Abstract
Hypoxia-inducible factor-1 (HIF-1) is a regulator of metabolic adaptation to hypoxia. It is now appreciated that HIF-1α accumulation is achieved under normoxic conditions by various factors, such as TNF-α. Here, it was our intention to gain insight into the signaling mechanisms used by TNF-α to stimulate HIF-1α. In tubular LLC-PK1 or human embryonic kidney cells, TNF-α induced accumulation of HIF-1α protein but not HIF-1α mRNA. Blocking nuclear factor (NF)-κB with sulfasalazine or expression of an IκB superrepressor attenuated HIF-1α accumulation, whereas transfection of active p50/p65-NF-κB subunits mimicked a TNF-α response. Experiments with actinomycin D and cycloheximide also pointed to a transcriptional and translational process in facilitating the TNF-α response. Interestingly, and in contrast to established hypoxic signaling concepts, TNF-α elicited HIF-1α accumulation in a ubiquitinated form that still bound the von Hippel-Lindau (pVHL) protein. These data indicate that HIF-1α accumulation by TNF-α demands the NF-κB pathway, preserves ubiquitination of HIF-1α, and allows the HIF-1α-pVHL interaction.
INTRODUCTION
The hypoxia-inducible factor-1 (HIF-1) is a heterodimeric transcription factor that senses decreased oxygen availability and responds with enhanced transcription of classic hypoxia-inducible genes involved in angiogenesis, erythropoiesis, energy metabolism, and cell survival decisions. This makes HIF-1 a regulator of oxygen homeostasis (Semenza, 2002; Zhu et al., 2002). HIF-1 is composed of the helix-loophelix-Per-Arnt-Sim (bHLH)-PAS protein HIF-1α and the aryl hydrocarbon nuclear translocator (ARNT) known as HIF-1β (Wang and Semenza, 1995; Wang et al., 1995). In many cell types, the mRNA levels of both HIF-1α and HIF-1β seem to be permanently expressed. However, under normoxia, HIF-1α protein is kept at a low or undetectable level by continuous degradation via the 26S-proteasome, whereas HIF-1β protein is constitutively present.
Accumulation of HIF-1α that promotes active HIF-1 complex formation under hypoxia is known from observations that pointed to a crucial role of proly hydroxylases (HIFPHs) and the von Hippel-Lindau protein (pVHL) in HIF-1α degradation (Ivan et al., 2001; Jaakkola et al., 2001; Yu et al., 2001). Elegant work suggests that HIF-PHs sense oxygen and target proline residues at position 564 and/or 402 of HIF-1α to hydroxylate them (Ivan et al., 2001; Jaakkola et al., 2001; Yu et al., 2001). Proline hydroxylation seems to be necessary and sufficient for binding of pVHL to HIF-1α with concomitant degradation of HIF-1α by the ubiquitin/proteasome system. It is appreciated that hypoxia, transition metals such as CoCl2, and the iron chelator desferroxamine directly inhibit HIF-PHs with concomitant HIF-1α stabilization. However, recent data pointed out that HIF-1α expression could also be regulated at the translational level (Laughner et al., 2001). Furthermore, it emerges that stabilization and transactivation of HIF-1α are two separate processes that are regulated by hydroxylation at distinct residues. Hydroxylation of proline 564/402 affects protein accumulation, whereas hydroxylation of Asn803 is involved in transactivation. These modifications are regulated through partially overlapping and/or distinguishable pathways (Lando et al., 2002; Sang et al., 2002).
In addition to hypoxia, more recent evidence suggest that HIF-1 can be accumulated and activated during normoxia by growth factors, cytokines, hormones, and nitric oxide (El Awad et al., 2000; Sandau et al., 2001a). Reports on cytokines in HIF-1α stability regulation pointed to a role of interleukin (IL)-1β and tumor necrosis factor-α (TNF-α) (Thornton et al., 2000; Sandau et al., 2001b). However, details of HIF-1 regulation by TNF-α remain unclear, although the involvement of a reactive oxygen species (ROS)-sensitive pathway has been reported (Haddad and Land, 2001). Meanwhile, the role of ROS in stabilizing or destabilizing HIF-1α is controversial, and a unifying concept awaits clarification (Albina et al., 2001; Sandau et al., 2001b). However, considering the impact of cytokines during inflammation, one needs to know how cytokines contribute to HIF-1 regulation and whether mechanisms of HIF-1α accumulation share components established to operate during hypoxia.
A phosphatidylinositol-3-kinase (PI3K)/protein kinase B (Akt)/FKBP-rapamycin-associated protein (FRAP) pathway emerged as being crucial in IL-1β-, insulin-, and epithelial growth factor-evoked HIF-1 responses (Zhong et al., 2000; Stiehl et al., 2002; Treins et al., 2002). However, the PI3K pathway seems to be cell type-specific and may participate in the hypoxic response (Alvarez-Tejado et al., 2002; Arsham et al., 2002). For TNF-α, we noticed that the general kinase inhibitor genistein and more specifically, the PI3K inhibitors wortmannin and LY 294002 blocked HIF-1α accumulation (Sandau et al., 2001b).
Here, we determined signaling mechanisms that provoke HIF-1α accumulation in response to TNF-α. Unexpectedly, TNF-α-elicited HIF-1α accumulation was associated with ubiquitination of HIF-1α and an intact pVHL-HIF-1α association, which is distinct from those mechanisms established for hypoxia. We went on by using nuclear factor (NF)-κB inhibitors, overexpression of a dominant IκB versus active p50/p65 proteins, actinomycin D, and cycloheximide (CHX) application to demonstrate a crucial role of a transcriptionally/translationally active NF-κB pathway in TNF-α signaling. We conclude that in contrast to hypoxia, TNF-α uses a distinct signaling pathway to accumulate HIF-1α.
MATERIALS AND METHODS
Materials
Chemicals were of the highest grade of purity and were commercially available. Specifically, medium, fetal calf serum (FCS), and supplements were purchased from PAA Laboratories (Linz, Austria). Methionine-free medium was from PromoCell (Heidelberg, Germany). Recombinant human TNF-α, MG132, sulfasalazine, actinomycin D, DAPI, CHX, anti-actin antibody, and FITC-labeled anti-mouse secondary antibodies were from Sigma (St. Louis, MO). A protein assay kit was from Bio-Rad. Ni-NTA-agarose was from Qiagen (Hilden, Germany). The anti-HA mAb came from Babco (Richmond, CA). Anti-mouse dynal beads were from Dynal (Great Neck, NY). Protease inhibitor cocktails came from Roche (Mannheim, Germany). Nitrocellulose membrane, ECL detection system, and horseradish peroxidase (HRP)-labeled anti-mouse or anti-rabbit secondary antibodies were from Amersham Life Science (Arlington Heights, IL). [35S]methionine, multitest slides, and coverslips were from ICN Biomedicals (Cleveland, OH), and FluorSave mounting medium was from Calbiochem (La Jolla, CA). The CoolSNAP CCD camera was from Roper Scientific Photometrics (Tucson, AZ), and the MetaMorph software package came from Universal Imaging (West Chester, PA). The peqGOLD RNAPure kit was from Peqlab Biotechnologie (Erlangen, Germany), and primers were from MWG-Biotech (Ebeusberg, Germany). The pCMV-HIS6-ubiquitin plasmid was given by Dr. D. Bohmann (European Molecular Biology Laboratory, Heidelberg, Germany). Plasmids pHIF-1α and pCMV-HA-pVHL were kindly provided by Dr. P.J. Ratcliffe (Institute of Molecular Medicine, John Radcliffe Hospital, Oxford, UK). HIF-1α antibody, pVHL antibody Ig32, Advantage RT-for-PCR kit, AdvanTaq PCR kit, and plasmids pCMV-IκBα and pCMV-IκBαM were purchased from Becton Dickinson (Mountain View, CA). Plasmids pRSV-NF-κB1 (p50) and pRSV-RelA (p65) were constructed by Dr. G. Nabel and Dr. N. Perkins and supplied by the AIDS Research and Reference Program (McKesson BioServices Corporation, Rockville, MD).
Cell Culture
Human embryonic kidney (HEK293) cells were cultured in DMEM with 4.5 g/l d-glucose. Proximal tubular LLC-PK1 cells (obtained from Prof. D. Dietrich, Konstanz, Germany) were cultured in DMEM with 1 g/l d-glucose. Medium was supplemented with 10% FCS, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were transferred two times per week, and medium was changed before experiments. Cells were kept in a humidified atmosphere of 5% CO2 in air at 37°C.
Indirect Immunofluorescence and Fluorescence Microscopy
LLC-PK1 cells grown on multitest slides were stimulated with 500 ng/ml TNF-α for 16 h or 1 μM MG132 for 4 h. Cells were fixed with 4% paraformaldehyde for 5 min and permeabilized with 4% paraformaldehyde/0.2% Triton X-100 for 5 min at room temperature. To block nonspecific antibody binding, slides were incubated for 30 min at room temperature with 5% milk/PBS. Slides were incubated successively with the HIF-1α antibody (1:100 in 1% milk/PBS) at 4°C overnight, and FITC-labeled secondary antibodies (1:100 in 1% milk/PBS) at 37°C for 1 h. Finally, 0.2 μg/ml DAPI/PBS was added at room temperature for 2 min. Slides were washed three times for 5 min with PBS and rinsed briefly with distilled water. Coverslips were mounted to the multitest slides with intermediate FluorSave mounting medium. Slides were examined by an Axioskop fluorescence microscope. Photographs were taken with a CoolSNAP CCD camera, and images were created by the MetaMorph software package.
Cell Transfection
HEK293 cells (2 × 106) were plated in 10-cm dishes, and 4 × 105 LLC-PK1 cells were seeded in 6-cm dishes 1 day before transfection. At a rate of 60% confluence, cells were transfected with plasmids by use of the calcium phosphate precipitation method (Sambrook et al., 1989). Briefly, plasmids in the presence of 125 mM CaCl2 and HBS buffer (25 mM HEPES, 140 mM NaCl, 0.75 mM Na2HPO4, pH 7.05) were incubated for 30 min at room temperature and added dropwise to cells. At 16 h later, medium was changed, and incubations continued for another 8-h period before cell stimulation.
HIF-1α Ubiquitination Assay
HEK293 cells were cotransfected with 0.3 μg of a plasmid encoding full-length HIF-1α (pHIF-1α) and 3 μg of pCMV-HIS6-ubiquitin plasmid encoding HIS-tagged ubiquitin. After stimulation with 500 ng/ml TNF-α, 1 μM MG132, or 100 μM CoCl2 for the indicated times, cells were scraped off and centrifuged. Lysis buffer A (300 μl) (50 mM Tris, 150 mM NaCl, 8 M urea, pH 7.5) was added to each pellet and immediately vortexed three times for 15 s. After centrifugation at 15,000 × g for 30 min, lysates were transferred into fresh tubes. Five hundred micrograms of protein from the supernatant was mixed with 100 μl Ni-NTA-agarose (1:1 resuspended in lysis buffer A) and incubated, while rolling, at room temperature for 1 h. Then, beads were pelleted by centrifugation at 1000 × g for 5 min, washed three times with 200 μl lysis buffer A, resuspended with 50 μl of 2× sample buffer (125 mM Tris/HCl, 2% SDS, 10% glycerin, 1 mM DTT, 0.002% bromphenol blue, pH 6.9), and heated at 95°C for 10 min. Beads were removed by centrifugation. Proteins were separated electrophoretically on 7.5% SDS gels, followed by Western analysis using HIF-1α antibodies.
HIF-1α-pVHL Coimmunoprecipitation
HEK293 cells were cotransfected with 3 μg pHIF-1α and 1 μg pCMV-HA-pVHL encoding HA-tagged full-length pVHL. Cells were stimulated with 500 ng/ml TNF-α, 1 μM MG132, or 100 μM CoCl2 for the indicated times. Cells were scraped off from the dishes and collected. To each cell pellet, 300 μl lysis buffer B (50 mM Tris, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 mM PMSF, protease inhibitor cocktail, pH 7.5) was added, followed by immediate vortexing (3 × 15 s). After centrifugation (15,000 × g for 30 min), supernatants were transferred into fresh tubes. The supernatant (1 mg protein) was supplied with 1 μg anti-HA antibody and incubated at 4°C for 1 h. Thereafter, 20 μl anti-mouse dynal beads were added, and incubations continued at 4°C overnight. Beads were collected, washed three times with 100 μl lysis buffer B, supplemented with 50 μl of 2 × sample buffer, and boiled at 95°C for 10 min. Beads were removed by centrifugation, and supernatants were loaded on 7.5% SDS-gels. Western blot analysis was performed by use of the HIF-1α antibody.
Western Blot Analysis
HIF-1α was quantified by Western analysis. Briefly, cells were incubated, scraped off, lysed in 300 μl (for HEK293 cells) or 150 μl (for LLC-PK1 cells) lysis buffer B, sonicated, and centrifuged (15000 × g, 15 min). Protein (80 μg) was added to the same volume of 2 × SDS-PAGE sample buffer and boiled for 5 min. Proteins were resolved on 7.5% SDS-polyacrylamide gels. Gels were washed with blotting buffer (25 mM Tris, 192 mM glycine, 20% methanol, pH 8.3) for 5 min, proteins were blotted onto nitrocellulose by a semidry transfer cell, and unspecific binding sites were blocked with 5% milk/TTBS (50 mM Tris/HCl, 140 mM NaCl, 0.05% Tween-20, pH 7.2) for 1 h. The HIF-1α antibody (1:1000 in 1% milk/TTBS) was added and incubated overnight at 4°C. Then, nitrocellulose membranes were washed three times for 5 min with TTBS. For protein detection, blots were incubated with an HRP-labeled goat anti-mouse secondary antibody (1:2000 in 1% milk/TTBS) for 1 h and washed three times for 15 min with TTBS, followed by ECL detection.
35S-Radioisotopic Labeling
Cells were starved for 1 h in serum- and methionine-free medium, followed by replacement with methionine-free medium containing 10% FCS and 100 μCi/ml [35S]methionine for 8 h in the presence of 100 μM CoCl2, 1 μM MG132, or 500 ng/ml TNF-α. Cells were then washed with PBS and lysed with buffer B. HIF-1α was coimmunoprecipitated from lysates containing 2 mg of total protein by use of 2 μg anti-pVHL antibody Ig32 as described above. After 10% SDS-PAGE, the gel was dried and exposed to film.
Semiquantitative RT-PCR
LLC-PK1 cells (2 × 106) were seeded 1 day before experiments. The following day, medium was changed, and if applicable, inhibitors were preincubated for 30 min. Cells were stimulated with 500 ng/ml TNF-α for 8 h. Total RNA was isolated by use of the peqGOLD RNAPure kit. The reverse transcription was completed with an Advantage RT-for-PCR kit using hexamer random primers. PCR was performed with an AdvanTaq PCR kit. The following primer pairs were selected: HIF-1α, 5′-CTCAAAGTCGGACAGCCTCA-3′, 5′-CCCTGCAGTAGGTTTCTGCT-3′; actin, 5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′, 5′-CTAGAAGCATTTGCGGTCGACGATGGAGGG-3′. The amplification program was as follows: 95°C, 30 s; 56°C, 30 s; 72°C, 1 min; 20 cycles; 72°C, 10 min. RT-PCR products were separated on 2% agarose gels and visualized with ethidium bromide.
Statistical Analysis
Each experiment was performed at least three times, and representative data are shown.
RESULTS
TNF-α Provoked Accumulation and Ubiquitination of HIF-1α
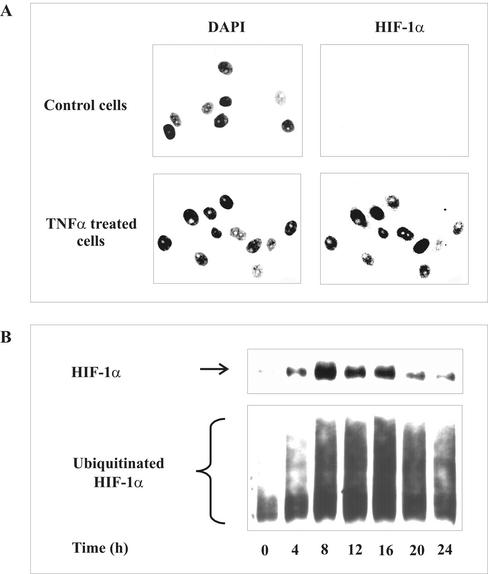
In previous studies, we have shown that tubular LLC-PK1, HEK293 cells, and HepG2 cells accumulate HIF-1α and/or transactivate HIF-1 in response to hypoxia, chemical hypoxia, and TNF-α. Therefore, we used these cell lines to examine mechanisms of TNF-α-evoked HIF-1α accumulation. In a first set of experiments, we activated LLC-PK1 cells with 500 ng/ml TNF-α for 16 h to follow subcellular localization of HIF-1α (Figure 1A). Immunofluorescence analysis highlighted HIF-1α immunoreactivity exclusively in the nucleus on the basis of DAPI counterstaining. As expected, HIF-1α was absent, i.e., below the detection limit in control cells.
Figure 1.
HIF-1α accumulation and ubiquitination by TNF-α. (A) LLC-PK1 cells were stimulated with 500 ng/ml TNF-α for 16 h or remained untreated. Nuclei were visualized by DAPI staining. HIF-1α was detected by an anti-HIF-1α mAb and FITC-labeled anti-mouse secondary antibody. (B) HEK293 cells were cotransfected with 3 μg pCMV-HIS6-ubiquitin and 0.3 μg pHIF-1α expression plasmids for 16 h. Medium was changed and incubations continued for another 8-h period before stimulation with 500 ng/ml TNF-α for times indicated. HIF-1α and ubiquitinated HIF-1α were determined by Western analysis as described in MATERIALS AND METHODS. Each experiment was performed at least three times, and representative data are shown.
To follow HIF-1α accumulation and ubiquitination, we cotransfected HEK293 cells with 0.3 μg pHIF-1α and 3 μg pCMV-HIS6-ubiquitin expression plasmids. After the addition of 500 ng/ml TNF-α, we noticed HIF-1α accumulation in a time-dependent manner with low, although significant protein accumulation at 4 h (Figure 1B). A maximum increase was seen between 8 and 16 h, with declining amounts of protein thereafter. Somewhat surprisingly, ubiquitinated HIF-1α showed a similar response. In controls, ubiquitination was low. The amount of ubiquitination increased in parallel to HIF-1α accumulation, reached maximal values between 8 and 16 h, and declined thereafter. The identical time response seen for HIF-1α accumulation and ubiquitination suggests that this is based on the presence of the protein, i.e., the HIF-1α target, rather than reflecting an enhanced ubiquitination reaction. In any case, accumulation of HIF-1α and increased ubiquitination of the protein is in contrast to mechanisms established for hypoxic signaling, in which HIF-1α accumulation results from decreased ubiquitination.
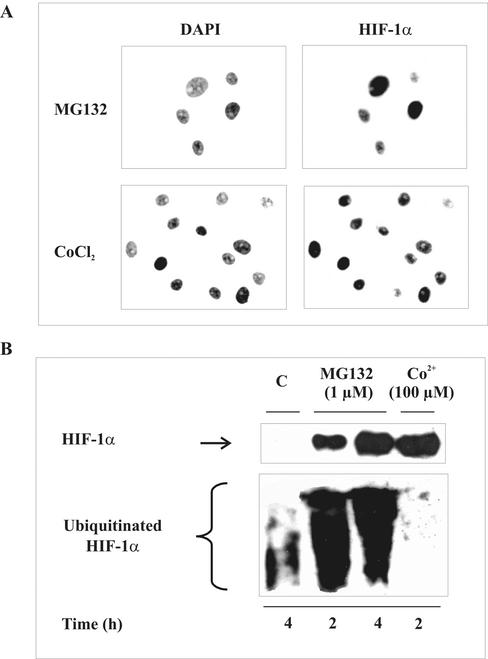
To follow up the idea that HIF-1α accumulation may indeed be compatible with increased ubiquitination, we used the proteasome inhibitor MG132, and as a further established control, we chose CoCl2, known to mimic hypoxia. As expected, exposing LLC-PK1 cells to 1 μM MG132 or 100 μM CoCl2 revealed a strong HIF-1α immunoreactivity (Figure 2A). Nuclear protein accumulation was proven by DAPI counterstaining.
Figure 2.
HIF-1α stabilization and ubiquitination by MG132 and CoCl2. (A) LLC-PK1 cells were stimulated with 1 μM MG132 or 100 μM CoCl2 for 4 h. HIF-1α and DAPI staining were performed as described in Figure 1A. (B) HEK293 cells were cotransfected with 3 μg pCMV-HIS6-ubiquitin and 0.3 μg pHIF-1α to follow HIF-1α and ubiquitinated HIF-1α accumulation as described in Figure 1B. HEK293 cells were stimulated with 1 μM of the proteasome inhibitor MG132 or 100 μM CoCl2 for times indicated. Each experiment was performed at least three times, and representative data are shown.
Therefore, we repeated examination in HEK293 cells and determined HIF-1α accumulation and ubiquitination (Figure 2B). Not surprisingly, both MG132 and CoCl2 provoked massive HIF-1α accumulation to roughly equal amounts on the basis of Western analysis. However, whereas CoCl2 decreased basal ubiquitination, MG132 dramatically increased ubiquitination of HIF-1α. Thus, MG132 and TNF-α share the ability to accumulate HIF-1α despite ubiquitination. Considering that ubiquitination of HIF-1α requires the interaction with pVHL with the further prerequisite of HIF-1α proline hydroxylation, we looked into the HIF-1α-pVHL interaction.
TNF-α Left the HIF-1α-pVHL Interaction Intact
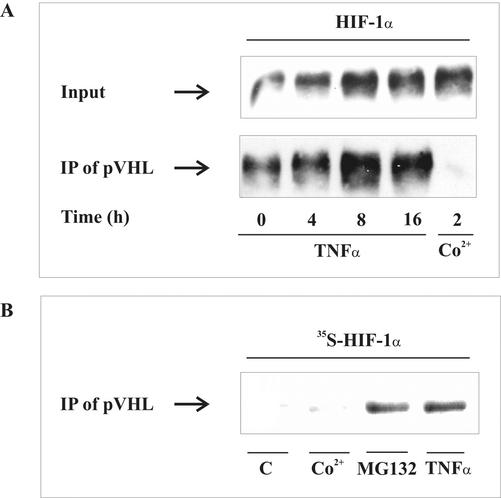
For these experiments, HEK293 cells were cotransfected with 1 μg pCMV-HA-pVHL and 3 μg pHIF-1α expression plasmids. At 24 h after transfection, HEK293 cells were stimulated with 500 ng/ml TNF-α or 100 μM CoCl2, and the HIF-1α-pVHL interaction was assessed by coimmunoprecipitation (Figure 3A). The presence of HIF-1α in the lysate used for immunoprecipitation was followed by Western analysis (input control) and revealed increasing amount of HIF-1α after TNF-α- or CoCl2-stimulation.
Figure 3.
Interaction between HIF-1α and pVHL. (A) HEK293 cells were cotransfected with 1 μg pCMV-HA-pVHL and 3 μg pHIF-1α expression plasmids. After medium was changed at 16 h, incubations continued for 8 h before stimulation with 500 ng/ml TNF-α or 100 μM CoCl2 for times indicated. Coimmunoprecipitation was performed with an anti-HA mAb that recognized pVHL- and anti-mouse antibody-coated magnetic beads. Accumulation of HIF-1α in cell lysates (input) and immunoprecipitates (IP) were determined by Western analysis using an anti-HIF-1α mAb as described in MATERIALS AND METHODS. (B) Autoradiography of endogenous [35S]methionine-labeled HIF-1α that coimmunoprecipitated with pVHL. LLC-PK1 cells were treated for 8 h with 500 ng/ml TNF-α, 1 μM MG132, or 100 μM CoCl2 or remained unstimulated (C). For details, see MATERIALS AND METHODS. Each experiment was performed at least three times, and representative data are shown.
As noticed in previous experiments, TNF-α evoked the strongest HIF-1α accumulation at 8-16 h. When pVHL was immunoprecipitated followed by HIF-1α Western analysis, we noticed a significant pVHL-HIF-1α interaction in controls. More interestingly, in parallel with HIF-1α accumulation, we observed a dramatically increased amount of HIF-1α that coprecipitated with pVHL. Only CoCl2-evoked HIF-1α accumulation was associated with a decreased pVHL-HIF-1α interaction, because no HIF-1α coprecipitated with pVHL under these conditions. To study the pVHL-HIF-1α interaction in a more physiological environment not making use of overexpressed proteins, we used a more sensitive radioactive approach (Figure 3B). LLC-PK1 cells were incubated with [35S]methionine to label HIF-1α during its expression under the impact of TNF-α, CoCl2, or MG132. When pVHL was immunoprecipitated, we identified radioactive HIF-1α in the precipitates. As expected, in controls and CoCl2-treated samples, no HIF-1α coimmunoprecipitated with pVHL, whereas in TNF-α- and MG132-exposed samples, pVHL contained endogenous, radioactive HIF-1α. We must conclude that accumulation of HIF-1α in response to TNF-α differs from the situation seen with CoCl2 with respect to not only ubiquitination but also the pVHL interaction. To obtain more information on the signaling pathways used by TNF-α, we considered NF-κB of importance.
Inhibition of NF-κB Attenuated TNF-α-Evoked HIF-1α Accumulation
NF-κB is a prototype of transcription factor known to regulate an impressive number of genes. We began by using the transcriptional inhibitor actinomycin D to check for a transcriptional activity in facilitating the TNF-α response (Figure 4A). Actinomycin D, a universal inhibitor of mRNA synthesis, dose-dependently attenuated HIF-1α accumulation in response to TNF-α. Inhibition was evident at a concentration of 50 ng/ml and more pronounced at 100-200 ng/ml. To rule out the possibility that actinomycin D simply depleted HIF-1α mRNA, thereby suppressing the TNF-α response, we used actinomycin D in combination with MG132 (Figure 4B). At all concentrations tested, actinomycin D left accumulation of HIF-1α in response to MG132 unaltered. This indicates that under the experimental conditions used, actinomycin D did not alter HIF-1α accumulation when blocking protein degradation, although it attenuated the TNF-α response. We can assume that actinomycin D indeed blocked transcription in association with NF-κB action.
Figure 4.
Actinomycin D attenuated TNF-α-stimulated HIF-1α accumulation. LLC-PK1 cells were preincubated for 30 min with the transcription inhibitor actinomycin D. Thereafter, cells were stimulated with (A) 500 ng/ml TNF-α or (B) 3 μM MG132 for 8 h. HIF-1α was determined by Western analysis. For details, see MATERIALS AND METHODS. Each experiment was performed at least three times, and representative data are shown.
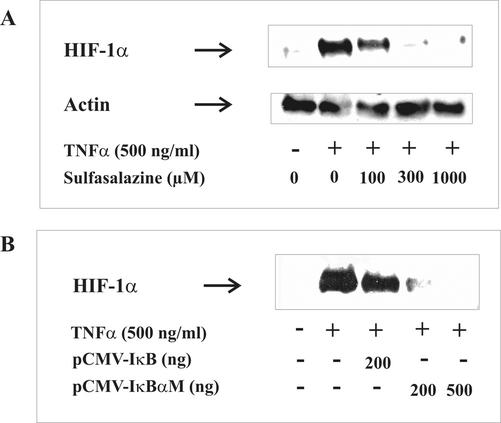
In the following experiment, we used sulfasalazine, a specific inhibitor for IκB degradation (Figure 5A). Sulfasalazine dose-dependently attenuated HIF-1α accumulation in response to TNF-α.
Figure 5.
Inhibition of NF-κB attenuated TNF-α-stimulated HIF-1α accumulation. (A) LLC-PK1 cells were preincubated for 30 min with the NF-κB inhibitor sulfasalazine at the indicated concentrations. (B) Cells were transfected with pCMV-IκBα and pCMVIκBαM expression plasmids for 24 h. Thereafter, cells were stimulated with 500 ng/ml TNF-α for 16 h. HIF-1α was determined by Western analysis. For details, see MATERIALS AND METHODS. Each experiment was performed at least three times, and representative data are shown.
Although 100 μM sulfasalazine was effective to some extent, a concentration of 300 μM of the NF-κB inhibitor abrogated HIF-1α accumulation completely. To confirm the requirement of NF-κB and thus avoid any drug-related side effects, we transiently transfected cells with a pCMV-IκBαM plasmid known to express the superrepressor IκB. This IκB mutant cannot be phosphorylated and thus degraded by the proteasome and thereby blocks NF-κB activation. Transfection of LLC-PK1 cells with pCMV-IκBαM blocked HIF-1α accumulation completely (Figure 5B). However, a control plasmid (pCMV-IκBα) that expresses a form of IκB that still can be phosphorylated and degraded by the proteasome showed only a very minor impact on TNF-α-elicited HIF-1α accumulation. To sum up, inhibitor studies suggested a role of NF-κB in the signal transduction pathways used by TNF-α to accumulate HIF-1α. To more directly address the involvement of NF-κB, we sought to transfect cells with active p50- and p65-NF-κB subunits and to study HIF-1α accumulation.
Transient Transfection of Cells with Active p50/p65 Caused HIF-1α Accumulation
LLC-PK1 cells were cotransfected with pRSV-NF-κB1 (p50) and pRSV-RelA (p65) expression plasmids. After transfection for 8 h, the medium was changed, and cells were left for another 16-h period (Figure 6). Thereafter, Western analysis showed HIF-1α accumulation in cells cotransfected with pRSV-NF-κB1 and pRSV-RelA.
Figure 6.
Cotransfection of p50/p65 NF-κB subunits promoted HIF-1α accumulation. LLC-PK1 cells were cotransfected with indicated amount of pRSV-NF-κB1/pRSV-RelA expression plasmids. After transfection for 16 h, medium was changed, and incubations continued for 8 h. As the positive control, cells were treated with 500 ng/ml TNF-α for 8 h. HIF-1α was detected by Western analysis as described in MATERIALS AND METHODS. Each experiment was performed at least three times, and representative data are shown.
HIF-1α accumulation was comparable to the effect achieved by TNF-α treatment. Apparently, activation of NF-κB transmits a signal that accounts for HIF-1α accumulation. Considering that NF-κB is a transcription factor, it appeared rational to check for HIF-1α mRNA and thus to gain further insight into TNF-α signal transmission.
TNF-α Did Not Alter the mRNA of HIF-1α
In this set of experiments, it was our intention to unravel any impact on the mRNA of HIF-1α, either under conditions associated with HIF-1α protein accumulation or under conditions correlated with its absence. LLC-PK1 cells were transfected with 500 ng pCMV-IκBαM plasmid or cotransfected with 500 ng pRSV-NF-κB1/pRSV-RelA plasmids overnight to express p50/p65. TNF-α (500 ng/ml) in the absence or presence of the NF-κB inhibitor sulfasalazine (500 μM), preincubated for 30 min, was supplied for 8 h (Figure 7). Total RNA was isolated, and the mRNA of HIF-1α was quantified by RT-PCR using actin as an internal control.
Figure 7.
HIF-1α mRNA under the impact of TNF-α. LLC-PK1 cells were transfected with 500 ng pIκBαM (IκB-mutant) or with 500 ng pRSV-NF-κB1/pRSV-RelA expression plasmids as indicated in Figures 4 and 5. Sulfasalazine (500 μM) was preincubated for 30 min. Cells were then stimulated with 500 ng/ml TNF-α for 8 h. Total RNA was isolated, and 1 μg total RNA was used for reverse transcription. PCR products were visualized with ethidium bromide on 2% agarose gels as described in MATERIALS AND METHODS. Each experiment was performed at least three times, and representative data are shown.
Visualization and quantification of RT-PCR products revealed no changes. We must assume that TNF-α left the mRNA level of HIF-1α unaltered, although HIF-1α protein accumulated. Blocking NF-κB or using active p50/p65 was without any impact on the HIF-1α mRNA.
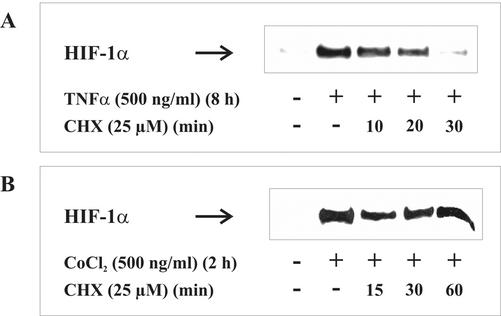
TNF-α Caused HIF-1α Protein Synthesis
To obtain additional information on pathways used by TNF-α to accumulate HIF-1α, we determined HIF-1α expression. To this end, we used CHX to block protein synthesis (Figure 8).
Figure 8.
CHX affects TNF-α-induced HIF-1α accumulation. LLCPK1 cells were stimulated with (A) 500 ng/ml TNF-α for 8 h or (B) 100 μM CoCl2 for 2 h. Thereafter, the protein translation inhibitor CHX was added (25 μM), and incubations continued for 10-60 min. HIF-1α was detected by Western analysis using the anti-HIF-1α mouse mAb as described in MATERIALS AND METHODS. Each experiment was performed at least three times, and representative data are shown.
CHX was incubated up to 30 min after HIF-1α accumulation that was achieved with 500 ng/ml TNF-α for 8 h. Application of CHX reduced HIF-1α protein significantly to a basal level during 30 min (Figure 8A). In contrast, in cells exposed to CoCl2, HIF-1α level remained constant >60 min despite the lack of ongoing protein synthesis. This observation is consistent with previous studies showing that CoCl2 had no effect on HIF-1α synthesis but blocked its proteasomal degradation. Together, these results suggest that TNF-α increases HIF-1α protein levels through a translation-dependent pathway.
DISCUSSION
Mechanisms of TNF-α-versus Hypoxia-evoked HIF-1α Accumulation
Here, we provide evidence that TNF-α accumulates HIF-1α protein but not HIF-1α mRNA by a pathway that seems to be distinct from the one known for hypoxia. Accumulation of HIF-1α by TNF-α is associated with ubiquitination of HIF-1α and an intact pVHL-HIF-1α interaction. By pharmacological intervention with NF-κB activation, transfection of a dominant noncleavable IκB-mutant, transfection of active p50/p65-NF-κB subunits, and actinomycin D and cycloheximide supplementation, we provide evidence that a transcriptional/translational activity of NF-κB signaling is a prerequisite for HIF-1α accumulation in response to TNF-α. It is concluded that TNF-α uses a thus far unappreciated signaling pathway for HIF-1α accumulation under normoxia.
In recent years, a number of breakthroughs and elegant studies have allowed detailed mechanistic insights in oxygen sensing and HIF-1α stabilization (Ivan et al., 2001; Jaakkola et al., 2001; Yu et al., 2001). Under normoxia, the continuous hydroxylation of HIF-1α by prolyl hydroxylases at proline 564 and/or 402 ensures the interaction of HIF-1α with the pVHL protein. Binding of this E3-ligase complex guarantees ubiquitination and subsequent degradation of HIF-1α via the 26S-proteasome degradation machinery. As a result, the protein level of HIF-1α is kept at a low, often undetectable level. Considering that HIF-PHs require molecular oxygen and Fe2+ for catalysis, it seems logical that HIF-PH activity is impaired under hypoxia or by iron chelation. As a result, hydroxylation of HIF-1α is decreased, binding of pVHL to HIF-1α is ruined, degradation of HIF-1α is hindered, and consequently, HIF-1α accumulates. On the basis of our results, this scenario does not apply for TNF-α-evoked HIF-1α accumulation. Under conditions of TNF-α treatment and HIF-1α accumulation, the interaction of HIF-1α and pVHL remains intact. The increased amount of HIF-1α that coimmunoprecipitated with pVHL (Figure 3) may simply reflect increased amounts of HIF-1α under these experimental conditions, rather than an increased pVHL-HIF-1α interaction. For control reasons, we applied the hypoxia-mimicking agent CoCl2, which supports current concepts on HIF-1α accumulation on the basis of a largely attenuated pVHL-HIF-1α interaction. Clearly, TNF-α behaved differently.
Having established that TNF-α left the interaction of HIF-1α with its putative E3-ubiquitin ligase intact, one would predict that ubiquitination of HIF-1α still occurs. Indeed, TNF-α favored accumulation of a ubiquitinated form of HIF-1α (Figure 1). Again, this is distinct from hypoxic signaling. Because these findings were somewhat puzzling, it was our intention to use other agonists, known for their impact on HIF-1α accumulation, that may share properties of the TNF-α signaling system. Because the proteasome system is known to degrade HIF-1α, several studies used proteasome inhibitors, such as MG132, to stabilize HIF-1α protein (Palmer et al., 2000; Hur et al., 2001). As expected, MG132 provoked accumulation of HIF-1α in its ubiquitinated form and more importantly, revealed nuclear localization. In some analogy to protein appearance in the nucleus, it has been noticed that MG132 promoted binding of HIF-1 to its DNA recognition sites (Salceda and Caro, 1997). Although MG132 stabilizes HIF-1α and causes DNA binding, it does not cause transactivation of HIF-1 (Salceda and Caro, 1997; Kallio et al., 1999). As pointed out earlier, this phenomenon is most likely caused by a nonspecific toxic effect of this compound, because proteasome inhibitors both produced a decrease in basal expression of luciferase activity and inhibited stimulation of its expression (Salceda and Caro, 1997). We made the same observation (data not shown), which unfortunately excluded the use of MG132 in reporter assays. However, TNF-α is known to transactivate HIF-1 in HepG2 cells, with concomitant up-regulation of classic hypoxia-responsive genes (Hellwig-Bürgel et al., 1999; Sandau et al., 2001b), which suggests that a ubiquitinated form of HIF-1α is transcriptionally active.
The Role of NF-κB in TNF-α-evoked HIF-1α Accumulation
The transcription factor NF-κB is a classic component downstream of TNF-α-receptor activation. NF-κB activation is complex. However, it is clear that IκB kinase activation promotes phosphorylation of IκB, with subsequent dissociation from p50/p65 and proteasomal degradation of the ubiquitinated IκB inhibitor. Although details on IκB kinase activation remain unclear, it is generally believed that in many cases, an active p50/p65 heterodimer complex moves to the nucleus to drive gene activation. Our initial results using the inhibitor sulfasalazine (Wahl et al., 1998; Weber et al., 2000) are compatible with a role of NF-κB in the TNF-α pathway. To circumvent potential side effects of drugs, we substantiated a role of NF-κB by transfecting LLC-PK1 cells with an IκB-mutant that can no longer be phosphorylated and degraded, thus acting as a superrepressor of NF-κB and consequently blocking HIF-1α accumulation. Additional experiments (Figure 6) made use of transfecting active p50- and p65-NF-κB subunits, which provoked HIF-1α accumulation without the further addition of TNF-α. The sum of these examinations provided experimental evidence that TNF-α uses the NF-κB pathway to accumulate HIF-1α. The facts that active p50/p65 mimicked the effect of TNF-α and that actinomycin D attenuated HIF-1α accumulation in response to TNF-α strongly suggests that transcriptional activation of NF-κB is involved. At this point, any gene product awaits identification. However, simple up-regulation of HIF-1α mRNA can be ruled out.
Although they did not provide detailed mechanistic insights, several groups noticed the ability of TNF-α to affect the HIF-1 system. In HepG2 cells, TNF-α caused a moderate activation of HIF-DNA binding under normoxic conditions, whereas in hypoxia, TNF-α strongly increased HIF-1 activity compared with the effect of hypoxia alone (Hellwig-Bürgel et al., 1999). In line with our observations, no changes in the HIF-1α mRNA were noticed in HepG2 cells in response to TNF-α. However, in fibroblasts, the HIF-1α mRNA increased after addition of TNF-α or IL-1, although protein accumulation has not been examined (Thornton et al., 2000). Furthermore, in primary inflammatory cells, TNF-α but not IL-1β induced HIF-1α protein accumulation. The capacity to enhance HIF-1α accumulation was also noticed in inflammatory, TNF-α-stimulated peritoneal neutrophils but not in a variety of cell lines (Albina et al., 2001). Mechanistically, it has been proposed that oxygen radical formation in response to TNF-α facilitates HIF-1α accumulation (Haddad and Land, 2001). Again, this observation is fully compatible with our observation on the role of NF-κB activation, taking into account that reactive oxygen species are implicated in IκB kinase and thus NF-κB activation (Napoli et al., 2001).
On the basis of our findings, we assume that TNF-α regulates HIF-1α protein expression through a translation-dependent pathway rather than attenuating proteasomal degradation (Figure 8). A PI3K-Akt-FRAP-dependent pathway exists to accelerate HIF-1α synthesis in HER2 signaling and insulin signaling (Laughner et al., 2001; Treins et al., 2002). Our previous work pointed to the involvement of PI3K in TNF-α-stimulated HIF-1α accumulation (Sandau et al., 2001b). Further studies are needed to define a potential signaling interaction between PI3K-Akt-FRAP and NF-κB pathways, with the important note that Akt was recently identified as a downstream target of NF-κB (Meng et al., 2002). Interestingly, Chan et al. (2002) reported that active Akt provoked accumulation of hydroxylated HIF-1α, most likely because of increased protein translation of decreased protein degradation. The notion of a stable and hydroxylated form of HIF-1α is compatible with our observation that TNF-α-accumulated HIF-1α is ubiquitinated and thus remains bound to pVHL.
More recently, pharmacological approaches pointed to the involvement of the MAPK cascade, in particular ERK1/2 and p38 in HIF-1α accumulation (Richard et al., 1999; Gorlach et al., 2001). Especially p38 activation is a common signal of downstream TNF-α receptor activation (Baud and Karin, 2001). Although biological consequences are obscure, several of these transduction pathways may turn out to be relevant for the TNF-α-HIF-1α signaling circuit.
Our study adds new information to the concepts of HIF-1α regulation by TNF-α under normoxia, identifies a distinct pathway for HIF-1α accumulation, and points to NF-κB as an important signaling component in facilitating the TNF-α response. Further studies are needed to address how NF-κB mediates protein synthesis. One may speculate whether 4E-BP1 and eIF-4E are involved, as previously suggested for HER2 signaling (Laughner et al., 2001). The ability of TNF-α to accumulate HIF-1α may imply a role of HIF-1 during inflammation, which broadens the sphere of HIF-1 action.
Acknowledgments
We are grateful to Dr. D. Bohmann (European Molecular Biology Laboratory, Heidelberg, Germany) and Dr. P.J. Ratcliffe (Institute of Molecular Medicine, John Radcliffe Hospital, Oxford, UK) for providing reagents. Dr. G. Nabel and Dr. N. Perkins are acknowledged for sharing reagents with the AIDS Research and Reference Program. The technical assistance of Sandra Christmann and Andrea Trinkaus is highly appreciated. This work was supported by grants from the Deutsche Forschungsgemeinschaft (BR999) and the Fonds der Chemischen Industrie and European Community (QLK6-CT-2000-00064).
References
- Albina, J.E., Mastrofrancesco, B., Vessella, J.A., Louis, C.A., Henry, W.L., Jr., and Reichner, J.S. (2001). HIF-1 expression in healing wounds: HIF-1alpha induction in primary inflammatory cells by TNF-alpha. Am. J. Physiol. Cell Physiol. 281, C1971-C1977. [DOI] [PubMed] [Google Scholar]
- Alvarez-Tejado, M., Alfranca, A., Aragones, J., Vara, A., Landazuri, M.O., and del Peso, L. (2002). Lack of evidence for the involvement of the phosphoinositide 3-kinase/akt pathway in the activation of hypoxia-inducible factors by low oxygen tension. J. Biol. Chem. 277, 13508-13517. [DOI] [PubMed] [Google Scholar]
- Arsham, A.M., Plas, D.R., Thompson, C.B., and Simon, M.C. (2002). Phosphatidylinositol 3-kinase/Akt signaling is neither required for hypoxic stabilization of HIF-1alpha nor sufficient for HIF-1-dependent target gene transcription. J. Biol. Chem. 277, 15162-15170. [DOI] [PubMed] [Google Scholar]
- Baud, V., and Karin, M. (2001). Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 11, 372-377. [DOI] [PubMed] [Google Scholar]
- Chan, D.A., Sutphin, P.D., Denko, N.C., and Giaccia, A.J. (2002). Role of prolyl hydroxylation in oncogenically stabilized hypoxiainducible factor-1alpha. J. Biol. Chem. 277, 40112-40117. [DOI] [PubMed] [Google Scholar]
- El Awad, B., Kreft, B., Wolber, E.M., Hellwig-Burgel, T., Metzen, E., Fandrey, J., and Jelkmann, W. (2000). Hypoxia and interleukin-1beta stimulate vascular endothelial growth factor production in human proximal tubular cells. Kidney Int. 58, 43-50. [DOI] [PubMed] [Google Scholar]
- Gorlach, A., Diebold, I., Schini-Kerth, V.B., Berchner-Pfannschmidt, U., Roth, U., Brandes, R.P., Kietzmann, T., and Busse, R. (2001). Thrombin activates the hypoxia-inducible factor-1 signaling pathway in vascular smooth muscle cells: role of the p22(phox)-containing NADPH oxidase. Circ. Res. 89, 47-54. [DOI] [PubMed] [Google Scholar]
- Haddad, J.J., and Land, S.C. (2001). A non-hypoxic, ROS-sensitive pathway mediates TNF-alpha-dependent regulation of HIF-1alpha. FEBS Lett. 505, 269-274. [DOI] [PubMed] [Google Scholar]
- Hellwig-Bürgel, T., Rutkowski, K., Metzen, E., Fandrey, J., and Jelkmann, W. (1999). Interleukin-1beta and tumor necrosis factor-alpha stimulate DNA binding of hypoxia-inducible factor-1. Blood 94, 1561-1567. [PubMed] [Google Scholar]
- Hur, E., Chang, K.Y., Lee, E., Lee, S.K., and Park, H. (2001). Mitogen-activated protein kinase kinase inhibitor PD98059 blocks the transactivation but not the stabilization or DNA binding ability of hypoxia-inducible factor-1alpha. Mol. Pharmacol. 59, 1216-1224. [DOI] [PubMed] [Google Scholar]
- Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J.M., Lane, W.S., and Kaelin, W.G., Jr. (2001). HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464-468. [DOI] [PubMed] [Google Scholar]
- Jaakkola, P., Mole, D.R., Tian, Y.M., Wilson, M.I., Gielbert, J., Gaskell, S.J., von Kriegsheim, A., Hebestreit, H.F., Mukherji, M., Schofield, C.J., et al. (2001). Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468-472. [DOI] [PubMed] [Google Scholar]
- Kallio, P.J., Wilson, W.J., O'Brien, S., Makino, Y., and Poellinger, L. (1999). Regulation of the hypoxia-inducible transcription factor 1alpha by the ubiquitin-proteasome pathway. J. Biol. Chem. 274, 6519-6525. [DOI] [PubMed] [Google Scholar]
- Lando, D., Peet, D.J., Whelan, D.A., Gorman, J.J., and Whitelaw, M.L. (2002). Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295, 858-861. [DOI] [PubMed] [Google Scholar]
- Laughner, E., Taghavi, P., Chiles, K., Mahon, P.C., and Semenza, G.L. (2001). HER2 (neu) signaling increases the rate of hypoxiainducible factor 1alpha (HIF-1alpha) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol. Cell. Biol. 21, 3995-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, F., Liu, L., Chin, P.C., and D'Mello, S.R. (2002). Akt is a downstream target of NF-kappa B. J. Biol. Chem. 277, 29674-29680. [DOI] [PubMed] [Google Scholar]
- Napoli, C., de Nigris, F., and Palinski, W. (2001). Multiple role of reactive oxygen species in the arterial wall. J. Cell Biochem. 82, 674-682. [DOI] [PubMed] [Google Scholar]
- Palmer, L.A., Gaston, B., and Johns, R.A. (2000). Normoxic stabilization of hypoxia-inducible factor-1 expression and activity: redox-dependent effect of nitrogen oxides. Mol. Pharmacol. 58, 1197-1203. [DOI] [PubMed] [Google Scholar]
- Richard, D.E., Berra, E., Gothie, E., Roux, D., and Pouyssegur, J. (1999). p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1alpha (HIF-1alpha) and enhance the transcriptional activity of HIF-1. J. Biol. Chem. 274, 32631-32637. [DOI] [PubMed] [Google Scholar]
- Salceda, S., and Caro, J. (1997). Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. J. Biol. Chem. 272, 22642-22647. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Sandau, K.B., Fandrey, J., and Brüne, B. (2001a). Accumulation of HIF-1 alpha under the influence of nitric oxide. Blood 97, 1009-1015. [DOI] [PubMed] [Google Scholar]
- Sandau, K.B., Zhou, J., Kietzmann, T., and Brune, B. (2001b). Regulation of the hypoxia-inducible factor 1alpha by the inflammatory mediators nitric oxide and tumor necrosis factor-alpha in contrast to desferroxamine and phenylarsine oxide. J. Biol. Chem. 276, 39805-39811. [DOI] [PubMed] [Google Scholar]
- Sang, N., Fang, J., Srinivas, V., Leshchinsky, I., and Caro, J. (2002). Carboxyl-terminal transactivation activity of hypoxia-inducible factor 1 alpha is governed by a von Hippel-Lindau protein-independent, hydroxylation-regulated association with p300/CBP. Mol. Cell. Biol. 22, 2984-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza, G.L. (2002). HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol. Med. 8, S62-S67. [DOI] [PubMed] [Google Scholar]
- Stiehl, D.P., Jelkmann, W., Wenger, R.H., and Hellwig-Burgel, T. (2002). Normoxic induction of the hypoxia-inducible factor 1alpha by insulin and interleukin-1beta involves the phosphatidylinositol 3-kinase pathway. FEBS Lett. 512, 157-162. [DOI] [PubMed] [Google Scholar]
- Thornton, R.D., Lane, P., Borghaei, R.C., Pease, E.A., Caro, J., and Mochan, E. (2000). Interleukin 1 induces hypoxia-inducible factor 1 in human gingival and synovial fibroblasts. Biochem. J. 350, 307-312. [PMC free article] [PubMed] [Google Scholar]
- Treins, C., Giorgetti-Peraldi, S., Murdaca, J., Semenza, G.L., and Van Obberghen, E. (2002). Insulin stimulates hypoxia-inducible factor 1 through a phosphatidylinositol 3-kinase/target of rapamycin-dependent signaling pathway. J. Biol. Chem. 277, 27975-27981. [DOI] [PubMed] [Google Scholar]
- Wahl, C., Liptay, S., Adler, G., and Schmid, R.M. (1998). Sulfasalazine: a potent and specific inhibitor of nuclear factor kappa B. J. Clin. Invest. 101, 1163-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.L., and Semenza, G.L. (1995). Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270, 1230-1237. [DOI] [PubMed] [Google Scholar]
- Wang, G.L., Jiang, B.H., Rue, E.A., and Semenza, G.L. (1995). Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92, 5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, C.K., Liptay, S., Wirth, T., Adler, G., and Schmid, R.M. (2000). Suppression of NF-kappaB activity by sulfasalazine is mediated by direct inhibition of IkappaB kinases alpha and beta. Gastroenterology 119, 1209-1218. [DOI] [PubMed] [Google Scholar]
- Yu, F., White, S.B., Zhao, Q., and Lee, F.S. (2001). HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. USA 98, 9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, H., Chiles, K., Feldser, D., Laughner, E., Hanrahan, C., Georgescu, M., Simons, J.W., and Semenza, G.L. (2000). Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase /PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 60, 1541-1545. [PubMed] [Google Scholar]
- Zhu, H., Jackson, T., and Bunn, H.F. (2002). Detecting and responding to hypoxia. Nephrol. Dial. Transplant. 17(suppl 1), 3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]