Abstract
During hepatic wound healing, activation of key effectors of the wounding response known as stellate cells leads to a multitude of pathological processes, including increased production of endothelin-1 (ET-1). This latter process has been linked to enhanced expression of endothelin-converting enzyme-1 (ECE-1, the enzyme that converts precursor ET-1 to the mature peptide) in activated stellate cells. Herein, we demonstrate up-regulation of 56- and 62-kDa ECE-1 3′-untranslated region (UTR) mRNA binding proteins in stellate cells after liver injury and stellate cell activation. Binding of these proteins was localized to a CC-rich region in the proximal ECE-1 3′ UTR base pairs (the 56-kDa protein) and to a region between 60 and 193 base pairs in the ECE-1 3′ UTR mRNA (62 kDa). A functional role for the 3′ UTR mRNA/protein interaction was established in a series of reporter assays. Additionally, transforming growth factor-β1, a cytokine integral to wound healing, stimulated ET-1 production. This effect was due to ECE-1 mRNA stabilization and increased ECE-1 expression in stellate cells, which in turn was a result of de novo synthesis of the identified 56- and 62-kDa ECE-1 3′ UTR mRNA binding proteins. These data indicate that liver injury and the hepatic wound healing response lead to ECE-1 mRNA stabilization in stellate cells via binding of 56- and 62-kDa proteins, which in turn are regulated by transforming growth factor-β. The possibility that the same or similar regulatory events are present in other forms of wound healing is raised.
INTRODUCTION
The response to hepatic wounding is characterized by the activation of hepatic stellate cells (also lipocytes, fat-storing cells), critical effectors of the wound healing process (Ballardini et al., 1988; Friedman and Arthur, 1989; Kent et al., 1976; Maher and McGuire, 1990). During stellate cell activation, these cells exhibit features of myofibroblasts, producing increased quantities of extracellular matrix proteins as well as smooth muscle α-actin. A further characteristic of stellate cell activation is enhanced production of the vasoconstrictive peptide, endothelin-1 (ET-1), which contributes to perpetuation of the fibrogenic process (Gandhi et al., 1998; Rockey and Chung, 1996) as well as in control hepatic vascular tone (Bauer et al., 1994; Kawada et al., 1993; Rockey and Weisiger, 1996).
Each of the three known endothelin isoforms (-1, -2, -3) arise by proteolytic processing of large precursors (∼200 amino acid residues). Intermediates, termed big ET-1, -2, and -3 (38–41 aa) are excised from prepropeptides at sites containing paired basic amino acids. Big endothelins, which have little or no biological activity (Yanagisawa, 1994), are cleaved at Trp-21-Val/Ile-22 to produce mature 21-residue, biologically active peptides. The enzyme responsible for the specific cleavage at Trp-21 has been termed endothelin-converting enzyme (ECE); it is a neutral membrane-bound metalloprotease with Mr = 120 kDa, belonging to the endo-peptidase-24.11 family found in brain (Ohnaka et al., 1993; Turner and Murphy, 1996). The production of ET-1 seems to be regulated at the level of preproET-1 as well as by ECE. The mechanism by which ET-1 overproduction in the injured liver occurs is linked to increased ECE-1 expression and presumed posttranscriptional modulation of ECE-1 mRNA (Shao et al., 1999).
The transforming growth factor-β (TGF-β) superfamily consists of multiple family members, including the highly homologous isoforms of TGF-β (TGF-β1–5) and the related bone morphogenetic proteins activins, and inhibins (Massague et al., 1992). The TGF-βs have a wide variety of biological actions, including in cell growth, differentiation, and fibrogenesis (form review, see Massague et al., 1992). TGF-β1 in particular has emerged as a key component of the fibrogenic response to wounding and is upregulated during many different types of wound healing, including in the liver (Munger et al., 1999; Nakatsukasa et al., 1990; Sanderson et al., 1995) (for review, see (Border and Noble, 1994). In the liver, the importance of TGF-β1 has been emphasized by the demonstration of its direct effects on stellate cells, presumably at the level of extracellular matrix gene transcription (Armendariz-Borunda et al., 1992).
Posttranscriptional processes, including mRNA stabilization, play an important role in gene expression (Beelman and Parker, 1995; Burd and Dreyfuss, 1994; Dreyfuss et al., 1996; Jacobson and Peltz, 1996). Stability of most mRNAs is determined by sequences in their 3′-untranslated regions (3′ UTRs). AU-rich elements (AREs) and poly(C) regions found in the 3′ UTR are among the most common stability determinants identified in mammalian cells (You et al., 1992; Levine et al., 1993; Wang et al., 1995; Holcik and Liebhaber, 1997; Peng et al., 1998; Czyzyk-Krzeska and Bendixen, 1999; Laroia et al., 1999; Paulding and Czyzyk-Krzeska, 1999). Functionally, AREs and poly(C) regions seem to mediate mRNA deadenylation and cleavage of mRNA itself. mRNA stability is generally controlled by the interaction between these specific RNA sequences and RNA binding proteins. Several AU-specific binding proteins and poly(C) binding proteins have been described and characterized Wang et al., 1995; DeMaria and Brewer, 1996; Ma et al., 1997; Fan and Steitz, 1998; Czyzyk-Krzeska and Bendixen, 1999). Interaction of RNA binding proteins and 3′ UTR sequences is thought to lead to configurational changes that prevent mRNA degradation and decrease mRNA turnover. Most RNA binding proteins seem to be cytoplasmic proteins of 30–70 kDa (Hamilton et al., 1993; Zhang et al., 1993 Gueydan et al., 1999; Tillmann-Bogush et al., 1999).
Given previous data demonstrating enhanced expression and stabilization of ECE-1 mRNA in stellate cells during hepatic wound healing, we aimed to identify regulatory regions in the ECE-1 3′ UTR that might mediate this process. In addition, the prominence of ET-1 and TGF-β in wound healing led us to hypothesize that these two factors are linked and to specifically investigate the possibility that TGF-β might regulate production of ET-1 in vivo during the wounding process. We have used a hepatic wound healing model not only to dissect the molecular and cellular regulatory events controlling ET-1 synthesis in vivo during wounding but also molecular mechanisms by which TGF-β controls ET-1 production.
MATERIALS AND METHODS
Liver Injury and Hepatic Fibrosis
Male Sprague Dawley rats (450–500 g; Harlan, Indianapolis, IN) were maintained on standard chow and water ad libitum. Hepatic fibrogenesis was induced by gavage with carbon tetrachloride (1.0 ml/kg mixed with corn oil) on consecutive weeks or by ligation of the common bile duct (Proctor and Chatamra, 1982 Kountouras et al., 1984). Bile duct ligation (BDL) resulted in periportal expansion of biliary duct cells with concomitant periductular and lobular matrix deposition. Repeated administration of carbon tetrachloride led to extensive portal-portal, portal-central, and central-central bridging fibrosis, also typical of severe hepatic wounding. In some experiments, rats undergoing bile duct ligation received a soluble TGF-β receptor (generated by fusion of rabbit type II TGF-β receptor with Fc region of human IgG1; Smith et al., 1999; this construct inhibits biological activity of TGF-β1 and TGF-β3) or matched IgG1 control (each 5 mg/kg intraperitonally) 1 d before bile duct ligation and 4 d later. Animal care and experimental procedures were approved by the Duke University Medical Center Institutional Animal Care and Use Committee as set forth in the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health.
Cell Isolation and Culture
Nonparenchymal cells were isolated as described previously (de Leeuw et al., 1984). Briefly, stellate and endothelial cells were isolated by perfusion in situ with 20 mg/100 ml pronase (Boehringer Mannheim Biochemicals, Indianapolis, IN) and 7.3 mg/100 ml collagenase (Crescent Chemical, Hauppauge, NY). The cell suspension was layered on a discontinuous density gradient of 8.2 and 15.6% accudenz (Accurate Chemical & Scientific, Westbury, NY). The resulting upper layer consists of >95% stellate cells. Endothelial cells were further purified by centrifugal elutriation (18 ml/min flow). Purity was assessed by specific markers for each stellate cells and endothelial cells. Cells were plated in modified medium 199, containing 20% serum (10% horse serum and 10% calf serum; Invitrogen, Carlsbad, CA), 4 mg/100 ml streptomycin, and 0.25 mg/100 ml amphotericin at a density of ∼1 × 106 cells/ml. Cultures were incubated at 37°C in a humidified incubator (containing 95% O2 and 2.5% CO2), and the medium was changed at every 24 h. Cell viability was >80% in all cultures used.
ET-1 Radioimmunoassay
ET-1 in conditioned medium was measured using a radioimmunoassay kit according to manufacturer specifications (Peninsula Laboratories, Palo Alto, CA). In brief, samples were incubated with 100 μl of rabbit anti-ET-1 antibody for 12 h at 4°C. Then 100 μl of 125I-ET-1 (10,000 cpm) was added and incubated for 12 h at 4°C. After incubation with 100 μl of goat anti-rabbit IgG and normal rabbit serum for 90 min at room temperature, the mixture was added to buffer and centrifuged at 3000 rpm for 20 min; 125I-ET-1 in the precipitate was measured (Beckman 5500, Beckman Coulter, Palo Alto, CA). Counts were normalized to total protein content for each sample (Bio-Rad, Hercules, CA). The ET-1 antibody used in the radioimmunoassay kit exhibited <5% cross-reactivity with unlabeled ET-3, <3% cross-reactivity with unlabeled ET-2, and 3% cross-reactivity with big ET-1. Inter- and intraassay variability of the radioimmunoassay system were each <5%.
ECE-1 3′ UTR cDNAs and Oligonucleotide Mutation
Based on the published rat ECE-1 sequence (Shimada et al., 1994), oligodeoxynucleotide primers encoding the proximal 675 base pairs of the ECE-1 UTR (distal to the stop codon) were synthesized (Operon Technologies, Alameda, CA). 5′ and 3′ sequences for sense and antisense primers were 5′-TAAGGGCTGAAGCGCAGA-3′ and 5′-GCCTGATTTATGAGCATG-3′, respectively. Different cDNA fragments encoding specific portions of the ECE-1 3′ UTR (431, 361, 307, 190, 60, or 46 base pairs, respectively) were generated by restriction digestion (Nsi I, DraI, AvaII, Csp45 I, SmaI, or PstI). Oligonucleotides and mutants were synthesized (Operon Technologies) and cloned into pGEM3+. All cDNA fragments were sequenced by the dideoxy chain termination method and found to have 100% homology to published sequences.
Preparation of Cell Cytoplasmic and Nuclear Extracts
Cells were resuspended in lysis buffer containing 10 mM Tris-Cl (pH 7.4), 10 mM MgCl2, 3 mM NaCl, and 0.4% Nonidet P-40 (NP-40). After incubation on ice for 10 min, cells were centrifuged at 120,000 × g for 10 min, and the supernatant containing the cytoplasmic fraction was used to calculate protein concentration. Preparation of nuclear extracts was performed by resuspending cells in phosphate-buffered saline and sonication followed by centrifugation at 1500 × g for 5 min and washing. The resulting cell pellet was resuspended in buffer containing 0.01 M HEPES (pH 7.6), 1.5 mM MgCl2, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg/ml aprotinin, 5 μg/ml leupeptin, 5 μg/ml pepstatin, 1 mM sodium vanadate, and 2 mM KCl. After incubation on ice for 15 min, samples were resuspended in 10% NP-40 and centrifuged at 120,000 × g for 1 min. The pellet was dissolved in high salt buffer containing 2 mM KCl, 0.42 M NaCl, 0.01 M HEPES (pH 7.6), 1.5 mM MgCl2, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 5 μg/ml leupeptin, 5 μg/ml pepstatin, 1 mM sodium vanadate, 25% glycerol, and 0.2 mM EDTA. Finally, after centrifugation at 120,000 × g for 5 min, supernatants were collected and used for study.
RNA Isolation and RNase Protection Assay
Total RNA was isolated from cell lysates (Tri-reagent; Molecular Research, Cincinnati, OH); RNA concentration and purity were determined spectrophotometrically (A260/280), and the integrity of all samples was documented by visualization of 18S and 28S ribosomal bands after electrophoresis through a 1% agarose/formaldehyde gel. ECE-1 (270 base pairs) cDNA was cloned as described previously. Radiolabeled probes were synthesized by transcription of appropriate plasmid cDNA with T7 RNA polymerase in the presence of [α-32P]CTP (Amersham Biosciences, Piscataway, NJ). Total RNA (10 μg) was incubated with 1.0 × 106 cpm of 32P-labeled cRNA, denatured at 78°C for 15 min, and hybridized at 55°C for 12–16 h. Unhybridized RNA was digested and protected hybrids were denatured and separated by electrophoresis through a 5% polyacrylamide/urea sequencing gel. Dried gels were exposed to x-ray film (X-OMat AR-5; Eastman Kodak, Rochester, NY), and scanning densitometry was used to quantitate autoradiographic signals. All RNA samples were also probed with a cDNA encoding 316 base pairs of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Ambion, Austin, TX) to verify the integrity of mRNA in each sample and to control internally for the amount of mRNA present in an individual assay. tRNA and lung RNA were used as a negative and positive control RNAs, respectively.
Mobility Shift Assay and UV Cross-Linking
32P-labeled 3′ UTR RNAs encoding ECE-1 3′ UTR fragments were generated by in vitro transcription as described above. Radiolabeled RNA transcripts (200,000 cpm/reaction) were incubated with cytoplasmic extracts (40 μg) in 15 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.24 mM EDTA, 12 mM HEPES (pH 7.8), 0.5% NP-40, 5% glycerol, and yeast tRNA at 30°C for 30 min. After addition of ribonuclease T1 (100 U) at 30°C for 20 min, heparin (150 μg) was added to the reaction and the reaction was incubated on ice for 10 min. Samples were loaded on a 6% nondenaturing acrylamide gel. For UV cross-linking, after addition of heparin, samples were irradiated with 254-nm UV light for 10 min at a distance of 10 cm (UVP, San Gabriel, CA). Ribonuclease T1 (1 U) was added and samples were resolved by 10% SDS-PAGE. Dried gels were exposed to x-ray film.
Nuclear Run-On Assay
After washing with phosphate-buffered saline, cultured cells were lysed and incubated on ice for 10 min with lysis buffer containing 10 mM Tris-Cl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol, 0.5 mM PMSF, 1 μg/ml leupeptin, and 1 μg/ml aprotonin. The samples were centrifuged for 5 min at 500 × g and the pellet was resuspended (1–2 × 107 nuclear/0.1 ml) in storage buffer containing 20 mM Tris-Cl (pH 8.0), 20% glycerol, 60 mM NaCl, 0.1 mM EDTA, and 1 mM dithiothreitol, and followed by storage in liquid nitrogen. Transcription assays were performed in 200 μl of reaction buffer containing 62.5 mM Tris-Cl (pH 8.0), 10 mM MgCl2, 200 mM KCl, 50 U RNasin, 1 mM CTP, 1 mM GTP, 1 mM ATP, and 0.1 mCi of [α-32P]UTP at room temperature for 1 h. After incubation, the reaction mixture was digested with 10 U of DNase I and followed by 40 μg of proteinase K. The newly synthesized RNA was isolated with 1 ml of Tri-reagent. RNA samples containing 1 × 106 cpm were added to 0.5 ml (final volume) of hybridization buffer containing 50 mM PIPES (pH 6.8), 1 mM EDTA, 0.2% SDS, 2.5× Denhardt's solution, 50 μg of denatured salmon sperm DNA, and 200 μg of tRNA. The samples were then hybridized to ECE-1, GAPDH, and pGEM 3z cDNAs immobilized on nitrocellulose membranes for 3 d at 65°C. Membranes were washed twice for 10 min at room temperature in 0.1% SDS-2× SSC and signals were detected by exposure to X-film (X-OMat AR-5; Eastman Kodak) for 5 d.
Reporter Construct, Transient and Stable Transfection
The proximal 675-base pair fragment of the ECE-1 3′ UTR was cloned into the C-terminal portion of a pcDNA-Luciferase plasmid construct (kindly provided by Kevin Claffey, Center for Vascular Biology, University of Connecticut, Storrs, CT) or the C-terminal portion of an enhanced green fluorescent protein (GFP) vector (Clontech, Palo Alto, CA). Constructs were transfected into stellate cells or NIH 3T3 fibroblasts by using Lipofectin (Invitrogen) following the manufacturer's instructions. Luciferase activity was determined 48 h after transfection as manufacturer's instructions (Promega, Madison, WI). For stable transfection, GFP fluorescent images were analyzed from day 1 to day 12 after cell growth in medium containing serum and G418.
Immunoblot
Cultured cells were homogenized in 20 mM Tris-HCl buffer (pH 7.4) containing 5 mM MgCl2 and 0.1 mM PMSF, 20 μM pepstatin A and 20 μM leupeptin. The homogenate was centrifuged at 1000 × g for 10 min, and the resulting supernatant was centrifuged at 100,000g for 60 min. Protein content in the supernatant was quantitated (Bio-Rad) and equal quantities of protein (30 μg) were separated by SDS-PAGE under reducing conditions and transferred to nitrocellulose (Bio-Rad). Blots were incubated with anti-ECE-1 primary antibody B61/104 (1:400) (kindly provided by Dr. Thomas Sub-kowski, BASF, Ludwigshafen, Germany) and detected by chemiluminescence (ECL kit; Amersham Biosciences).
ECE-1 Enzymatic Activity
ECE-1 enzymatic activity was determined as described previously (Ohnaka et al., 1993). In brief, microsomal membrane fractions (30 μg) were preincubated with 0.1 M sodium phosphate buffer (pH 6.8) containing 0.5 M NaCl and protease inhibitors at 37°C for 15 min before addition of 1 μM big ET-1. The reaction was incubated at 37°C for 2 h in siliconized tubes and terminated by adding 50 μl of 5 mM EDTA. The mixture was then processed to detect immunoreactive ET-1.
Statistics
Data are expressed as mean ± SE and n refers to the numbers of individual experiments performed. Differences among groups were determined using one-way analysis of variance followed by the Newman-Keuls procedure. The 0.05 level of probability was used as the criterion of significance.
RESULTS
ECE-1 3′ UTR RNA–Protein Complexes Are Present in Stellate Cells Only after Liver Injury
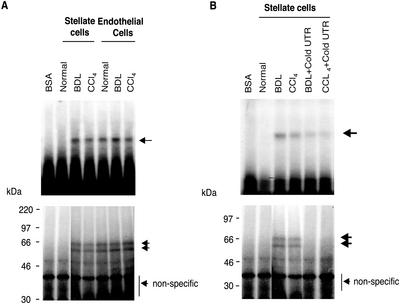
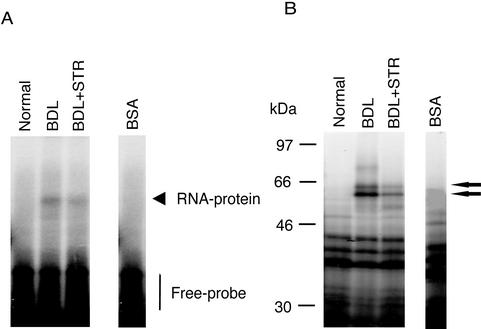
We have previously reported that increased ET-1 production in stellate cells after liver injury results (in part) from increased ECE-1 expression (Shao et al., 1999) and that the mechanism involves enhanced ECE-1 mRNA stability. We therefore postulated that specific ECE-1 3′ UTR binding proteins were responsible for ECE-1 mRNA stabilization. To investigate this possibility, we attempted to detect RNA binding proteins within the proximal 675 base pairs of the ECE-1 3′ UTR. We focused on this fragment because it contains multiple putative binding elements, including three AUUUA repeats, three ACCCCA repeats, and eight CCCU repeats (Shimada et al., 1994). Electromobility shift assays and UV cross-linking experiments, performed using stellate and endothelial cell cytoplasmic extracts from both normal and injured livers, reproducibly revealed an RNA–protein complex in the cytoplasmic fraction from normal and injured populations of endothelial cells, but only in stellate cells after liver injury (either BDL or carbon tetrachloride) (Figure 1). The results of UV cross-linking revealed that RNA binding protein sizes were ∼56 and 62 kDa (Figure 1). We also performed RNA protein binding assays by using nuclear fractions, but failed to identify RNA–protein complexes in either cell type (our unpublished data). The results identify the proximal 675-base pair region of the ECE-1 3′ UTR as potentially important in regulation of ECE-1 mRNA stability in stellate cells after liver injury. This result is highly relevant from a pathophysiological standpoint, because liver injury resulting in activation of stellate cells is typified by increases in ET-1 production (Shao et al., 1999).
Figure 1.
ECE-1 3′ UTR RNA–protein complexes are detected in stellate cells only after liver injury. Stellate and endothelial cells were isolated from normal rats and from those 8 d after bile duct ligation or after eight doses of carbon tetrachloride. Cells were lysed and cytoplasmic extracts were incubated with a 32P-labeled 675-base pair fragment of the ECE-1 3′ UTR as described in MATERIALS AND METHODS. (A) RNA–protein complexes were analyzed by gel mobility assay (top). Bottom, cytoplasmic extracts bound to 32P-labeled probe were exposed to UV irradiation and subjected to 10% SDS-PAGE. Arrows indicate an RNA–protein complex. Bovine serum albumin was used as a negative control. The figure is representative of four different experiments. (B) A 100-fold molar excess of cold specific 3′ UTR was added as competitors. RNA–protein complexes were analyzed by gel mobility assay, shown in the top panel. The bottom panel shows UV cross-linking. Arrows indicate RNA–protein complexes. Bovine serum albumin was used as a negative control. The figure is representative of four different experiments.
We confirmed the specificity of cytoplasmic protein binding to the cloned 675 base pairs of ECE-1 3′ UTR, by adding molar excess nonradiolabeled ECE-1 3′ UTR as a specific competitor (Figure 1B). RNA protein binding activity was significantly inhibited by addition of nonradiolabeled 3′ UTR; and additionally, binding by 56- and 62-kDa proteins was eliminated.
Identification of 56- and 62-kDa Protein Binding Regions
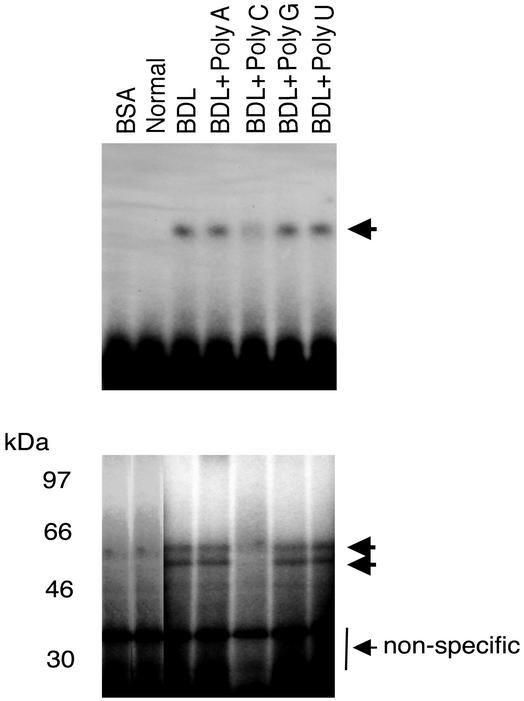
Because AREs and poly(C) regions mediate 3′ UTR stabilization, we aimed to identify whether these regions play a similar role in ECE-1 3′ UTR protein binding. RNA protein binding activity was effectively abolished by excess poly(C) nucleotide but not with excess poly (A), (G), or (U) nucleotides (Figure 2). These data suggest that the two putative RNA binding proteins (∼56 and 62 kDa) bind to C-rich regions in the 675-base pair ECE-1 3′ UTR.
Figure 2.
RNA protein binding is dependent on CC-rich sequences. Stellate cells were isolated from normal rats and 8 d after bile duct ligation. Cellular cytoplasmic extracts were incubated with a 32P-labeled 675-base pair fragment of 3′ UTR as described in MATERIALS AND METHODS. Poly(A), (C), (G), or (U) (200 ng) was added to extracts and RNA–protein complexes were analyzed by gel mobility assay (top). The bottom panel reveals UV cross-linking. Arrows indicate RNA–protein complexes. Bovine serum albumin was used as a negative control. The figure is representative of three different experiments.
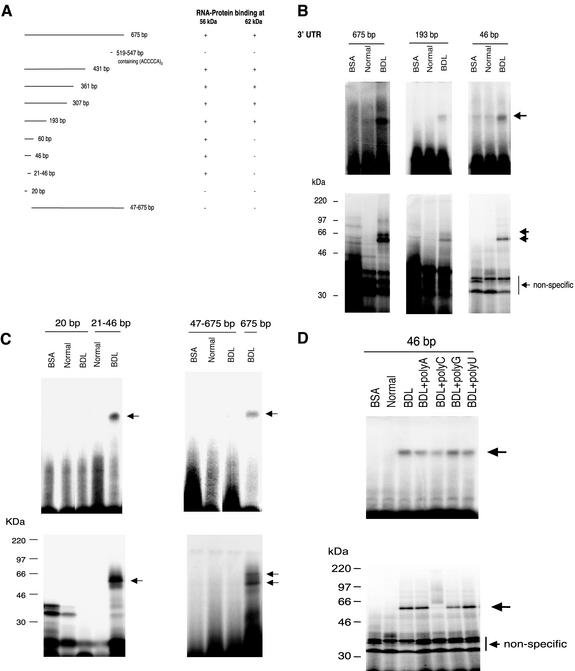
To identify specific regions responsible for protein binding within the proximal ECE-1 3′ UTR 675 base pairs, we isolated multiple 3′ UTR fragments (Figure 3A) and examined their ability to mediate protein binding. In electromobility shift assays and UV cross-linking experiments, progressively truncated fragments containing more than the first 193 base pairs distal to the stop codon possessed both 56- and 62-kDa protein binding activity. Importantly, the sequence from 519–547 base pairs (containing three ACCCCA) repeats did not possess binding activity for either protein. The proximal 46- and 60-base pair sequences retained 56-kDa protein binding activity, but did not exhibit 62-kDa binding activity (Figure 3, A and B). A fragment containing bp 21–46 retained binding activity, but the proximal 20-base pair fragment did not (Figure 3C). Interestingly, we were unable to detect protein binding by using the 47- to 675-base pair construct (or with other constructs that lacked the proximal fragment; our unpublished data), despite multiple attempts. Protein binding in the proximal 46-base pair area was inhibited by addition of excess poly(C) (Figure 3D), consistent with poly(C) competitive inhibition of protein binding in the full 675-base pair 3′ UTR fragment. The data suggest that the 56-kDa protein binds to one or more poly(C) regions in the 21- to 46-base pair segment of the ECE-1 3′ UTR and that the 62-kDa protein interaction is with one or more poly(C) regions between 60 and 193 base pairs. Furthermore, the data suggest that the binding of the 62-kDa protein is dependent on binding of the 56-kDa protein.
Figure 3.
RNA binding proteins bind different ECE-1 3′ UTR sequences. (A) Multiple 3′ UTR fragments were cloned as described in MATERIALS AND METHODS. Protein binding activity was determined by gel mobility assay and UV cross-linking as in MATERIALS AND METHODS and is reported diagrammatically. (B and C) Gel mobility assay (top panels) and UV cross-linking experiments (bottom panels) examining specific areas of the UTR are shown. Cellular cytoplasmic extracts were incubated with the indicated 32P-labeled fragments as described in MATERIALS AND METHODS. Arrows indicate RNA–protein complexes. Bovine serum albumin was used as a negative control. The figures are each representative of four different experiments. (D) Cellular cytoplasmic extracts were incubated with a 32P-labeled 46-base pair fragment of ECE-1 3′ UTR as described in MATERIALS AND METHODS. Poly(A), (C), (G), or (U) (200 ng) was added and RNA–protein complexes were analyzed by gel mobility assay (top). The lower panel shows UV cross-linking. Arrows indicate RNA–protein complex. Bovine serum albumin was used as a negative control. The figure is representative of six different experiments.
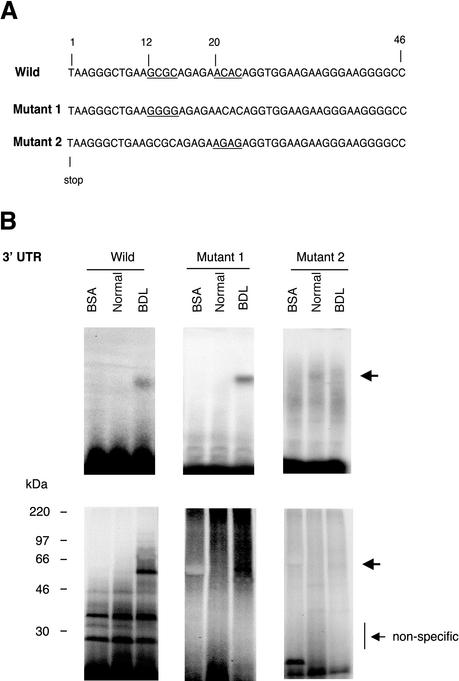
We next mutated the poly(C) region in the proximal ECE-1 3′ UTR 46-base pair fragment (Figure 4A). Examination of the ECE-1 3′ UTR revealed two sequences containing poly(C) sequences: GCGC and ACAC at 12 and 21 base pairs from the stop codon, respectively (Figure 4A). Substitution of the GCGC with GGGG (mutant 1) had no effect on protein binding activity, whereas substitution of the ACAC with AGAG (mutant 2) resulted in loss of RNA protein binding of the 56-kDa species (Figure 4B). These data, which are highly consistent with those shown in Figure 3C, indicate that the 56-kDa binding protein interacts with the ACAC poly(C) region in the proximal 46-base pair fragment.
Figure 4.
Identification of a specific binding region within the proximal 46-base pair ECE-1 UTR. (A) Wild-type 3′ UTR (46-base pair fragment) is shown at the top. The underlined areas were mutated (12–15 and 21–24 base pairs). Mutant 1 (Mut 1) was of GCGC (12–15 base pairs) to GGGG and mutant 2 (Mut 2) was of ACAC (21–24 base pairs) to AGAG. (B) Cellular cytoplasmic extracts were incubated with 32P-labeled 46 base pairs of wild-type, mutant 1, or mutant 2 of ECE-1 3′ UTR as described in MATERIALS AND METHODS. RNA–protein complexes were analyzed by gel mobility assay (top) and UV cross-linking (bottom). Arrows indicate RNA–protein complexes. Bovine serum albumin was used as a negative control. The figure is representative of three different experiments.
A Functional Role for ECE-1 3′ UTR Binding Proteins
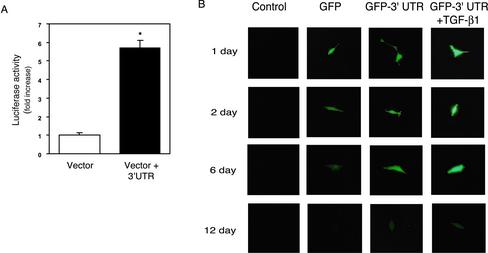
To evaluate a potential functional role for the ECE-1 3′ UTR as a stabilizing element, we introduced the 3′ UTR sequence into reporter constructs in stellate cells as well as in NIH 3T3 fibroblasts. We additionally chose to examine NIH 3T3 fibroblasts because of their great transduction efficiency and because in preliminary experiments, we verified that NIH 3T3 fibroblasts contain abundant quantities of each 56-and 62-kDa UTR binding proteins (our unpublished data). Cells were transiently transfected with a chimeric luciferase-ECE-1 3′ UTR construct in which the full-length 675-base pair 3′ UTR was cloned downstream of a cytomegalovirus-Luciferase construct. As shown in Figure 5A, luciferase activity in stellate cells transfected with the vector containing the 3′ UTR was significantly higher than that in cells transfected with vector alone. Identical results were found in NIH 3T3 cells transfected in a similar manner (our unpublished data). To further demonstrate a functional role for the 3′ UTR, we exposed stably transfected NIH 3T3 fibroblasts containing a chimeric ECE-1 3′ UTR-GFP construct to TGF-β1 (which we have shown to mediate ECE-1 mRNA stability; see below) (Figure 5B). GFP degradation in control cells began 2 d after transduction and continued throughout the course of the experiment. In contrast, GFP expression in cells containing the GFP-3′ UTR construct was substantially prolonged. Furthermore, exposure of cells containing the GFP-3′ UTR construct to TGF-β1 resulted in more intense and longer GFP expression than those not exposed to TGF-β1. Further controls consisted of constructs lacking putative protein binding regions (i.e., the 47- to 675-base pair, 60- to 193-base pair, and 193- to 675-base pair constructs); these revealed luciferase activity similar to the vector control (our unpublished data). The data indicate that the ECE-1 3′ UTR plays an important role in mRNA stabilization and moreover suggests that TGF-β1 could play a direct role in regulation of proteins that bind to the ECE-1 3′ UTR.
Figure 5.
A functional role for the ECE-1 3′ UTR. A cytomegalovirus-enhancer-driven luciferase (Luc)/GFP-3 UTR reporter construct containing the proximal 675 base pairs of ECE-1 UTR was introduced and transiently or stably transfected as in MATERIALS AND METHODS. (A) Luciferase activity of transiently transfected stellate cells (transfection for 24 h) is shown. Luciferase activity was normalized to total cellular protein (n = 6). (B) GFP degradation of stably transfected NIH 3T3 cells for up to 12 d is shown. The brightness of fluorescent images was captured at the indicated time points. Images in the first column show control cells without transfection; images in the second column show GFP plasmid transfection; images in the third column show GFP-3′ UTR plasmid transfection; and images in the fourth column show GFP-3′ UTR plasmid transfection in the presence of TGF-β1 (10 ng/ml). The data are representative of two separate experiments.
Effect of TGF-β on ET-1 Production in Liver Wound Healing
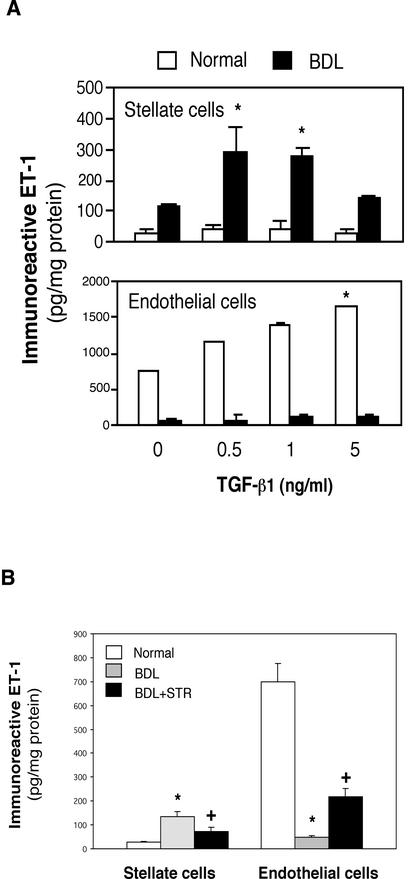
Potential sources of ET-1 in the liver include each sinusoidal endothelial and stellate cells (Shao et al., 1999), the latter, a key effector in the wounding response to liver injury. Furthermore, of factors that seem to be important in the fibrogenic response to injury, TGF-β1 is prominent. Therefore, we investigated the effect of TGF-β1 on ET-1 production in hepatic endothelial and stellate cells. TGF-β1 had no effect on ET-1 release in quiescent (i.e., those isolated from normal liver) stellate cells but caused significant increases in the release of ET-1 in activated (i.e., those from livers after injury induced by BDL) stellate cells (Figure 6A). Interestingly, higher TGF-β1 concentrations did not stimulate stellate cell release of ET-1. The effect of TGF-β1 on ET-1 release in normal and activated stellate cells contrasted sharply with that in endothelial cells in which TGF-β1 induced an increase in ET-1 release in normal endothelial cells but not in injured endothelial cells (Figure 6A). Similar results were elicited with each sinusoidal endothelial and stellate cells isolated in other models of injury and wounding (i.e., eight consecutive doses carbon tetrachloride injury; our unpublished data).
Figure 6.
Effect of TGF-β1 on ET-1 production in stellate and endothelial cells in vitro and in vivo. (A) Stellate and sinusoidal endothelial cells were isolated from normal rat livers or those injured by BDL as in MATERIALS AND METHODS. After adherence for 48 h, serum-free medium along with TGF-β1 was applied to isolated cells at the indicated concentrations was introduced for 24 h. ET-1 in conditioned supernatants was measured by radioimmunoassay as in MATERIALS AND METHODS and normalized to the total protein content in the monolayer. *p < 0.05 compared with medium without TGF-β1 (n = 5). Note that for stellate cells, ET-1 production by cells after BDL was significantly greater than in normal cells at all TGF-β concentrations. (B) Stellate and endothelial cells were isolated from normal rat livers, those after BDL, or after exposure to an STR during the induction of liver injury by BDL as in MATERIALS AND METHODS. After isolation, cells were allowed to adhere for 48 h, serum-free conditions were introduced for 24 h, and ET-1 in conditioned medium was measured by radioimmunoassay. *p < 0.05 compared with normal and +p < 0.05 compared with BDL (n = 8).
To further investigate whether ET-1 production in vivo could be mediated by TGF-β1, we induced liver injury in rats (by bile duct ligation for 8 d) and simultaneously inhibited TGF-β binding (TGF-β1 and -β3) with a soluble TGF-β receptor (STR) as described in MATERIALS AND METHODS. ET-1 production in freshly isolated endothelial cells from normal liver was 26-fold higher than that from normal stellate cells (Figure 6B). However, after liver injury induced by BDL, ET-1 production in freshly isolated stellate cells increased markedly, whereas it was significantly decreased in endothelial cells. Inhibition of TGF-β in vivo in rats with liver injury led to a significant decrease in ET-1 production in stellate cells (Figure 6B). We also investigated the effect of TGF-β1 on ET-1 production in vivo at earlier time points, including 4 d after liver injury and likewise found that suppression of the effect of TGF-β with the soluble TGF-β receptor inhibited ET-1 production in stellate cells (n = 4; our unpublished data). Importantly, exposure of rats to isotype matched (i.e., to the soluble TGF-β receptor) control IgG had no effect on ET-1 production.
TGF-β Controls ET-1 Production during Wounding by Regulation of ECE-1
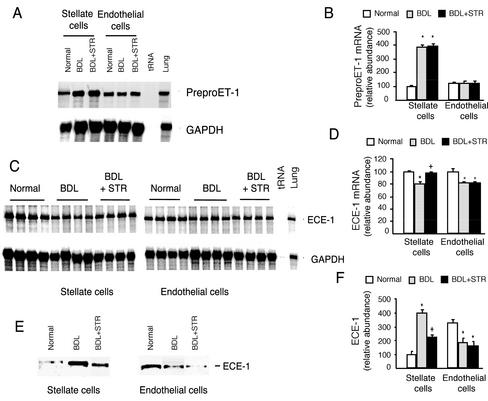
To explore the mechanism by which TGF-β modulates ET-1 production in liver injury, we examined both precursor ET-1 and ECE-1. Liver injury led to a fourfold increase in preproET-1 mRNA expression in stellate cells but had no significant effect on endothelial cell expression of preproET-1 mRNA (Figure 7, A and B). Inhibition of TGF-β with the soluble TGF-β receptor (Figure 7, A and B) had no effect on stellate or sinusoidal endothelial cell expression of preproET-1 mRNA. These data suggest that TGF-β does not play a direct role in regulation of precursor preproET-1 mRNA in stellate and endothelial cells after liver injury, but raise the possibility that other factors such as extracellular matrix, ET-1 itself and/or other cytokines lead to increases in preproET-1 mRNA in activated stellate cells.
Figure 7.
Effect of TGF-β1 on precursor ET-1 and ECE-1 in vivo during hepatic wound healing. Stellate and endothelial cells were isolated from three groups of rats as described in Figure 6. (A) Total cellular RNA was isolated and preproET-1 mRNA was detected by RNase protection assay as in MATERIALS AND METHODS. (B) Specific preproET-1 bands were normalized to the signal for GAPDH; the level of preproET-1 mRNA from normal stellate cells was set at 100. (C) Cellular ECE-1 mRNA was detected by RNase protection assay. (D) Specific ECE-1 bands were normalized to the signal for GAPDH, and ECE-1 mRNA from normal stellate cells was arbitrarily set at 100. (E) ECE-1 from the same cells was detected by immunoblot as in MATERIALS AND METHODS. (F) Specific bands were quantitated, the level of ECE-1 from normal stellate cells was arbitrarily set at 100, and data are presented graphically. *p < 0.05 compared with normal and +p < 0.05 compared with BDL (n = 4).
Because TGF-β had prominent effects on stellate (or endothelial) cell production of ET-1, but did not seem to affect precursor preproET-1 mRNA, we examined the effect of TGF-β1 on ECE-1. After liver injury, ECE-1 mRNA levels were reduced by 20% in stellate (and endothelial) cells (Figure 7, C and D). However, inhibition of TGF-β binding with soluble TGF-β receptor resulted in normalization of ECE-1 mRNA in stellate cells but not in endothelial cells. At the protein level, after liver injury, ECE-1 levels were increased by fourfold in stellate cells but decreased by 48% in endothelial cells (Figure 7, E and F). Furthermore, inhibition of TGF-β in vivo during liver injury led to a significant reduction in stellate cell ECE-1 (Figure 7, E and F), whereas it had little effect on endothelial cell ECE-1 expression. Finally, we found there was no change in ECE-1 enzymatic catalytic activity (conversion of big ET-1 to ET-1) in stellate cells after BDL-induced liver injury or after exposure of injured liver to the soluble TGF-β receptor (our unpublished data). These data suggest that TGF-β regulates ECE-1 protein expression (rather than preproET-1 levels or ECE-1 enzymatic activity), which in turn contributes to the observed production of ET-1 in each stellate and endothelial cells after liver injury (Figure 6).
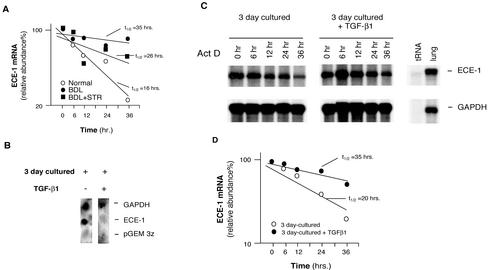
Given that ECE-1 mRNA became reduced while ECE-1 and protein expression was increased (Figure 7, C and D, and E and F), we hypothesized that ECE-1 mRNA must be stabilized after its synthesis, and moreover, that TGF-β plays a role in this process. Therefore, we examined posttranscriptional regulation of ECE-1 in vivo by inhibiting TGF-β signaling with the soluble TGF-β receptor. When TGF-β was inhibited in vivo, we found that stellate cell ECE-1 mRNA stability was shifted toward normal (Figure 8A). ECE-1 mRNA decay in quiescent stellate cells was more rapid than that in activated cells (half-life t1/2 16 vs. 35 h), and moreover, inhibition of TGF-β reduced ECE-1 mRNA t1/2, consistent with a role for TGF-β1 in ECE-1 mRNA stabilization. To further evaluate the transcriptional and/or posttranscriptional effect of TGF-β on ECE-1 mRNA, we performed nuclear run-on assays in isolated stellate cells (we used cells at 3 d in culture to mimic the early activation process). As shown in Figure 8B, TGF-β1 significantly inhibited, rather than stimulated, ECE-1 mRNA transcription. To further explore the role of TGF-β1 in control of ECE-1 mRNA stabilization, we performed experiments in which we exposed isolated stellate cells (again early in the activation process) to TGF-β1 and measured ECE-1 mRNA stability; ECE-1 mRNA stabilization was more pronounced (t1/2 = 35 h) in stellate cells stimulated with TGF-β1 than that (t1/2 = 20 h) of control cells (Figure 8, C and D).
Figure 8.
Transcriptional and posttranscriptional effects of TGF-β1 in stellate cell ECE-1 mRNA during liver injury and hepatic wound healing. (A) Stellate cells from normal livers, those from injured livers (BDL for 8 d), or those from injured livers exposed to the soluble TGF-β receptor were isolated and allowed to adhere to culture vessels for 48 h. Actinomycin D (10 μg/ml) was added to the medium and total cellular RNA was isolated at the indicated times. ECE-1 mRNA was detected by RNase protection assay; specific ECE-1 bands were normalized to the signal for GAPDH, and the data used to determine ECE-1 mRNA half-life t1/2 by interpolation as described previously (Ross, 1995) (n = 3). (B) Stellate cells were isolated from normal rats and were allowed to initiate their activation cascade (by culture in the presence of serum containing medium for 3 d, a model that mimics in vivo activation of stellate cells). Culture medium with or without of TGF-β1 (5 ng/ml) was introduced for 24 h and nuclei were isolated. Nuclear transcription assay was performed as described in MATERIALS AND METHODS. (C) Isolated stellate cells (as in B) were used; culture medium with or without TGF-β1 (5 ng/ml) was introduced for 24 h and then actinomycin D (10 μg/ml) was added. Total cellular RNA was isolated at the indicated times. RNA (10 μg/sample) was hybridized with 32P-labeled cRNAs (ECE-1 and GAPDH) as described in MATERIALS AND METHODS and an RNase protection assay is shown. (D) Specific ECE-1 bands were scanned, quantitated, and normalized to the signal for GAPDH, and the data are presented graphically.
Because TGF-β seemed to stabilize ECE-1 mRNA in vivo, we explored the possibility that this was a result of stimulation of putative ECE-1 mRNA 3′ UTR binding proteins by TGF-β. Thus, we examined RNA–protein complexes in stellate cell lysates (as in Figure 1) and the effect of TGF-β inhibition with the soluble TGF-β receptor. After induction of liver injury by bile duct ligation, RNA–protein complexes were identified in stellate cell lysates (as in Figure 1) and inhibition of TGF-β1 abrogated this protein expression (Figure 9). These data demonstrate that in vivo, TGF-β modulates expression of ECE-1 UTR binding proteins and suggest that the mechanism by which TGF-β modulates expression of ECE-1 is via modulation of the expression of these binding proteins.
Figure 9.
TGF-β–mediated induction of 56- and 62-kDa ECE-1 UTR binding proteins during wounding. Stellate cells were isolated from normal rats and those after BDL as in Figure 2. (A) Cell cytosol was isolated and incubated with 32P-labeled ECE-1 3′ UTR RNA as in MATERIALS AND METHODS, and RNA–protein complexes were resolved on a 6% native acrylamide gel. (B) After incubation of cytosol with 32P-labeled RNA, the RNA–protein samples were exposed to UV irradiation and then resolved by SDS-PAGE. Bovine serum albumin is used as a negative control for each set of experiments. Arrows indicate positions of RNA binding proteins. The data shown are representative of four separate experiments.
Mechanisms Underlying TGF-β–induced ECE-1 Stabilization and ET-1 Production in Stellate Cells
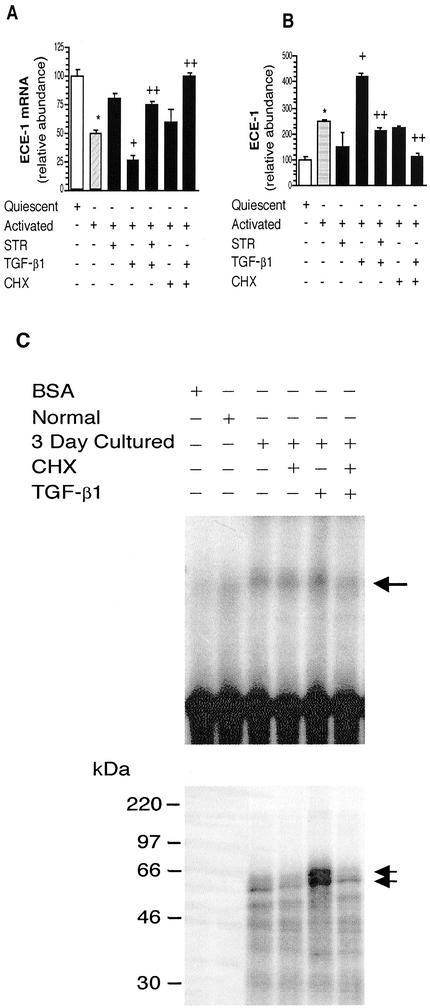
To further explore the mechanisms by which TGF-β1 exerts its effects on stellate cell ECE-1 expression, we used a culture based in which stellate cells are isolated from normal livers and allowed to undergo spontaneous activation (induced by serum and plastic substratum). Cells from normal livers grown in very early (24–48 h) culture mimic the normal quiescent phenotype, whereas those after more prolonged culture mimic the injured activated phenotype (Friedman, 1993). This model was chosen specifically to allow study of the requirement for de novo protein synthesis. TGF-β1 had no effects on preproET-1 mRNA or ECE-1 mRNA expression in quiescent stellate cells, consistent with data presented in Figures 6 and 7 (our unpublished data). In contrast, analogous to the situation after in vivo activation (Figure 7, C and D), exogenous TGF-β1 led to a reduction in ECE-1 expression, and as predicted, inhibition of TGF-β with the soluble TGF-β receptor inhibited this effect (Figure 10, A and B). Furthermore, exposure of activated cells to the soluble TGF-β receptor reduced ECE-1 levels to those found in quiescent stellate cells, a finding consistent with known endogenous production of TGF-β1 in activated stellate cells (Bissell et al., 1995). Also analogous to the situation after in vivo activation, ECE-1 protein levels in culture-activated stellate cells were elevated (Figure 10B). Furthermore, TGF-β1 stimulated ECE-1 protein expression, whereas the soluble TGF-β receptor inhibited this effect (Figure 10B). Finally, inhibition of protein synthesis with cycloheximide abolished the effects of TGF-β1 on ECE-1 mRNA (Figure 10A) as well as protein expression (Figure 10B), suggesting that the effect of TGF-β1 on ECE-1 in activated stellate cells is dependent on new protein synthesis.
Figure 10.
TGF-β–stimulated ET-1 production after wounding requires protein synthesis. A culture model that mimics in vivo activation of stellate cells was used in this set of experiments. (A) Quiescent (i.e., cells from normal livers, cultured for 2 d) or activated (i.e., cells from normal livers but cultured for 7 d in the presence of serum) were exposed to serum-free medium containing TGF-β1 (5 ng/ml), STR (50 μg/ml), CHX (63 ng/ml), or the combinations identified for 24 h. Cellular ECE-1 mRNA was quantitated by RNase protection assay as in MATERIALS AND METHODS, specific ECE-1 bands were normalized to the signal for GAPDH; the level for ECE-1 mRNA from quiescent stellate cells was arbitrarily set at 100. (B) ECE-1 was detected by immunoblot as in methods after exposure of cells to the conditions described in A, specific bands were quantitated, and the level of ECE-1 from quiescent cells was arbitrarily set at 100. *p < 0.05 compared with quiescent cells and +p < 0.05 compared with activated cells, and ++p < 0.05 compared with activated cells with TGF-β1 (n = 4). (C) Cell culture medium containing TGF-β1 (10 ng/ml), CHX (63 ng/ml), or a combination was introduced (for 24 h). Cytoplasmic extracts were incubated with 32P-labeled 675-base pair ECE-1 3′ UTR. RNA–protein complexes were analyzed by gel mobility assay in the top panel. The bottom panel showed UV cross-linking. Arrows indicate RNA–protein complex. Bovine serum albumin was used as a negative control. The figure is representative of three different experiments.
We further evaluated TGF-β1's ability to modulate expression of ECE-1 3′ UTR binding proteins. In isolated stellate cells, normal cells did not express ECE-1 3′ UTR binding proteins; however, binding proteins were expressed after activation. Notably, TGF-β1 significantly increased expression of both 56- and 62-kDa complexes (Figure 10C) and the formation of RNA–protein complexes was inhibited by addition of cycloheximide (CHX), an inhibitor of protein synthesis. These data suggest that TGF-β leads to increased ECE-1 mRNA stability through new synthesis of RNA binding proteins.
DISCUSSION
We have highlighted posttranscriptional modification of ECE-1 mRNA in stellate cells during hepatic wound healing, an event that is due to up-regulation of ECE-1 3′ UTR binding proteins. This finding is consistent with data emphasizing that regulation of mRNA turnover serves as an important mechanism for controlling the fate of cytoplasmic mRNA and consequently, gene and protein expression (Ross, 1995; McGary et al., 1997; Zehner et al., 1997). Among the most common elements found in mRNAs that mediate protein binding are AREs and poly(C) regions found in the 3′ UTRs of many mRNAs (Wang et al., 1995; Holcik and Liebhaber, 1997; Xu et al., 1997; Czyzyk-Krzeska and Bendixen, 1999; Paulding and Czyzyk-Krzeska, 1999; Tillmann-Bogush et al., 1999); indeed, in the system described in this study, poly(C) regions found in the ECE-1 3′ UTR are the target of mRNA binding proteins.
Our findings, in the context of other recent work in stellate cells, emphasizes the concept that wound healing as a general biological phenomenon, may alter gene and protein expression via unique regulatory processes. For example, stellate cell type collagen I mRNA is stabilized after wound healing and contributes to enhanced type I collagen mRNA and protein expression (Stefanovic et al., 1997, 1999). In these experiments, increased mRNA stability was a result of αCP binding to poly(C)-rich areas of the type I collagen 3′ UTR mRNA (Focht and Adams, 1984; Herget et al., 1989; Stefanovic et al., 1997, 1999). Thus, our work has extended the concept that hepatic wound healing is associated with changes in mRNA stability by identifying TGF-β, a critical cytokine involved in diverse forms of wounding (Border and Noble, 1994; Sanderson et al., 1995), as a regulator of 3′ UTR binding proteins that contribute to mRNA stability during this process.
An important finding of this work was that poly(C) elements, rather than AREs, mediated protein binding to the ECE-1 3′ UTR. AREs, in particular, those containing AUUUA pentamers, have been studied extensively (Levine et al., 1993; Chen and Shyu, 1994; Chen et al., 1995; Chung et al., 1996; Fan and Steitz, 1998; Peng et al., 1998). For example, a number of binding proteins (such as HuC, HuD, HuR, and Hel-N1 expressed in differentiated neurons) have been shown to interact with ARE of the c-fos 3′ UTR mRNA and to stabilize c-fos mRNA; likewise, a variety of other AU-rich binding proteins have been identified in different cell types (Zhang et al., 1993; DeMaria and Brewer, 1996; Ma et al., 1997; Laroia et al., 1999). It is notable that the proximal portion of the ECE-1 3′ UTR mRNA includes a classic ARE, containing three AUUUA pentamers. However, we were unable to demonstrate binding of stabilizing proteins to this area, suggesting that ECE-1 mRNA stabilization in this system is ARE independent. Because no other AREs (i.e., other than those identified in the proximal sequence examined in our study) are found in the ECE-1 3′ UTR mRNA, it is unlikely that AREs are involved in ECE mRNA stabilization.
The differential effects of TGF-β on the central cellular sources of ET-1, endothelial and stellate cells, in the hepatic wound healing model were notable. In normal liver endothelial cells, TGF-β1 stimulated ET-1 production in a “linear” dose-response manner, yet had little effect on endothelial cells from injured liver (Figure 6A). This finding suggests that TGF-β could be an important regulator of ET-1 in the normal liver. Interestingly, TGF-β1 did not have an effect on stellate cells from normal livers, but rather had prominent effects on stellate cells from injured livers. Interestingly, stellate cell synthesis of ET-1 was stimulated in a nonlinear manner. The mechanism underlying this phenomenon is not understood at present. However, because TGF-β1 acts locally, it may not be necessary for very high concentrations of TGF-β1 to be present in situ. Not only was ET-1 production by sinusoidal endothelial cells markedly decreased after liver injury but also it did not seem to be affected TGF-β1. The decrease in ET-1 synthesis in sinusoidal endothelial cells was presumably due to decreased ECE-1 production (Figure 7E); further evidence supporting the important role of ECE-1 was that neither liver injury nor TGF-β1 had significant effects on the precursor ET-1 synthetic pathway in sinusoidal endothelial cells (Figure 7A).
The mechanism for the differential effect of TGF-β in endothelial and stellate cells from normal and injured livers is unknown. We have considered the possibility that changes in receptor (i.e., the TGF-β type II receptor complex) density or affinity for TGF-β1 on stellate or endothelial cells account for the differences. For example, increased binding of TGF-β1 has been documented during stellate cell activation in vivo and in culture (Friedman et al., 1994). This could explain increased responsiveness of injured stellate cells to TGF-β1, but does not explain lack of a response to normal cells, because these cells clearly possess TGF-β receptor. Alternatively, signal transduction pathways may be altered after injury. Current evidence indicates that TGF-β signals through each the p38 mitogen-activated protein kinase or SMAD pathways (Derynck et al., 1998; Datto et al., 1999; Hanafusa et al., 1999). In preliminary studies (Shao and Rockey, unpublished observation), divergent TGF-β signaling pathways after hepatic injury and wound healing have been identified in each stellate and sinusoidal endothelial cells, making it attractive to speculate that divergent signaling pathways account for the differences in responses by sinusoidal endothelial and stellate cells.
The importance of TGF-β was further assessed in vivo by using a soluble TGF-β receptor that binds to TGF-β and inhibits its function. Although these experiments (Figures 6B and 7) clearly verified the important regulatory effect of TGF-β on stellate cells in vivo during liver injury, they further emphasized the complexity of the regulatory pathways. For example, we found that precursor preproET-1 mRNA was stimulated in injured stellate cells, but that this effect was not linked to TGF-β (Figure 7, A and B). However, consistent with an important role for TGF-β in regulation of ECE-1 expression, inhibition of TGF-β reduced ECE-1 levels. Notably, TGF-β does not seem to be the only factor involved, because inhibition of TGF-β only partially abrogated the up-regulation of ECE-1 after injury in stellate cells (Figure 7, E and F). In contrast to the effect of TGF-β on stellate cells, its effect on sinusoidal endothelial cells was less prominent. Indeed, the soluble TGF-β receptor had little effect on either sinusoidal endothelial cell precursor preproET-1 or ECE-1, suggesting that factors other than TGF-β are important inhibitors of ECE-1 expression in injured sinusoidal endothelial cells (Figure 7F). In aggregate, these data emphasize the importance of TGF-β in the regulation of stellate cell ET-1 synthesis in the injured liver, and likewise support the contention that other putative factors in the wounding system also affect both stellate and endothelial cell ET-1 synthesis.
Our data suggest that factors other than TGF-β are likely to be important in regulation of ET-1 in the injured liver. For example, the wounding milieu is complex and the wounding response is regulated by interplay among the various systems. Indeed, other cytokines and the extracellular matrix are prominent in many forms of wounding and are likely to be important regulators of ET-1 in this environment. Nonetheless, TGF-β has been reported to lead to increased production of ET-1 in other systems (Kurihara et al., 1989; Kanse et al., 1991; Schnermann et al., 1996,Eakes and Olson, 1998), consistent with its effect in liver wounding. In these reports and in systems in which other factors stimulate ET-1 synthesis, the effect has been largely via modulation of precursor ET-1 (i.e., preproET-1 mRNA expression), primarily at the level of preproET-1 mRNA transcription. To our knowledge, a system in which TGF-β plays a major role in regulation of ET-1 via effects on ECE-1 has not described.
The current data suggest that ET-1 synthesis may be compartmentalized (i.e., in endothelial and stellate cells). Classic ET biology assigns ET-1 production to endothelial cells; endothelial derived ET-1 in turn has important paracrine effects on smooth muscle cells (Yanagisawa et al., 1988; Goto and Warner, 1995). Furthermore, ET-1 is thought to act in a paracrine, or autocrine manner, physically its source of synthesis. In the current report, we have used an in vivo model to explore cell systems important in ET production in the intact organ and have demonstrated an important deviation from the canonical synthetic paradigm. In this model, ET-1 production shifted from the sinusoidal endothelial cell in the normal liver to the hepatic stellate cell after injury (Figure 6A). Although relative ET-1 production by stellate and endothelial cells seemed to be similar after injury (Figure 6A), from a quantitative standpoint, the bulk of ET-1 produced in the healing wound is likely to be stellate cell derived. Striking proliferation of stellate cells after liver injury results in a dramatic increase in the total number of stellate cells (Geerts et al., 1991); therefore, the overall increase in production of ET-1 in the injured liver (Pinzani et al., 1996) is likely to be a result of stellate rather than endothelial cell synthesis and release of ET-1. Furthermore, localization of ET-1 in the injured liver further seems to assign its production to stellate cells (Pinzani et al., 1996). Inhibition of soluble TGF-β inhibited total hepatic ET-1 production, is highly consistent with this conclusion. The transition of ET-1 production from the endothelial cell to a nonendothelial compartment after injury is consistent with emerging data in other forms of wound healing in which nonendothelial elements such as fibroblasts and smooth muscle cells can produce endothelins (Sung et al., 1994). This “switch” in the cellular source of ET-1, from endothelial cells in normal tissue to mesenchyme-derived cells after injury seems to be characteristic in particular of pathological situations.
We found that new protein synthesis is required for the up-regulation of ECE-1 by TGF-β. The requirement for new protein synthesis and the induction of 56- and 62-kDa RNA binding proteins (Figure 10, A–C) by TGF-β strongly supports the contention that TGF-β helps control the ECE-1 synthetic pathway after liver injury (and perhaps in other forms of injury in which ET-1 is stimulated). Further investigation in which the 56- and 62-kDa RNA binding proteins are identified and the mechanism by which TGF-β controls their synthesis will therefore be required.
The findings reported herein have important therapeutic implications for wound healing. Most studies involving TGF-β and wound healing have presumed that TGF-β is directly responsible for reduced fibrogenesis in effector cells such as stellate cells. Although TGF-β clearly has direct effects on stellate cell fibrogenesis in cell culture systems, its effect is unlikely to be this simple in vivo; indeed, we have clearly shown important indirect effects of TGF-β on the endothelin system. Moreover, because enhanced production of ET-1 is common to many forms of wound healing, including pulmonary fibrosis, heart failure, and renal fibrosis (Forbes et al., 1996; Kakugawa et al., 1996; Karam et al., 1996; Kohan, 1997; Park et al., 1997; Cho et al., 2000), all of which are also characterized by elevated production of TGF-β, we speculate that in vivo, TGF-β modulates the wound healing response additionally by regulating the production of ET-1. Given the emerging importance that ET-1 itself plays in the wound healing response (i.e., as a stimulator of each cellular proliferation, fibrogenesis, and wound contraction), our data emphasize a novel relationship between TGF-β and ET-1 in wound healing.
Acknowledgments
This work was presented in part at the 50th Annual Meeting of the American Association for the Study of Liver Diseases (Dallas, Texas, 1999) and was supported by National Institutes of Health grants R01 DK50574 and R01 DK57830 and the Burroughs Wellcome Fund (to D.C.R.).
Abbreviations used: BDL, bile duct ligation; CHX, cycloheximide; ECE-1, endothelin-converting enzyme-1; ET, endothelin; STR, soluble transforming growth factor-β receptor; TGF-β1, transforming growth factor-β1; 3′ UTR, 3′ untranslated region.
References
- Armendariz-Borunda, J., Katayama, K., and Seyer, J.M. (1992). Transcriptional mechanisms of type I collagen gene expression are differentially regulated by interleukin-1β, tumor necrosis factor α, and transforming growth factor β in Ito cells. J. Biol. Chem. 267, 14316-14321. [PubMed] [Google Scholar]
- Ballardini, G., Fallani, M., Biagini, G., Bianchi, F.B., and Pisi, E. (1988). Desmin and actin in the identification of Ito cells and in monitoring their evolution to myofibroblasts in experimental liver fibrosis. Virchows Arch. B Cell Pathol. 56, 45-49. [DOI] [PubMed] [Google Scholar]
- Bauer, M., Zhang, J.X., Bauer, I., and Clemens, M.G. (1994). Endothelin-1 as a regulator of hepatic microcirculation: sublobular distribution of effects and impact on hepatocellular secretory function. Shock 1, 457-465. [PubMed] [Google Scholar]
- Beelman, C.A., and Parker, R. (1995). Degradation of mRNA in eukaryotes. Cell 81, 179-183. [DOI] [PubMed] [Google Scholar]
- Bissell, D.M., Wang, S.S., Jarnagin, W.R., and Roll, F.J. (1995). Cell-specific expression of transforming growth factor-β in rat liver. Evidence for autocrine regulation of hepatocyte proliferation. J. Clin. Investig. 96, 447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Border, W.A., and Noble, N.A. (1994). Transforming growth factor β in tissue fibrosis. N. Engl. J. Med. 331, 1286-1292. [DOI] [PubMed] [Google Scholar]
- Burd, C.G., and Dreyfuss, G. (1994). Conserved structures and diversity of functions of RNA-binding proteins. Science 265, 615-621. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., and Shyu, A.B. (1994). Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol. Cell. Biol. 14, 8471-8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.Y., Xu, N., and Shyu, A.B. (1995). mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 15, 5777-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, J.J., Hocher, B., Herbst, H., Jia, J., Ruehl, M., Hahn, E.G., Riecken, E.O., and Schuppan, D. (2000). An oral endothelin A receptor antagonist blocks collagen synthesis and deposition in advanced rat liver fibrosis. Gastroenterology 118, 1169-1178. [DOI] [PubMed] [Google Scholar]
- Chung, S., Jiang, L., Cheng, S., and Furneaux, H. (1996). Purification and properties of HuD, a neuronal RNA-binding protein. J. Biol. Chem. 271, 11518-11524. [DOI] [PubMed] [Google Scholar]
- Czyzyk-Krzeska, M.F., and Bendixen, A.C. (1999). Identification of the poly(C) binding protein in the complex associated with the 3′ untranslated region of erythropoietin messenger RNA. Blood 93, 2111-2120. [PubMed] [Google Scholar]
- Datto, M.B., Frederick, J.P., Pan, L., Borton, A.J., Zhuang, Y., and Wang, X.F. (1999). Targeted disruption of Smad3 reveals an essential role in transforming growth factor β-mediated signal transduction. Mol. Cell. Biol. 19, 2495-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw, A.M., McCarthy, S.P., Geerts, A., and Knook, D.L. (1984). Purified rat liver fat-storing cells in culture divide and contain collagen. Hepatology 4, 392-403. [DOI] [PubMed] [Google Scholar]
- DeMaria, C.T., and Brewer, G. (1996). AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 271, 12179-12184. [DOI] [PubMed] [Google Scholar]
- Derynck, R., Zhang, Y., and Feng, X.H. (1998). Smads: transcriptional activators of TGF-β responses. Cell 95, 737-740. [DOI] [PubMed] [Google Scholar]
- Dreyfuss, G., Hentze, M., and Lamond, A.I. (1996). From transcript to protein. Cell 85, 963-972. [DOI] [PubMed] [Google Scholar]
- Eakes, A.T., and Olson, M.S. (1998). Regulation of endothelin synthesis in hepatic endothelial cells. Am. J. Physiol. 274, G1068-G1076. [DOI] [PubMed] [Google Scholar]
- Fan, X.C., and Steitz, J.A. (1998). Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17, 3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focht, R.J., and Adams, S.L. (1984). Tissue specificity of type I collagen gene expression is determined at both transcriptional and post-transcriptional levels. Mol. Cell. Biol. 4, 1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes, R.D., Cernacek, P., Zheng, S., Gomersall, M., and Guttmann, R.D. (1996). Increased endothelin expression in a rat cardiac allograft model of chronic vascular rejection. Transplantation 61, 791-797. [DOI] [PubMed] [Google Scholar]
- Friedman, S.L. (1993). Seminars in medicine of the Beth Israel Hospital, Boston. The cellular basis of hepatic fibrosis. Mechanisms and treatment strategies. N. Engl. J. Med. 328, 1828-1835. [DOI] [PubMed] [Google Scholar]
- Friedman, S.L., and Arthur, M.J. (1989). Activation of cultured rat hepatic lipocytes by Kupffer cell conditioned medium. Direct enhancement of matrix synthesis and stimulation of cell proliferation via induction of platelet-derived growth factor receptors. J. Clin. Investig. 84, 1780-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, S.L., Yamasaki, G., and Wong, L. (1994). Modulation of transforming growth factor β receptors of rat lipocytes during the hepatic wound healing response. Enhanced binding and reduced gene expression accompany cellular activation in culture and in vivo. J. Biol. Chem. 269, 10551-10558. [PubMed] [Google Scholar]
- Gandhi, C.R., Nemoto, E.M., Watkins, S.C., and Subbotin, V.M. (1998). An endothelin receptor antagonist TAK-044 ameliorates carbon tetrachloride-induced acute liver injury and portal hypertension in rats. Liver 18, 39-48. [DOI] [PubMed] [Google Scholar]
- Geerts, A., Lazou, J.M., De Bleser, P., and Wisse, E. (1991). Tissue distribution, quantitation and proliferation kinetics of fat-storing cells in carbon tetrachloride-injured rat liver. Hepatology 13, 1193-1202. [PubMed] [Google Scholar]
- Gueydan, C., Droogmans, L., Chalon, P., Huez, G., Caput, D., and Kruys, V. (1999). Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem. 274, 2322-2326. [DOI] [PubMed] [Google Scholar]
- Goto, K., and Warner, T.D. (1995). Molecular pharmacology. Endothelin versatility. Nature 375, 539-540. [DOI] [PubMed] [Google Scholar]
- Hamilton, B.J., Nagy, E., Malter, J.S., Arrick, B.A., and Rigby, W.F. (1993). Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J. Biol. Chem. 268, 8881-8887. [PubMed] [Google Scholar]
- Hanafusa, H., Ninomiya-Tsuji, J., Masayama, N., Nishita, M., Fujisawa, J., Shibuya, H., Matsumoto, K., and Nishida, E. (1999). Involvement of the p38 mitogen-activated protein kinase pathway in transforming growth factor beat induced gene expression. J. Biol. Chem. 274, 27161-27167. [DOI] [PubMed] [Google Scholar]
- Herget, T., Burba, M., Schmoll, M., Zimmermann, K., and Starzinski-Powitz, A. (1989). Regulated expression of nuclear protein(s) in myogenic cells that binds to a conserved 3′ untranslated region in pro α1(I) collagen cDNA. Mol. Cell. Biol. 9, 2828-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik, M., and Liebhaber, S.A. (1997). Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis and trans components. Proc. Natl. Acad. Sci. USA 94, 2410-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, A., and Peltz, S.W. (1996). Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 65, 693-739. [DOI] [PubMed] [Google Scholar]
- Kakugawa, Y., Paraskevas, S., Metrakos, P., Giaid, A., Qi, S.J., Duguid, W.P., and Rosenberg, L. (1996). Alterations in pancreatic microcirculation and expression of endothelin-1 in a model of chronic pancreatitis. Pancreas 13, 89-95. [DOI] [PubMed] [Google Scholar]
- Kanse, S.M., Takahashi, K., Lam, H.C., Rees, A., Warren, J.B., Porta, M., Molinatti, P., Ghatei, M., and Bloom, S.R. (1991). Cytokine stimulated endothelin release from endothelial cells. Life Sci. 48, 1379-1384. [DOI] [PubMed] [Google Scholar]
- Karam, H., Heudes, D., Bruneval, P., Gonzales, M.F., Loffler, B.M., Clozel, M., and Clozel, J.P. (1996). Endothelin antagonism in end-organ damage of spontaneously hypertensive rats. Comparison with angiotensin-converting enzyme inhibition and calcium antagonism. Hypertension 28, 379-385. [DOI] [PubMed] [Google Scholar]
- Kawada, N., Tran-Thi, T.A., Klein, H., and Decker, K. (1993). The contraction of hepatic stellate (Ito) cells stimulated with vasoactive substances. Possible involvement of endothelin 1 and nitric oxide in the regulation of the sinusoidal tonus. Eur. J. Biochem. 213, 815-823. [DOI] [PubMed] [Google Scholar]
- Kent, G., Gay, S., Inouye, T., Bahu, R., Minick, O.T., and Popper, H. (1976). Vitamin A-containing lipocytes and formation of type III collagen in liver injury. Proc. Natl. Acad. Sci. USA 73, 3719-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohan, D.E. (1997). Endothelins in the normal and diseased kidney. Am. J. Kidney Dis. 29, 2-26. [DOI] [PubMed] [Google Scholar]
- Kountouras, J., Billing, B.H., and Scheuer, P.J. (1984). Prolonged bile duct obstruction: a new experimental model for cirrhosis in the rat. Br. J. Exp. Pathol. 65, 305-311. [PMC free article] [PubMed] [Google Scholar]
- Kurihara, H., Yoshizumi, M., Sugiyama, T., Takaku, F., Yanagisawa, M., Masaki, T., Hamaoki, M., Kato, H., and Yazaki, Y. (1989). Transforming growth factor-β stimulates the expression of endothelin mRNA by vascular endothelial cells. Biochem. Biophys. Res. Commun. 159, 1435-1440. [DOI] [PubMed] [Google Scholar]
- Laroia, G., Cuesta, R., Brewer, G., and Schneider, R.J. (1999). Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284, 499-502. [DOI] [PubMed] [Google Scholar]
- Levine, T.D., Gao, F., King, P.H., Andrews, L.G., and Keene, J.D. (1993). Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol. Cell. Biol. 13, 3494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W.J., Chung, S., and Furneaux, H. (1997). The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 25, 3564-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher, J.J., and McGuire, R.F. (1990). Extracellular matrix gene expression increases preferentially in rat lipocytes and sinusoidal endothelial cells during hepatic fibrosis in vivo. J. Clin. Investig. 86, 1641-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague, J., Cheifetz, S., Laiho, M., Ralph, D.A., Weis, F.M., and Zentella, A. (1992). Transforming growth factor-β. Cancer Surv. 12, 81-103. [PubMed] [Google Scholar]
- McGary, E.C., Rondon, I.J., and Beckman, B.S. (1997). Post-transcriptional regulation of erythropoietin mRNA stability by erythropoietin mRNA-binding protein. J. Biol. Chem. 272, 8628-8634. [DOI] [PubMed] [Google Scholar]
- Munger, J.S., et al. (1999). The integrin αvβ6 binds and activates latent TGFβ1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell 96, 319-328. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa, H., Nagy, P., Evarts, R.P., Hsia, C.C., Marsden, E., and Thorgeirsson, S.S. (1990). Cellular distribution of transforming growth factor-β1 and procollagen types I, III, and IV transcripts in carbon tetrachloride-induced rat liver fibrosis. J. Clin. Investig. 85, 1833-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnaka, K., Takayanagi, R., Nishikawa, M., Haji, M., and Nawata, H. (1993). Purification and characterization of a phosphoramidon-sensitive endothelin-converting enzyme in porcine aortic endothelium. J. Biol. Chem. 268, 26759-26766. [PubMed] [Google Scholar]
- Park, S.H., Saleh, D., Giaid, A., and Michel, R.P. (1997). Increased endothelin-1 in bleomycin-induced pulmonary fibrosis and the effect of an endothelin receptor antagonist. Am. J. Respir. Crit. Care Med. 156, 600-608. [DOI] [PubMed] [Google Scholar]
- Paulding, W.R., and Czyzyk-Krzeska, M.F. (1999). Regulation of tyrosine hydroxylase mRNA stability by protein-binding, pyrimidine-rish sequence in the 3′-untranslated region. J. Biol. Chem. 274, 2532-2538. [DOI] [PubMed] [Google Scholar]
- Peng, S.S., Chen, C.Y., Xu, N., and Shyu, A.B. (1998). RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17, 3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzani, M., Milani, S., De Franco, R., Grappone, C., Caligiuri, A., Gentilini, A., Tosti-Guerra, C., Maggi, M., Failli, P., Ruocco, C., and Gentilini, P. (1996). Endothelin 1 is overexpressed in human cirrhotic liver and exerts multiple effects on activated hepatic stellate cells. Gastroenterology 110, 534-548. [DOI] [PubMed] [Google Scholar]
- Proctor, E., and Chatamra, K. (1982). High yield micronodular cirrhosis in the rat. Gastroenterology 83, 1183-1190. [PubMed] [Google Scholar]
- Rockey, D.C., and Chung, J.J. (1996). Endothelin antagonism in experimental hepatic fibrosis. Implications for endothelin in the pathogenesis of wound healing. J. Clin. Investig. 98, 1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey, D.C., and Weisiger, R.A. (1996). Endothelin induced contractility of stellate cells from normal and cirrhotic rat liver: implications for regulation of portal pressure and resistance. Hepatology 24, 233-240. [DOI] [PubMed] [Google Scholar]
- Ross, J. (1995). mRNA stability in mammalian cells. Microbiol. Rev. 59, 423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson, N., Factor, V., Nagy, P., Kopp, J., Kondaiah, P., Wakefield, L., Roberts, A.B., Sporn, M.B., and Thorgeirsson, S.S. (1995). Hepatic expression of mature transforming growth factor β1 in transgenic mice results in multiple tissue lesions. Proc. Natl. Acad. Sci. USA 92, 2572-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnermann, J.B., Zhu, X.L., Shu, X., Yang, T., Huang, Y.G., Kretzler, M., and Briggs, J.P. (1996). Regulation of endothelin production and secretion in cultured collecting duct cells by endogenous transforming growth factor-β. Endocrinology 137, 5000-5008. [DOI] [PubMed] [Google Scholar]
- Shao, R., Yan, W., and Rockey, D.C. (1999). Regulation of endothelin-1 synthesis by endothelin-converting enzyme-1 during wound healing. J. Biol. Chem. 274, 3228-3234. [DOI] [PubMed] [Google Scholar]
- Shimada, K., Takahashi, M., and Tanzawa, K. (1994). Cloning and functional expression of endothelin-converting enzyme from rat endothelial cells. J. Biol. Chem. 269, 18275-18278. [PubMed] [Google Scholar]
- Smith, J.D., Bryant, S.R., Couper, L.L., Vary, C.P., Gotwals, P.J., Koteliansky, V.E., and Lindner, V. (1999). Soluble transforming growth factor-β type II receptor inhibits negative remodeling, fibroblast transdifferentiation, and intimal lesion formation but not endothelial growth. Circ. Res. 84, 1212-1222. [DOI] [PubMed] [Google Scholar]
- Stefanovic, B., Hellerbrand, C., and Brenner, D.A. (1999). Regulatory role of the conserved stem-loop structure at the 5′ end of collagen α1(I) mRNA. Mol. Cell. Biol. 19, 4334-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic, B., Hellerbrand, C., Holcik, M., Briendl, M., Aliebhaber, S., and Brenner, D.A. (1997). Posttranscriptional regulation of collagen α1(I) mRNA in hepatic stellate cells. Mol. Cell. Biol. 17, 5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, C.P., Arleth, A.J., Storer, B.L., and Ohlstein, E.H. (1994). Angiotensin type 1 receptors mediate smooth muscle proliferation and endothelin biosynthesis in rat vascular smooth muscle. J. Pharmacol. Exp. Ther. 271, 429-437. [PubMed] [Google Scholar]
- Tillmann-Bogush, M., Heaton, J.H., and Gelehrter, T.D. (1999). Cyclic nucleotide regulation of PAI-1 mRNA stability. Identification of cytosolic proteins that interact with an a-rich sequence. J. Biol. Chem. 274, 1172-1179. [DOI] [PubMed] [Google Scholar]
- Turner, A.J., and Murphy, L.J. (1996). Molecular pharmacology of endothelin converting enzymes. Biochem. Pharmacol. 51, 91-102. [DOI] [PubMed] [Google Scholar]
- Wang, X., Kiledjian, M., Weiss, I.M., and Liebhaber, S.A. (1995). Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human α-globin mRNA stability [published erratum in Mol. Cell. Biol. (1995) 15, 2331]. Mol. Cell. Biol. 15, 1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, N., Chen, C.Y., and Shyu, A.B. (1997). Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell. Biol. 17, 4611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa, M. (1994). The endothelin system. A new target for therapeutic intervention. Circulation 89, 1320-1322. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, M., Kurihara, H., Kimura, S., Tomobe, Y., Kobayashi, M., Mitsui, Y., Yazaki, Y., Goto, K., and Masaki, T. (1988). A novel potent vasoconstrictor peptide produced by vascular endothelial cells [see comments]. Nature 332, 411-415. [DOI] [PubMed] [Google Scholar]
- You, Y., Chen, C.Y., and Shyu, A.B. (1992). U-rich sequence-binding proteins (URBPs) interacting with a 20-nucleotide U-rich sequence in the 3′ untranslated region of c-fos mRNA may be involved in the first step of c-fos mRNA degradation. Mol. Cell. Biol. 12, 2931-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehner, Z.E., Shepherd, R.K., Gabryszuk, J., Fu, T.F., Al-Ali, M., and Holmes, W.M. (1997). RNA-protein interactions within the 3 'untranslated region of vimentin mRNA. Nucleic Acids Res. 25, 3362-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Wagner, B.J., Ehrenman, K., Schaefer, A.W., DeMaria, C.T., Crater, D., DeHaven, K., Long, L., and Brewer, G. (1993). Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 13, 7652-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]