PubMed] [Google Scholar]
- Stephens, J.C. et al. (2001). Haplotype variation and linkage disequilibrium in 313 human genes. Science 293, 489-493. [DOI] [PubMed] [Google Scholar]
- Szafraniec, K., Borts, R.H., and Korona, R. (2001). Environmental stress and mutational load in diploid strains of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98, 1107-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson, I., and Bodmer, W. (1999). Selection, the mutation rate and cancer: ensuring that the tail does not wag the dog. Nat. Med. 5, 11-12. [DOI] [PubMed] [Google Scholar]
- Tomlinson, I.P., Novelli, M.R., and Bodmer, W.F. (1996). The mutation rate and cancer. Proc. Natl. Acad. Sci. USA 93, 14800-14803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T.L. et al. (2002). Prevalence of somatic alterations in the colorectal cancer cell genome. Proc. Natl. Acad. Sci. USA 99, 3076-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]