Abstract
In Saccharomyces cerevisiae, mitochondrial fusion requires at least two outer membrane proteins, Fzo1p and Ugo1p. We provide direct evidence that the dynamin-related Mgm1 protein is also required for mitochondrial fusion. Like fzo1 and ugo1 mutants, cells disrupted for the MGM1 gene contain numerous mitochondrial fragments instead of the few long, tubular organelles seen in wild-type cells. Fragmentation of mitochondria in mgm1 mutants is rescued by disrupting DNM1, a gene required for mitochondrial division. In zygotes formed by mating mgm1 mutants, mitochondria do not fuse and mix their contents. Introducing mutations in the GTPase domain of Mgm1p completely block mitochondrial fusion. Furthermore, we show that mgm1 mutants fail to fuse both their mitochondrial outer and inner membranes. Electron microscopy demonstrates that although mgm1 mutants display aberrant mitochondrial inner membrane cristae, mgm1 dnm1 double mutants restore normal inner membrane structures. However, mgm1 dnm1 mutants remain defective in mitochondrial fusion, indicating that mitochondrial fusion requires Mgm1p regardless of the morphology of mitochondria. Finally, we find that Mgm1p, Fzo1p, and Ugo1p physically interact in the mitochondrial outer membrane. Our results raise the possibility that Mgm1p regulates fusion of the mitochondrial outer membrane through its interactions with Fzo1p and Ugo1p.
INTRODUCTION
Mitochondrial fusion and division play important roles in controlling the specialized shape, number, and distribution of mitochondria in many cell types (Tyler, 1992; Bereiter-Hahn and Voth, 1994). In the yeast S. cerevisiae, mitochondrial fusion and division are highly regulated during growth, mating, and sporulation and control the structure and function of this essential organelle (Hermann and Shaw, 1998; Yaffe, 1999; Jensen et al., 2000; Shaw and Nunnari, 2002).
Mitochondrial fusion in yeast requires the Fzo1 protein (Hermann et al., 1998; Rapaport et al., 1998; Sesaki and Jensen, 1999) and the Ugo1 protein (Sesaki and Jensen, 2001). Fzo1p is a homolog of Drosophila fuzzy onions protein, which is required for mitochondrial fusion during fly spermato-genesis (Hales and Fuller, 1997). Fzo1p is located in the mitochondrial outer membrane (OM) with an N-terminal GTPase domain facing the cytosol (Hermann et al., 1998; Rapaport et al., 1998). Ugo1p is also an OM protein with a single transmembrane domain (Sesaki and Jensen, 2001). Cells disrupted for FZO1 or UGO1 contain many small mitochondrial fragments instead of the few tubular mitochondria seen in wild-type cells (Hermann et al., 1998; Rapaport et al., 1998; Sesaki and Jensen, 2001). Defects in mitochondrial fusion in these mutants have been directly demonstrated using a mating assay. In addition to fusion, Fzo1p and Ugo1p are also important for mitochondrial maintenance of mitochondrial DNA (mtDNA). fzo1 and ugo1 mutants lose mtDNA, but the mechanism by which mtDNA is lost in these mutants is not understood and appears to be a secondary consequence of mitochondrial morphology defect (Bleazard et al., 1999; Sesaki and Jensen, 1999)
The fragmentation of mitochondria in fzo1 and ugo1 mutants depends on mitochondrial division (Bleazard et al., 1999; Sesaki and Jensen, 1999, 2001). Mitochondrial division is mediated by Dnm1p, a dynamin-related GTPase (Gammie et al., 1995; Bleazard et al., 1999; Sesaki and Jensen, 1999). Cells disrupted for DNM1 contain a single mitochondrion consisting of a network of interconnected tubules. fzo1 dnm1 and ugo1 dnm1 double mutants contain nearly wild-type, tubular-shaped mitochondria, suggesting that mitochondrial shape and number are normally regulated by a balance between division and fusion (Sesaki and Jensen, 1999, 2001). Like mitochondrial fragmentation, the loss of mtDNA in fzo1 and ugo1 mutants requires Dnm1p, and fzo1 dnm1, and ugo1 dnm1 double mutants maintain mtDNA (Bleazard et al., 1999; Sesaki and Jensen, 1999, 2001).
Taking advantage of the mtDNA phenotype of fzo1 mutants, we developed a genetic screen for mutants defective in mitochondrial fusion, which yielded ugo1 mutants (Sesaki and Jensen, 2001). Using this screen, we also identified five mgm1 mutants, leading us to investigate the role of Mgm1p in mitochondrial fusion. mgm1 was originally identified as a mutant that was unable to maintain mtDNA (Jones and Fangman, 1992). Later studies show that mgm1 mutants also lose normal mitochondrial morphology and contain mitochondrial fragments, which often aggregate into clusters within the yeast cells (Guan et al., 1993; Shepard and Yaffe, 1999). These clumped mitochondria are not efficiently inherited from mother to daughter cells (Shepard and Yaffe, 1999). Mgm1p contains a predicted GTP-binding motif that is homologous to the GTPase dynamin (Jones and Fangman, 1992), and this GTPase domain is essential for the function of Mgm1p (Shepard and Yaffe, 1999). There are two species of Mgm1p, a 90- and a 100-kDa form, but the relationship between these two forms is unknown (Shepard and Yaffe, 1999). Mgm1p has been localized to mitochondria, but its submitochondrial location is controversial. Although Shepard and Yaffe (1999) localized Mgm1p to the OM, Wong et al. (2000) found Mgm1p in the intermembrane space, peripherally associating with the inner membrane (IM). Therefore, the actual location of Mgm1p remains unclear.
Because the phenotypes of mgm1 mutants are similar to those of mutants defective in mitochondrial fusion, such as fzo1 (Hermann et al., 1998; Rapaport et al., 1998) and ugo1 (Sesaki and Jensen, 2001), the ability to fuse mitochondria was examined in mgm1 and mgm1 dnm1 mutants (Wong et al., 2000). Supporting Mgm1p's involvement in mitochondrial fusion, temperature-sensitive mgm1-5 mutants were unable to fuse mitochondria during yeast mating. However, mgm1-5 dnm1 double mutants fused their mitochondria at the restrictive temperature (Wong et al., 2000). Therefore, it is not clear whether Mgm1p is involved in mitochondrial fusion, or in mitochondrial shape. In this article, using complete disruptions of the MGM1 open reading frame, we show that both mgm1Δ and mgm1Δ dnm1Δ cells are defective in mitochondrial fusion, demonstrating that Mgm1p is essential for mitochondrial fusion. We further show that the Mgm1p is required for both OM and IM fusion. Consistent with its essential role in OM fusion, the majority of Mgm1p is associated with the OM and physically interacts with Fzo1p and Ugo1p.
MATERIALS AND METHODS
Strains, Media, and Genetic Methods
Yeast strains used in this study are listed in Table 1. Yeast cells were grown in media including YEPD (YEP medium containing 2% glucose), YEPGE (YEP medium containing 2% glycerol and 2% ethanol), YEPRaf (YEP medium containing 2% raffinose), SRaf (synthetic medium containing 2% raffinose), SRafSuc (synthetic medium containing 2% raffinose and 2% sucrose), and SGalSuc (synthetic medium containing 2% galactose and 2% sucrose). Standard genetic techniques were used (Adams et al., 1997).
Table 1.
Yeast Strains
| Strain | Genotype (plasmid) | Source |
|---|---|---|
| FY833 | MATahis3 leu2 lys2 trp1 ura3 | Winston et al. (1995) |
| FY844 | MATα his3 leu2 lys2 trp1 ura3 | Winston et al. (1995) |
| YRJ1278 | MATa his3 leu2 trp1 ura3 fzo1::kanMX4 rho0 | Sesaki and Jensen (2000) |
| YRJ1282 | MATahis3 leu2 lys2 trp1 ura3 fzo1::kanMX4 (pTRP1-His6-HA-FZO1) | This study |
| YRJ1287 | MATahis3 leu2 lys2 trp1 ura3 rho0 | Sesaki and Jensen (2000) |
| YRJ1289 | MATa his3 leu2 lys2 trp1 ura3 dnm1::kanMX4 | Sesaki and Jensen (2000) |
| YRJ1290 | MATα his3 leu2 lys2 trp1 ura3 dnm1::kanMX4 | Sesaki and Jensen (2000) |
| YRJ1291 | MATahis3 leu2 lys2 trp1 ura3 ugo1::HIS3 dnm1::kanMX4 | Sesaki and Jensen (2000) |
| YRJ1292 | MATα his3 leu2 lys2 trp1 ura3 ugo1::HIS3 dnm1::kanMX4 | Sesaki and Jensen (2000) |
| YRJ1294 | MATahis3 leu2 lys2 trp1 ura3 ugo1::HIS3 (pTRP1-myc-UGO1) | Sesaki and Jensen (2000) |
| YRJ1347 | MATa his3 leu2 trp1 ura3 fzo1::kanMX4 dnm1::kanMX4 | This study |
| YRJ1348 | MATα his3 leu2 trp1 ura3 fzo1::kanMX4 dnm1::kanMX4 | This study |
| YRJ1355 | MATα his3 leu2 lys2 trp1 ura3 mmm1::HIS3 | This study |
| YRJ1356 | MATahis3 leu2 lys2 trp1 ura3 mmm1::HIS3 | This study |
| YRJ1383 | MATahis3 leu2 lys2 trp1 ura3 mgm1::kanMX4 rho0 | This study |
| YRJ1384 | MATα his3 leu2 lys2 trp1 ura3 mgm1::kanMX4 rho0 | This study |
| YRJ1398 | MATa his3 leu2 trp1 ura3 mgm1::kanMX4 dnm1::kanMX4 | This study |
| YRJ1493 | MATα his3 leu2 trp1 ura3 mgm1::kanMX4 dnm1::kanMX4 | This study |
| YRJ1554 | MATa ade2 his3 leu2 trp1 ura3 mgm1::HIS3 dnm1::kanMX4 | This study |
| YRJ1555 | MATα ade2 his3 leu2 trp1 ura3 mgm1::HIS3 dnm1::kanMX4 | This study |
| YRJ1556 | MATahis3 leu2 lys2 trp1 ura3 dnm1::kanMX4 mgm1::MGM1-TRP1-kanMX4 | This study |
| YRJ1557 | MATα his3 leu2 lys2 trp1 ura3 dnm1::kanMX4 mgm1::MGM1-TRP1-kanMX4 | This study |
| YRJ1558 | MATahis3 leu2 lys2 trp1 ura3 dnm1::kanMX4 mgm1::mgm1K223A-TRP1-kanMX4 | This study |
| YRJ1559 | MATα his3 leu2 lys2 trp1 ura3 dnm1::kanMX4 mgm1::mgm1K223A-TRP1-kanMX4 | This study |
| YRJ1560 | MATahis3 leu2 lys2 trp1 ura3 dnm1::kanMX4 mgm1::mgm1S224N-TRP1-kanMX4 | This study |
| YRJ1561 | MATα his3 leu2 lys2 trp1 ura3 dnm1::kanMX4 mgm1::mgm1S224N-TRP1-kanMX4 | This study |
Plasmids
pSS1, a CEN-TRP1 plasmid expressing OM45-GFP was constructed as follows. pKC2, a CEN-LEU2 plasmid carrying OM45-GFP (Cerveny et al., 2001) was digested with PvuI. The OM45-GFP fragment and XhoI/NotI-digested pRS314 (Sikorski and Hieter, 1989) were transformed into yeast and pSS1 was formed by homologous recombination (Oldenburg et al., 1997).
pHS70, a CEN-URA3 plasmid expressing CFP fused to the C termini of Yta10p under the control of the GAL1 promoter was constructed as follows. YTA10 was PCR amplified from yeast genomic DNA (Hoffman and Winston, 1987) using oligos 720 (5′-GGGCTCGAGATGATGATGTGGCAACG-3′) and 721 (5′-GGGTCTAGAATTTGTTGCTGCAGGTG-3′). The PCR fragment was digested with XhoI and XbaI and subcloned into XhoI/XbaI-digested pGAL1-COX4-CFP (HS52; Sesaki and Jensen, 2001).
pHS71, a CEN-URA3 plasmid expressing YFP fused to the C termini of Yta10p under the control of the GAL1 promoter was constructed as follows. YFP was PCR amplified from pEYFP (Clontech Laboratories, Inc.) by using oligos 355 (5′-TGCTCTAGAATGGTGAGCA AGGGC-3′) and 498 (5′-CCGGATCCTTACTTGTACAGCTCGTC-3′). The PCR fragment was digested with XbaI and BamHI and cloned into XbaI/BamHI-digested pHS70.
pHS72, a CEN-URA3 plasmid expressing YFP fused to the first 40 amino acids of Tom72p under the control of the GAL1 promoter was constructed as follows. TOM72 was PCR amplified from yeast genomic DNA using oligos 549 (5′-GGGCTCGAGATGGCCGAAAA CTCCCTC-3′) and 548 (5′-GAAGCGGCCGCCCTGCTGCTTAATTTGCTG-3′). The PCR fragment was digested with XhoI and NotI and cloned into XhoI/NotI-digested pRS316GU (Nigro et al., 1992), forming pHS72–1. YFP was PCR amplified from pEYFP (Clontech Laboratories, Inc., Palo Alto, CA) by using oligos 347 (5′-AAGGAAAAAAGCGGCCGCATGGTGAGCA AGGGC-3′) and 498. The PCR fragment was digested with NotI and BamHI and cloned into NotI/BamHI-digested pHS72–1, forming pHS72.
pHS77, a CEN-TRP1 plasmid expressing His10-HA-Fzo1p was constructed as follows. The promoter of FZO1 was PCR amplified from yeast genomic DNA using oligos 904 (5′-GGGCTCGAGTGTTCCACTTTCTTGCG-3′) and 905 (5′-GGGGAATTCCGTTAAATGAGCCTACC-3′). The PCR fragment was digested with XhoI and EcoRI and cloned into XhoI/EcoRI-digested pRS314 (Sikorski and Hieter, 1989), forming pRS314-Fp. The HA epitope was PCR amplified from pGTEP (Tyers et al., 1992) using oligos OSM373 (5′-TTGAATTCATGCACCACCACCACCACCACCACCACCACCCTACCCATACGATGTTCCTG-3′) and 906 (5′-GGGGCGGCCGCGAGCAGCGTAATCTGGAACG-3). The PCR fragment was digested by EcoRI and NotI, and cloned into EcoRI/NotI-digested pRS314-Fp, forming pRS314-Fp- HA. The open reading frame of FZO1 was PCR amplified from yeast genomic DNA using oligos 907 (5′-GGGGCGGCCGCTCTGAAGGAAAACAACAATTC-3′) and 908 (5′-GGGCCGCGGAAAATGGACCTGCTTGG-3′). The PCR fragment was digested with NotI and SacII, and cloned into NotI/SacII-digested pRS314-Fp-HA, forming pHS77.
pHS73, a TRP1 plasmid expressing Mgm1p, was constructed as follows. MGM1 was PCR amplified from yeast genomic DNA using oligos 758 (5′-GGGCTCGAGGTGTCAGTAAATAACAGAG-3′) and 711 (5′-AATGCGGCCGCCTTAGATGAAGGGTATG-3′). The PCR fragment was digested with XhoI and NotI and cloned into XhoI/NotI-digested pRS304 (Sikorski and Hieter, 1989).
To introduce mutations in the GTPase domain of MGM on pHS73, site-directed mutagenesis (QuickChange, Stratagene, La Jolla, CA) was performed using the following mutagenic oligonucleotides 804 (5′-GGTTCACAATCGTCTGGTGCATCCTCAGTACTAGAATCC-3′) and 805 (5′-GGATTCTAGTACTGAGGATGCACCAGACGATTGTGA ACC-3′) for pHS74 carrying mgm1K223A, 806 (5′-CACAATCGTCTGGTAAAAACTCAGTACTAGAATCCATTG-3′) and 807 (5′-CAATGGATTCTAGTACTGAGTTTTTACCAGACGATTGTG-3′) for pHS75 carrying mgm1S224N, and 808 (5′-GGTTCCAACATGGTCGCAAGAAGACCCATTGAATTG-3′) and 809 (5′-CAATTCAATGGGTCTTCTTGCGACCATGTTGGAACC-3′) for pHS76 carrying mgm1T244A. The mutations were confirmed by DNA sequencing.
Gene Disruption
Complete disruptions of the MGM1 and ATP21genes were constructed by PCR-mediated gene replacement as described (Lawrence, 1991) into diploid strain FY833/844 (Winston et al., 1995). For mgm1::kanMX4, we used the kanMX4 gene from the pRS400 plasmid (Brachmann, 1998). Heterozygous diploids were sporulated and dissected to obtain MATa mgm1Δ strain YRJ1383 and MATα mgm1Δ strain YRJ1384. MATa mgm1Δ dnm1Δ strain YRJ1398 and MATα mgm1Δ dnm1Δ strain YRJ1493 were constructed by crossing MATa mgm1Δ strain YRJ1383 and MATα dnm1Δ strain YRJ1290 (Sesaki and Jensen, 2001). For atp21::HIS3 and mmm1::HIS3, the HIS3 gene from the pRS303 plasmid (Sikorski and Hieter, 1989) was used. Heterozygous diploids were sporulated and dissected to obtain MATa atp21Δ strain YRJ1564 and MATα atp21Δ strain YRJ1565. To create MATa mmm1Δ strain YRJ1355 and MATα mmm1Δ strain YRJ1356, the HIS3 gene from the pRS303 plasmid (Sikorski and Hieter, 1989) was used.
Chromosomal Integration of mgm1 GTPase Mutations
To create MGM1 dnm1Δ, mgm1K223 dnm1Δ, mgm1S224N dnm1Δ, and mgm1T244A dnm1Δ cells, pHS73, pHS74, pHS75, and pHS76 were digested with NdeI and transformed into MATa mgm1Δ dnm1Δ strain YRJ1398 and MATα mgm1Δ dnm1Δ strain YRJ1493. Integration of these plasmids into chromosomes was confirmed by PCR.
Microscopy
Cells were observed using a Zeiss Axioskop microscope (Thornwood, NY) with a 100× Plan-Neofluar objective. Fluorescence and differential interference contrast images were captured with a Hamamatsu Orca ER using Open Lab software version 3.0.8 (Improvision Inc., Lexington, MA).
Electron microscopy was performed as previously described (Reider et al., 1996). Briefly, cells were fixed in 3% glutaraldehyde and embedded in Spurrr's resin, and thin sections were cut on a Reichert Ultracut T ultramicrotome (Leica, Deerfield, IL). Samples were examined using a Philips EM420 electron microscope (FEI Co., Peabody, MA).
Mitochondrial Fusion Assay
Mitochondrial fusion during mating was observed as described (Azpiroz and Butow, 1995; Nunnari et al., 1997; Okamoto et al., 1998). MATa strains carrying pGAL1-YTA10-CFP (pHS70) and MATα strains carrying pGAL1-YTA10-YFP (pHS71) were grown to log phase in SGalSuc medium overnight, pelleted by centrifugation, washed, and resuspended in 1 ml of YEPD medium. Same OD600 units of MATa and MATα cells were mixed and collected by centrifugation. Cells were resuspended in YEPD medium at 0.025 OD600 units/μl and placed on a nitrocellulose membrane at 1.2 OD600 units/cm2. Excess solution was removed by placing the membrane on filter paper. The nitrocellulose membrane was then incubated on YEPD medium, adjusted to pH 4.5 using citric acid (Azpiroz and Butow, 1995), at 30°C for 3 h. Zygotes were examined by fluorescence microscopy.
For fusion of the mitochondrial OM, MATα strains carrying pGAL1-TOM72-YFP (pHS72) were grown to log phase in SRafSuc medium overnight, pelleted by centrifugation, and resuspended in SGalSuc medium to an OD600 of 0.2 for 2 h to induce TOM72-YFP expression. MATa strains carrying pGAL1-YTA10-CFP (pHS70) were grown to log phase in SGalSuc medium overnight. Cells were mated as described above.
Submitochondrial Fractionation
Mitochondria were isolated from wild-type cells (FY833) as previously described (Daum et al., 1982). Separation of OM and IM vesicles on sucrose gradients was performed as described (Ryan et al., 1994). For protease digestions, we resuspended 100 μg of mitochondria in 1 ml of 250 mM sucrose, 20 mM HEPES-KOH, pH 7.4. Mitochondria were treated with either 50 μg/ml trypsin for 20 min on ice, followed by the addition of 200 μg/ml soybean trypsin inhibitor, or 50 μg/ml proteinase K, followed by the addition of 1 mM phenylmethylsulfonylfluoride. To disrupt the mitochondrial OM, 100 μg of mitochondria were incubated in 1 ml of 20 mM HEPES-HCl, pH 7.4, for 40 min on ice.
Proteins were separated by SDS-PAGE (Laemmli, 1970) and transferred to Immobilon filters (Haid and Suissa, 1983). Filters were probed with antibodies to Mgm1p (Shepard and Yaffe, 1999), The β subunit of F1-ATPase, Tim23p (Emtage and Jensen, 1993), Om45p (Yaffe et al., 1989), Cox4p, and Mas2p (Jensen and Yaffe, 1988), all at 1:10,000 dilutions, or Tom37p (Gratzer et al., 1995) and Aac2p (a gift from N. Pfanner, Institut fur Biochemie and Molekularbiologie, Universitat Freiburg, Germany) at 1:5000 dilutions. Immune complexes were visualized using 1:10,000 dilution of HRP-conjugated secondary antibodies (Amersham Pharmacia Biotech, Piscataway, NJ) followed by chemiluminescence (SuperSignal; Pierce Chemical Co., Rockford, IL).
Immune Precipitation
Mitochondria were isolated from strain YRJ1282 expressing HA-Fzo1p from pHS77 and strain YRJ1294 expressing myc-Ugo1p from pHS57 (Sesaki and Jensen, 2001), as described before (Daum et al., 1982). For immune precipitations, 0.5 mg of mitochondria was solubilized in 0.5 ml of IP buffer (1% Triton X-100, 50 mM NaCl, 30 mM HEPES-KOH, pH 7.4) containing protease inhibitors (1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 mM phenylmethylsulfonylfluoride, 10 μM trans-epoxy-succinyl-L-leucylamido(4-guanidino)butane, 1 μg/ml chymostatin, 1 μg/ml pepstatin A) at 4°C with gentle agitation for 10 min. After centrifugation at 12,500 × g for 10 min, 100 μl of 50% slurry of antibody-coupled beads was added to the supernatant. We coupled the HA antibody (12CA5; Niman et al., 1983) or the myc antibody (9E10; Evan et al., 1985) to beads using Seize Immunoprecipitation Kit (Pierce) according to manufacturer's instructions. Samples were incubated at 4°C with gentle agitation for 6 h. Beads were washed three times with 0.4 ml of wash buffer (0.1% Triton X-100, 50 mM NaCl, 30 mM HEPES-KOH, pH 7.4) containing protease inhibitors. The bound proteins were eluted by 200 μl of 2× sample buffer (2% SDS, 20% glycerol, 10 μg/ml bromophenol blue, 200 mM Tris-HCl, pH 6.8) at 60°C for 5 min. We added 8 μl of 14.4 M β-mercaptoethanol to the eluates and boiled for 5 min. Proteins were separated by SDS-PAGE (Laemmli, 1970) and analyzed by immune blotting with antibodies to the myc epitope (PRB-150; Covance, Berkeley, CA), the HA epitope (12CA5; Niman et al., 1983), Mgm1p, OM45p, and Tim23p, all at 1:10,000 dilutions.
RESULTS
Disruption of the MGM1 Gene Causes Fragmentation of Mitochondria and Loss of mtDNA in a Dnm1p-dependent Manner
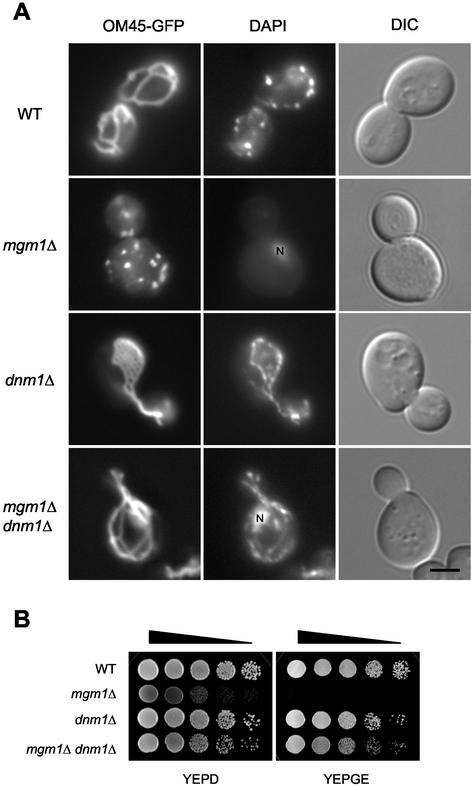
In our previous genetic screen, we isolated fzo1 and ugo1 mutants, both of which were shown to be defective in mitochondrial fusion (Sesaki and Jensen, 2001). In addition to fzo1 and ugo1 mutants, we also isolated 5 mgm1 mutants. To investigate whether Mgm1p functions in mitochondrial fusion, we disrupted MGM1, DNM1, and both MGM1 and DNM1 in yeast cells, and examined mitochondria and mtDNA in these mutant cells. Mitochondria were visualized using an OM-targeted GFP fusion protein, OM45-GFP (Cerveny et al., 2001). Wild-type cells contained 5–10 mitochondrial tubules with occasional branches. In contrast, mgm1Δ cells showed many small mitochondrial fragments, which often aggregated in the cell (Figure 1A). The mitochondria seen in mgm1Δ cells are similar to those seen in fzo1Δ and ugo1Δ cells (Hermann et al., 1998; Rapaport et al., 1998; Sesaki and Jensen, 2001). The fragmentation of mitochondria in fzo1Δ and ugo1Δ cells has been shown to result from a block in fusion along with continuing mitochondrial division. In dnm1Δ cells mitochondria form a single network consisting of interconnected tubules, due to ongoing fusion in the absence of mitochondrial division (Figure 1A; Bleazard et al., 1999; Sesaki and Jensen, 1999). In contrast, in the majority of mgm1Δ dnm1Δ cells (88%, n = 100), mitochondria were found to form elongated tubules with some branches, similar to those seen in wild-type cells. As seen in fzo1Δ dnm1Δ and ugo1Δ dnm1Δ cells (Sesaki and Jensen, 1999, 2001), these mitochondrial tubules were often collapsed to one side of the cell and appeared to be bundled.
Figure 1.
Disruption of MGM1 causes fragmentation of mitochondria and loss of mtDNA in a Dnm1p-dependent manner. (A) Wild-type (FY833), mgm1Δ (YRJ1383), dnm1Δ (YRJ1289), and mgm1Δ dnm1Δ (YRJ1398) cells expressing OM45-GFP were grown to log phase in SRaf medium, stained with 1 μg/ml DAPI, and viewed by DIC and fluorescence microscopy. N′ indicates nuclear DNA staining. Bar, 3 μm. (B) Wild-type, mgm1Δ, dnm1Δ, and mgm1Δ dnm1Δ cells were grown to log phase in YEPD medium. Cells were collected and resuspended in YEPD medium to an OD600 of 2. Cells were then diluted in 10-fold increments; 10 μl of each dilution was spotted onto YEPD and YEPGE media and then incubated at 30°C for 2 d (YEPD) and 6 d (YEPGE).
The loss of mtDNA in mgm1Δ cells was also rescued by disruption of DNM1. Wild-type and mutant cells were stained by 4′,6′-diamidino-2-phenylindole (DAPI) and tested for growth on nonfermentable carbon sources. We found that mgm1Δ cells lack mtDNA nucleoids (Figure 1A) and fail to grow on glycerol and ethanol-containing medium (YEPGE; Figure 1B). However, wild-type, dnm1Δ and mgm1Δ dnm1Δ cells all contained similar amounts of mtDNA nucleoids. Consistent with previous studies (Wong et al., 2000; Fekkes et al., 2000), mgm1Δ dnm1Δ cells were able to grow on YEPGE medium, demonstrating that the double mutants contain mtDNA. We note that mgm1Δ dnm1Δ cells, like fzo1Δ dnm1Δ and ugo1Δ dnm1Δ cells, grew more slowly than wild-type and dnm1Δ cells on both YEPD and YEPGE media. This growth appears to be due to lack of Mgm1p because mgm1Δ dnm1Δ cells contain normal amounts of mtDNA. Our results demonstrate that disruption of MGM1 causes fragmentation of mitochondria and loss of mtDNA in a Dnm1p-dependent manner. Because the phenotypes seen in mgm1Δ cells are almost identical to those seen in fzo1Δ and ugo1Δ mutants, which are defective in mitochondrial fusion (Hermann et al., 1998; Rapaport et al., 1998; Sesaki and Jensen, 2001), our data raise the possibility that Mgm1p likewise functions in mitochondrial fusion.
mgm1Δ and mgm1Δ dnm1Δ Cells Are Defective in Mitochondrial Fusion
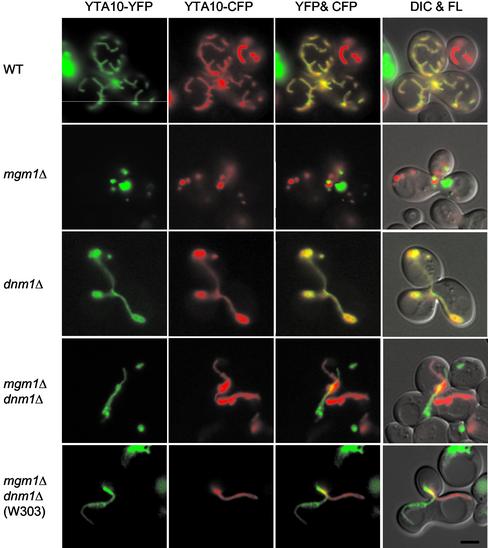
To determine directly whether Mgm1p is required for mitochondrial fusion, we examined the ability of mgm1Δ and mgm1Δ dnm1Δ mutants to fuse their mitochondria during yeast cell mating (Azpiroz and Butow, 1995; Nunnari et al., 1997; Okamoto et al., 1998; Sesaki and Jensen, 1999, 2001). The mitochondria in MATa cells were marked using an IM-targeted cyan fluorescent protein (YTA10-CFP) expressed from pHS70. In MATα cells, mitochondria were labeled by an IM-targeted yellow fluorescent protein (YTA10-YFP) carried on pHS71. Both plasmids express the fusion protein under the control of the inducible GAL1 promoter. MATa and MATα cells were pregrown in galactose-containing medium to induce the expression of the fusion proteins and transferred to glucose-containing medium to inhibit their further synthesis. Cells were mixed and mated on glucose-containing medium. If mitochondria fuse in the resulting zygotes, CFP and YFP fluorescence overlap because of the diffusion of the YTA10-CFP and YTA10-YFP proteins in the IM. If mitochondria fail to fuse, CFP and YFP are seen only in separate organelles.
We found that mgm1Δ and mgm1Δ dnm1Δ cells fail to fuse their mitochondria. Figure 2 shows representative examples of zygotes containing a medial diploid bud from each mating mixture. Fifty zygotes were examined for wild-type and each mutant. When two wild-type haploids mated, mitochondria in the zygote fused and mixed their IM contents, and the CFP and YFP fluorescence therefore overlapped. Mitochondrial fusion also occurred in zygotes formed between dnm1Δ mutants. In contrast, when two mgm1Δ cells were mated, mitochondrial fragments containing only CFP or YFP were observed. Although the disruption of DNM1 suppresses the fragmentation of mitochondria and the loss of mtDNA, mgm1Δ dnm1Δ double mutants were still unable to fuse their mitochondria. mgm1Δ dnm1Δ cells formed tubular mitochondria, but these tubules contained only CFP or YFP. In mgm1Δ dnm1Δ zygotes, we frequently found that two mitochondrial tubules, each derived from one parent, entered the diploid bud and were closely positioned in the middle of zygotes, but nonetheless these mitochondria did not fuse. Our results indicate that, whether mitochondria exist as fragments or tubules, cells lacking Mgm1p are defective in mitochondrial fusion.
Figure 2.
Mitochondrial fusion is defective in mgm1Δ and mgm1Δ dnm1Δ cells. MATa cells were transformed with plasmid pHS70, which expresses IM-targeted cyan fluorescent protein (YTA10-CFP) under the control of the GAL1 promoter. MATα cells were transformed with plasmid pHS71, which expresses IM-targeted yellow fluorescent protein (YTA10-YFP) from the GAL1 promoter. Cells were grown to log phase in SGalSuc medium overnight to induce the expression of the fusion proteins. MATa and α cells were mated for 3–3.5 h on YEPD medium, which inhibits further fusion protein expression. The distribution of YTA10-CFP and YTA10-YFP in representative zygotes containing a medial bud is shown. Zygotes formed by mating between wild-type cells (FY833 and FY834), mgm1Δ mutants (YRJ1383 and YRJ1384), dnm1Δ mutants (YRJ1289 and YRJ1290), mgm1Δ dnm1Δ mutants (YRJ1398 and YRJ1493), and mgm1Δ dnm1Δ mutants in W303 strain (YRJ1554 and YRJ1555) were examined by DIC and fluorescence (FL) microscopy. Partial overlaps between YFP and CFP are seen in mgm1Δ dnm1Δ zygotes because one tubule lies on the top of another in the same focal plane. Bar, 3 μm.
Our findings were unexpected because a previous study showed that mitochondria fuse normally in mgm1-5 dnm1Δ zygotes (Wong et al., 2000). mgm1-5 is a temperature-sensitive allele for MGM1. In a mating assay, mitochondria failed to fuse in mgm1-5 cells at the restrictive temperature, but mitochondria fusion was rescued in mgm1-5 dnm1Δ double mutants (Wong et al., 2000). To test if the difference between our results and those from previous studies results from the use of different yeast strains, we examined mitochondrial fusion in the W303 strain used in the previous study (Wong et al., 2000). When a null allele of MGM1, mgm1Δ, was constructed in W303, we found mitochondrial fusion to be defective. This fusion defect was seen in both mgm1Δ (our unpublished results) and mgm1Δ dnm1Δ cells (Figure 2). These results clearly demonstrate that Mgm1p is required for mitochondrial fusion in two different backgrounds.
Mutations in the GTPase Domain of Mgm1p Block Mitochondrial Fusion
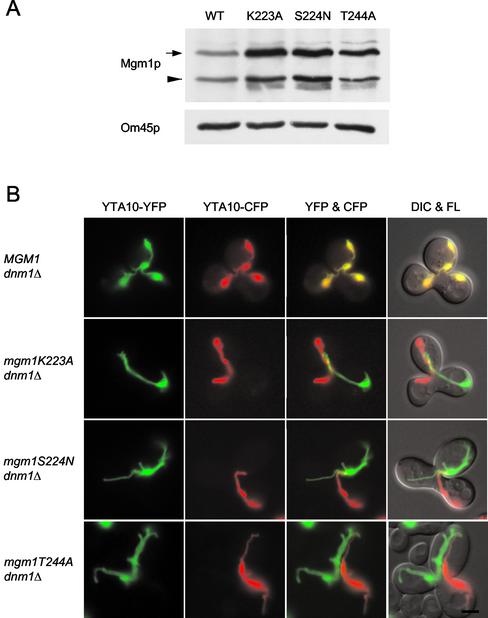
Mgm1p is a member of a family of proteins related to the dynamin GTPase. The MGM1 gene carrying a mutation in its GTPase domain is unable to rescue fragmentation of mitochondria and loss of mtDNA in temperature-sensitive mgm1 mutants (Shepard and Yaffe, 1999). These results suggest that the GTPase domain of Mgm1p is required for activity. To test this possibility, we mutated conserved residues in the GTPase domain among dynamin and dynamin-related GTPases and examined mitochondrial fusion. Equivalent mutations in dynamin inhibit its GTP binding and/or hydrolysis (Damke et al., 2001; Marks et al., 2001). Using site-directed mutagenesis, we changed three residues (lysine at residue 223 was changed to alanine, K223A; serine at residue 224 was changed to asparagine, S224N; threonine at residue 244 was changed to alanine, T244A), creating mgm1K223A, mgm1S224N, and mgm1T244A alleles, respectively. Plasmid constructs carrying wild-type and mutant alleles were integrated into the chromosome in mgm1Δ dnm1Δ cells.
We first confirmed that mutant versions of Mgm1p were expressed and localized to mitochondria. Mitochondria were isolated from cells expressing wild-type or mutant Mgm1p and analyzed by immune blotting using antibodies to Mgm1p. Yeast cells contain two species of Mgm1p, a 90- and a 100-kDa form (Shepard and Yaffe, 1999). We showed that neither species was expressed in mgm1Δ dnm1Δ cells (our unpublished results). As shown in Figure 3A, similar amounts of both forms of Mgm1p were found in wild-type and mutant mitochondria. Next, we examined mitochondria in the different cells using OM45-GFP. We found that mgm1K223A dnm1Δ, mgm1S224N dnm1Δ, and mgm1T244A dnm1Δ mutants contained tubular mitochondria similar to those seen in mgm1Δ dnm1Δ cells, indicating that inactivation of Mgm1p GTPase disrupts the function of Mgm1p (our unpublished results).
Figure 3.
Mutations in the GTPase domain of Mgm1p block mitochondrial fusion. (A) Expression of wild-type and mutant versions of Mgm1p in mitochondria. Mitochondria were isolated from wild-type (YRJ1556) and mutant cells (YRJ1558, YRJ1560, and YRJ1562), and analyzed by immune blotting with antibodies to Mgm1p and OM45p. An arrow and arrowhead indicate the 100- and 90-kDa forms of Mgm1p, respectively. (B) Mitochondrial fusion is defective in mgm1 GTPase mutants. MATa cells carrying plasmid pGAL-YTA10-CFP (pHS70) and MATα cells carrying plasmid pGAL1-YTA10-YFP (pHS71) were grown to log phase in SGalSuc medium overnight to induce the expression of the fusion proteins. MATa and α cells were mated for 3–3.5 h on YEPD medium. The distribution of YTA10-CFP and YTA10-YFP in representative zygotes containing a medial bud is shown. Zygotes formed by mating between MGM1 dnm1Δ cells (YRJ1556 and YRJ1557), mgm1K223A dnm1Δ mutants (YRJ1558 and YRJ1559), mgm1S224N dnm1Δ mutants (YRJ1560 and YRJ1561), and mgm1T244A dnm1Δ mutants (YRJ1562 and YRJ1563) were examined by DIC and fluorescence (FL) microscopy. Bar, 3 μm.
We found that a functional GTPase domain of Mgm1p is required for mitochondrial fusion. Using our mating assay, we examined 50 zygotes for wild-type and each mutant. As shown in Figure 3B, all MGM1 dnm1Δ zygotes fused their mitochondria. In contrast, no mitochondrial fusion occurred in homozygous zygotes of mgm1K223A dnm1Δ, mgm1S224N dnm1Δ, and mgm1T244A dnm1Δ mutants.
Mgm1p, Fzo1p, and Ugo1p Are Required for Fusion of the Mitochondrial OM
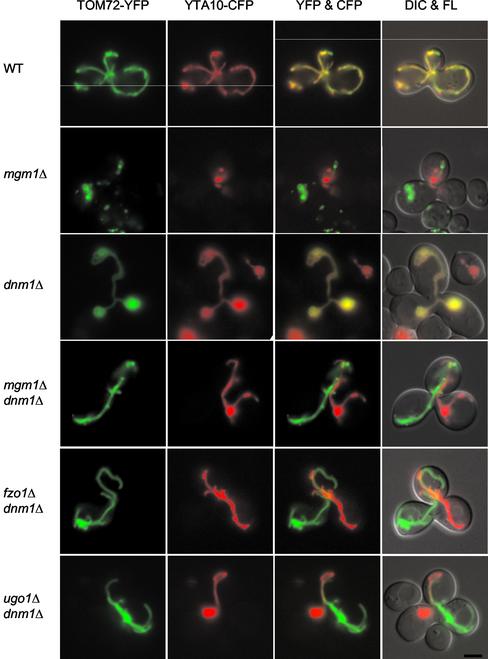
Our results shown in Figures 2 and 3 indicate that Mgm1p is required for mitochondrial fusion. However, because mitochondria have two membranes, an outer and inner membrane, and we used IM-localized YFP and CFP to assay fusion, it was not possible to determine if Mgm1p was required for OM fusion, IM fusion, or fusion of both membranes. To ask specifically if OM fusion is defective in mgm1Δ and mgm1Δ dnm1Δ mutants, we targeted YFP to the mitochondrial OM. In MATα cells, mitochondria were labeled by an OM-targeted YFP (TOM72-YFP) carried on pHS72. This construct expresses YFP fused to the first 40 amino acids derived from Tom72p, which includes the transmembrane domain (Bomer et al., 1996; Schlossmann et al., 1996), under the control of the GAL1 promoter. To confirm that TOM72-YFP is located in the mitochondrial OM, we isolated mitochondria from cells expressing the YFP fusion. When the mitochondria were treated with trypsin, TOM72-YFP was completely digested like the OM protein Tom37p (our unpublished results). The mitochondria in MATa cells were labeled using an IM-targeted CFP (YTA10-CFP) expressed from pHS70.
In mating between wild-type cells or between dnm1Δ cells, CFP and YFP fluorescence in zygotes overlapped in all the mitochondrial tubules (Figure 4). These results indicate that the OM of the mitochondria had fused. In contrast, when mgm1Δ cells were mated, we found mitochondrial fragments that contained only CFP or YFP, indicating that these mitochondria did not fuse. Similarly, in all mgm1Δ dnm1Δ zygotes, mitochondrial tubules contained only one of the two fluorescent markers. These results demonstrate that mgm1Δ and mgm1Δ dnm1Δ cells are defective in fusion of the mitochondrial OM.
Figure 4.
Fusion of the mitochondrial OM is defective in mgm1Δ, mgm1Δ dnm1Δ, fzo1Δ dnm1Δ, and ugo1Δ dnm1Δ cells. MATa cells containing plasmid pGAL1-YTA10-CFP (pHS70) were grown to log phase in SGalSuc medium overnight to induce the expression of IM-targeted YTA10-CFP. MATα cells containing plasmid pGAL1-TOM72-YFP (pHS72) were grown to log phase in SRaf medium overnight and then transferred to SGalSuc medium for 2 h to induce the expression of OM-targeted TOM72-YFP. MATa and α cells were mated for 3–3.5 h on YEPD medium. The distribution of YTA10-CFP and TOM72-YFP in representative zygotes containing a medial bud is shown. Zygotes formed by mating between wild-type cells (FY833 and FY834), mgm1Δ mutants (YRJ1383 and YRJ1384), dnm1Δ mutants (YRJ1289 and YRJ1290), mgm1Δ dnm1Δ mutants (YRJ1398 and YRJ1493), fzo1Δ dnm1Δ mutants (YRJ1347 and YRJ1348), and ugo1Δ dnm1Δ mutants (YRJ1291 and YRJ1292) were examined by DIC and fluorescence (FL) microscopy. Bar, 3 μm.
Because Fzo1p and Ugo1p are OM proteins required for mitochondrial fusion, they have been suggested to mediate fusion of the mitochondrial OM (Hermann et al., 1998; Rapaport et al., 1998; Sesaki and Jensen, 2001). To test this idea, we examined OM fusion in 50 zygotes formed by mating fzo1Δ dnm1Δ cells and 50 zygotes formed by mating ugo1Δ dnm1Δ mutants using TOM72-YFP and YTA10-CFP markers described above. As shown in Figure 4, both fzo1Δ dnm1Δ and ugo1Δ dnm1Δ cells failed to fuse their OMs and contained only YFP or CFP-labeled mitochondria, similar to mgm1Δ dnm1Δ zygotes. Our data indicate Fzo1p and Ugo1p, along with Mgm1p, are required for fusion of the mitochondrial OM. We note that because OMs do not fuse in cells lacking Mgm1p, Fzo1p, or Ugo1p, it is not possible to determine if these proteins are required for IM fusion. Their role in IM fusion therefore awaits further studies.
Mitochondrial IM Structure Is Altered in mgm1Δ Cells, but Normal in mgm1Δ dnm1Δ Cells
Although our data demonstrate that Mgm1p is required for mitochondrial fusion, it has been suggested that Mgm1p is involved in formation of IM structure and that fusion defects seen in mgm1 mutants is a secondary consequence of the altered IM (Wong et al., 2000). We examined the mitochondrial morphology of mgm1Δ cells by electron microscopy. As shown in Figure 5a, wild-type cells contained elongated tubular mitochondria, in which the IM frequently invaginated into the matrix, forming cristae. In contrast, mgm1Δ cells contained small round mitochondria, in which IM cristae were altered (Figure 5, c–e). These cristae were longer than those in wild-type cells and occasionally formed stacks. In addition, the number of cristae in mgm1Δ cells appeared to be reduced. The IM structure seen in mgm1Δ cells was similar to that seen in wild-type cells lacking mtDNA (rho0; Figure 5b). Although rho0 cells contained elongated mitochondrial tubules, the number of cristae in rho0 cells was less than in wild-type cells carrying mtDNA.
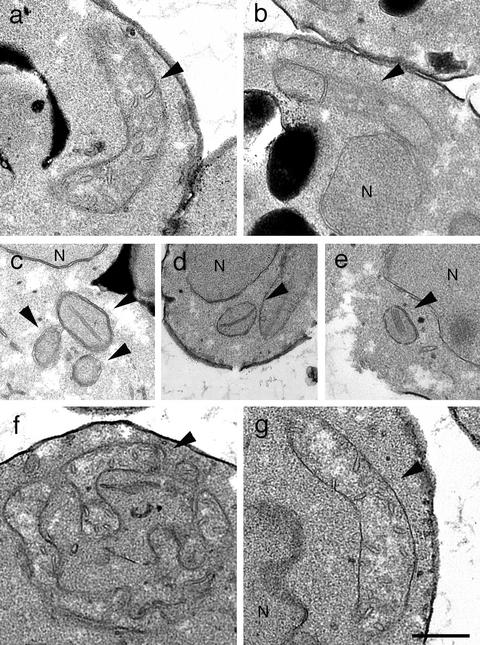
Figure 5.
The structure of the mitochondrial IM is altered in mgm1Δ cells, but is normal in mgm1Δ dnm1Δ cells. (a) Wild-type (FY833), (b) rho0 wild-type (YRJ1287), (c–e) mgm1Δ (YRJ1383), (f) dnm1Δ (YRJ1289), and (g) mgm1Δ dnm1Δ (YRJ1398) cells were grown to log phase in YEPRaf medium and fixed, and thin sections were examined by electron microscopy. Arrowheads and N′ indicate mitochondria and nuclei, respectively. Bar, 0.5 μm.
In dnm1Δ cells, mitochondria were more branched and interconnected, but their IM appeared normal in cristae structure (Figure 5f). Disruption of DNM1 restored both mitochondrial tubules and normal IM structure in mgm1Δ cells (Figure 5g). In mgm1Δ dnm1Δ cells, mitochondria formed tubular structures and displayed IM cristae distinguishable from those seen in wild-type cells. Because we showed that mgm1Δ dnm1Δ cells are still defective in mitochondrial fusion, the fusion defect cannot be due to an alteration of mitochondrial IM structure. We argue that the fusion defect is the direct result of a lack of Mgm1p.
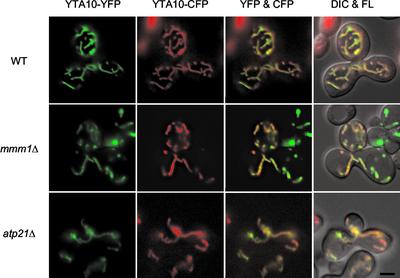
To further test the idea that alteration of IM structures affects mitochondrial fusion, we examined mitochondrial fusion in two mutants, in which IM structure is highly disorganized, mmm1Δ and atp21Δ. In mmm1Δ cells, the IM cristae collapse into large membrane sheets, and these sheets often form stacks (Hobbs et al., 2001). atp21Δ cells contain mitochondria whose IM excessively folds and forms onion-like structures (Paumard et al., 2002). Using the mating assay, we found that mitochondria fuse in both mmm1Δ and atp21Δ cells. As shown in Figure 6, zygotes formed between mmm1Δ cells contained partially fragmented mitochondria, and these mitochondria carried both YTA10-CFP and YTA10-YFP, demonstrating that mitochondria fused and mixed their contents. The mitochondrial shape seen in mmm1Δ zygotes was different from that seen in growing mmm1Δ cells (Burgess et al., 1994; Hobbs et al., 2001). It seems that mitochondria in mmm/Δ cells change their shape from large spheres to fragmented tubules during mating.
Figure 6.
mmm1Δ and atp21Δ mutants normally fuse mitochondria. MATa cells harboring plasmid pGAL-YTA10-CFP (pHS70), and MATα cells harboring plasmid pYTA10-YFP (pHS71) were grown to log phase in SGalSuc medium overnight to induce the expression of the fusion proteins. MATa and α cells were mated for 3–3.5 h on YEPD medium. The distribution of YTA10-CFP and YTA10-YFP in representative zygotes containing a medial bud is shown. Zygotes formed by mating between wild-type cells (FY833 and FY834), mmm1Δ mutants (YRJ1356 and YRJ1355), and atp21Δ mutants (YRJ1564 and YRJ1565) were examined. Bar, 3 μm.
In atp21Δ zygotes, mitochondria fused normally. We noticed that although overall patterns of YTA10-CFP and YTA10-YFP signals were very similar in homozygous zygotes of mmm1Δ and atp21Δ mutants, there were regions of mitochondria where CFP signals were stronger than YFP, or vice versa. This could be due to slow diffusion of IM proteins because IM structures in these mutants are remarkably complex (Hobbs et al., 2001; Paumard et al., 2002). Alternatively, it is also possible that mitochondrial fusion is less efficient in mmm1Δ and atp21Δ cells. Nevertheless, these results indicate that aberrant IM structures do not necessarily block mitochondrial fusion and further support that Mgm1p plays a direct role in mitochondrial fusion.
Mgm1p Is Associated with Both the Mitochondrial Outer and Inner Membranes
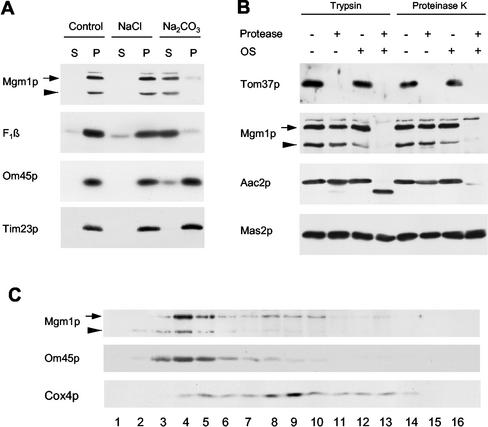
Although Mgm1p has been localized to mitochondria, its submitochondrial location is controversial. Shepard and Yaffe (1999) found Mgm1p in the mitochondrial OM, exposed to the cytosol. In contrast, Wong et al. (2000) suggested that Mgm1p is in the intermembrane space, peripherally associating with the IM. In an attempt to reconcile these differences, we first examined the association of Mgm1p with mitochondria. We treated mitochondria with 1.5 M sodium chloride or 0.1 M sodium carbonate (Figure 7A). The Mgm1p protein was extracted from the mitochondria with sodium carbonate, but not with sodium chloride, like the β subunit of the F1-ATPase, a peripheral membrane protein, confirming the previous observation (Wong et al., 2000). Conversely, the integral membrane proteins Om45p (Yaffe et al., 1989) and Tim23p (Emtage and Jensen, 1993) remained in the membrane fraction. To ask if Mgm1p is inside or outside of mitochondria, we treated intact mitochondria using trypsin or proteinase K. Consistent with the previous observation (Wong et al., 2000), we found that Mgm1p was not digested by either of these two proteases (Figure 7B). As controls, the OM protein Tom37p (Gratzer et al., 1995) was completely digested, whereas the IM protein Aac2p remained intact. When the mitochondrial OM was disrupted by osmotic shock, trypsin and proteinase K digested Mgm1p, Tom37p, and Aac2p, but not the matrix protein, Mas2p. Thus, Mgm1p was protected from proteases by the mitochondrial OM.
Figure 7.
Mgm1p is associated with the both mitochondrial OM and IM in the intermembrane space. (A) Mgm1p is a peripheral membrane protein. Mitochondria isolated from wild-type cells (FY833) were treated with 1.5 M sodium chloride, 0.1 M sodium carbonate, or buffer alone. Mitochondrial membranes were then separated into supernatant and pellet fraction by centrifugation at 100,000 × g for 60 min. Aliquots from each fraction were analyzed by immune blotting with antibodies to Mgm1p, Om45p (an integral membrane protein with a single transmembrane domain), Tim23p (an integral membrane protein with four transmembrane domains), and F1β (a peripheral membrane protein). An arrow and arrowhead indicate the 100- and 90-kDa forms of Mgm1p, respectively. (B) Mgm1p is located in the intermembrane space. Mitochondria, 100 μg, were first treated to disrupt the OM by osmotic shock (OS) or left intact and then incubated with 50 μg/ml trypsin or 50 μg/ml proteinase K for 20 min on ice. Proteins were analyzed by immune blotting with antibodies to Mgm1p, the OM protein, Tom37p, the IM protein, Aac2p, and the matrix protein, Mas2p. (C) Mgm1p is associated with the both mitochondrial OM and IM. Mitochondria, 5–10 mg, were resuspended at 10 mg/ml in SEH (20 mM HEPES-KOH, pH 7.4, 250 mM sucrose, 0.5 mM EDTA) containing protease inhibitors (1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 mM phenylmethylsulfonylfluoride, 10 μM trans-epoxy-succinyl-L-leucylamido(4-guanidino)butane, 1 μg/ml chymostatin, 1 μg/ml pepstatin A). After disrupting the OM by diluting into 9 vol of 20 mM HEPES-KOH, pH 7.4, with protease inhibitors for 30 min on ice, mitochondria were sonicated three times at setting 5 for 30 s with Model 60 Sonic Dismembrator (Fisher, Pittsburgh, PA). Unbroken mitochondria were pelleted by centrifugation at 32,000 × g for 20 min. The supernatant was centrifuged at 200,000 × g for 20 min to collect membrane vesicles. The membrane pellet was resuspended in 200 μl of 5 mM HEPES-KOH, pH 7.4, 10 mM KCl, and loaded on 11 ml of discontinuous sucrose gradients (40, 35.3, 30.7, 26% sucrose in 5 mM HEPES-KOH, pH 7.4, 10 mM KCl). After centrifugation at 142,555 × g for 16 h, fractions were collected and analyzed by immune blotting with antibodies to Mgm1p, OM45p, and Cox4p.
To ask which membrane Mgm1p is associated with, we prepared membrane vesicles from mitochondria and separated them into OM and IM fractions on sucrose gradients. As shown in Figure 7B, the majority of both 100-kDa (∼70%) and 90-kDa (∼100%) forms of Mgm1p cofractionated with the OM vesicle fraction, along with Om45p. However, ∼30% of the 100-kDa form of Mgm1p was also found in the IM vesicle fraction, along with Cox4p. These results, taken together, indicate that Mgm1p is peripherally associated with both the mitochondrial outer and inner membranes in the intermembrane space.
Mgm1p, Ugo1p, and Fzo1p Physically Interact
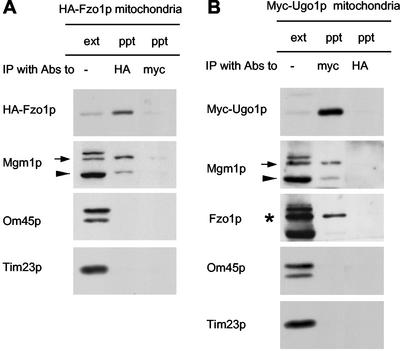
To test if Mgm1p interacts with Ugo1p and Fzo1p, we asked whether Mgm1p coimmune precipitated with Ugo1p and Fzo1p from detergent-solubilized mitochondria. We isolated mitochondria from cells expressing HA-Fzo1p and solubilized the mitochondria in buffer containing 1% Triton X-100. We then immune precipitated HA-Fzo1p using antibodies to the HA epitope. The pellet fraction was analyzed by immune blotting. As shown in Figure 8A, HA-Fzo1p was found in the pellet fraction. When we examined the pellet with antibodies to Mgm1p, we found that the 100-kDa form of Mgm1p coprecipitated along with HA-Fzo1p. About 5% of the 100-kDa form of Mgm1p was found in the pellet fraction. Although the 90-kDa form also coprecipitated, the efficiency was lower (∼0.1%). Other abundant mitochondrial proteins such as OM45 (an OM protein) and Tim23p (an IM protein) did not precipitate with HA-Fzo1p. As a control, we immune precipitated with antibodies to the myc epitope and found no HA-Fzo1p, Mgm1p, Om45p, or Tim23p in the pellet fractions. We also used 0.5% digitonin to solubilize mitochondria and obtained similar results (our unpublished results). These results indicate that Mgm1p physically interacts with Fzo1p and suggest that Mgm1p is a part of the fusion machinery located in the mitochondrial OM.
Figure 8.
Mgm1p, Fzo1p, and Ugo1p physically interact wit each other. Mitochondria were isolated from strain YRJ1282, which expresses HA-Fzo1p from pHS77 (A), or strain YRJ1294, which expresses myc-Ugo1p from plasmid pHS57 (B), and solubilized in 1% Triton X-100. Extracts were immune precipitated with antibodies to the HA epitope or to the myc epitope. Immunoprecipitates (ppt) were analyzed by immune blotting with antibodies to the HA epitope, the myc epitope, Mgm1p, OM45p, Tim23p, and Fzo1p and compared with 10% of the starting mitochondrial extract (ext). An asterisk, arrow, and arrowhead indicate Fzo1p, the 100- and 90-kDa forms of Mgm1p, respectively.
Mgm1p also associates with Ugo1p. Mitochondria were isolated from cells expressing myc-Ugo1p, solubilized in Triton X-100-containing buffer. myc-Ugo1p was then immune precipitated from the detergent-solubilized mitochondria with antibodies to the myc epitope. Immune blots showed that virtually all myc-Ugo1p was precipitated (Figure 8B). About 10% of the 100-kDa form of Mgm1p was found in the pellet fraction. Similar to our precipitation using HA-Fzo1p, only a small fraction of the 90-kDa form coprecipitated (∼0.1%). Furthermore, Ugo1p also interacts with Fzo1p. We found that ∼3% of Fzo1p coprecipitated along with Myc-Ugo1p (Figure 8C). In contrast, Om45p and Tim23p remained in the unbound fraction. As a control, we incubated mitochondria containing myc-Ugo1p with antibodies to the HA epitope and found no myc-Ugo1p or Mgm1p precipitated. Thus, Mgm1p, Ugo1p and Fzo1p, mitochondrial proteins required for outer membrane fusion, physically interact with each other.
DISCUSSION
We show that Mgm1p, together with the Fzo1 and Ugo1 proteins, is required for fusion of mitochondrial outer membranes. In a genetic screen that yielded mutants in FZO1 and UGO1, both shown to encode mitochondrial fusion components, we also isolated five different mgm1 mutants (Sesaki and Jensen, 2001). In addition, mgm1 mutants have several striking similarities to fzo1 and ugo1 mutants (Jones and Fangman, 1992; Guan et al., 1993; Hermann et al., 1998; Rapaport et al., 1998; Shepard and Yaffe, 1999; Wong et al., 2000; Sesaki and Jensen, 2001). mgm1, fzo1, and ugo1 cells all contain highly fragmented mitochondria and rapidly lose their mtDNA. The fragmentation of mgm1, fzo1, and ugo1 mitochondria is the result of ongoing organelle fission in the absence of fusion and can be suppressed in each mutant by disruption of DNM1, a gene required for the division of mitochondria (Bleazard et al., 1999; Sesaki and Jensen, 1999, 2001). Also arguing that Mgm1p functions in the same pathway as Fzo1p, we find that mgm1Δ fzo1Δ dnm1Δ triple mutants show no additional mitochondrial defects when compared with fzo1Δ dnm1Δ and mgm1Δ dnm1Δ double mutants. The double and triple mutants all contain similarly shaped tubular mitochondria (H. Sesaki, unpublished observation). In this report we find that cells lacking Mgm1p are defective in the fusion of their mitochondria and that the Mgm1 protein physically interacts with both Fzo1p and Ugo1p, arguing that the Mgm1 protein plays an essential role in mitochondrial fusion.
Mgm1p is related to several, large GTP-binding proteins. The first member of the family identified was dynamin, which is required for fission of the clathrin-coated plasma membrane invaginations during the formation of endocytic vesicles (McNiven, 1998; Schmid et al., 1998; Sever et al., 2000b). Supported by in vitro assembly studies, dynamin has been proposed to form ring-like structures around membrane invaginations (Takei et al., 1995). After a GTP-dependent constriction, the dynamin ring is proposed to pinch off the endocytic vesicle (Sweitzer and Hinshaw, 1998). Another member of the dynamin family, Dnm1p, is required for mitochondrial division and is located on the organelle surface (Bleazard et al., 1999; Labrousse et al., 1999; Sesaki and Jensen, 1999). By analogy to dynamin, Dnm1p may play a direct, mechanical role in the fission of mitochondria. However, our results show that another member of the dynamin family, Mgm1p, mediates mitochondrial membrane fusion, and Gu and Veerma (1996) shows that phragmoplastin, another dynamin-related protein, plays a role in fusion of Golgi-derived vesicles to the cell plate during plant cell division. It is possible that the dynamin-like proteins do not function as “pinchases,” but play a different role in membrane dynamics. For example, similar to the action of most G-proteins in the cell, dynamin-related proteins may act as regulatory molecules, recruiting and activating the fission or fusion machinery. Such a model has been suggested for dynamin (Sever et al., 1999, 2000a). However, it is important to note that, at least conceptually, membrane fusion and division are similar reactions running in opposite directions. Therefore, it may turn out that analogous molecules mediate both processes.
In matings between cells whose OM was marked by YFP, we find that Mgm1p, Fzo1p, and Ugo1p are all required for fusion of the mitochondrial OM. Moreover, the observation that all three proteins interact suggests that they are part of a fusion machine in the OM. Because Fzo1p and Ugo1p both contain domains facing the cytosol, they are candidates for proteins that mediate the docking and early fusion events between two mitochondria. Mgm1p, which is located inside the OM in the intermembrane space, may organize or activate Fzo1p and Ugo1p and regulate fusion of the OM. Interestingly, both Fzo1p and Mgm1p appear to be GTP-binding proteins. The GTPase domain of Fzo1p faces the cytosol, whereas that of Mgm1p is in the intermembrane space. It is not clear how these two G-proteins coordinate the fusion reaction and further studies are clearly needed to determine the roles of Mgm1p, Ugo1p, and Fzo1p.
Our coimmune precipitation studies demonstrated that Mgm1p, Fzo1p, and Ugo1p physically interact. Because small fractions of these proteins associate with each other at steady state, their associations could be transient and/or weak. Perhaps the small amounts of Mgm1p, Ugo1p, and Fzo1p that interact represent the small fractions of these proteins actually engaged in mitochondrial fusion at any given time. Similar short-lived interactions between SNARE proteins have been observed. About 1% of SNARE proteins assemble into SNARE complexes and this reflects transient assembly of the SNARE complex (Grote et al., 2000). It has been shown that the SNARE complex forms before and/or during fusion and disassembles immediately after fusion (Weber et al., 2000). By analogy, Mgm1p, Fzo1p, and Ugo1p may form protein complexes during mitochondrial fusion and actively dissociate after fusion. It is tempting to speculate that all three proteins assemble into the fusion machinery in the OM. Alternatively, it is also possible that pair-wise interactions of Mgm1p, Fzo1p, and Ugo1p take place at different steps of the fusion pathway.
Because mgm1, ugo1, and fzo1 mutants are defective in the fusion of the mitochondrial OM, it is not possible to determine if Mgm1p, Ugo1p, or Fzo11p plays a role in IM fusion. However, several observations raise the possibility that the two fusion events may be connected. First, direct observation of mitochondria fusing after cell mating suggests that the mitochondrial IM fuses immediately after OM fusion (Okamoto et al., 1998; H. Sesaki, unpublished observations). Second, our work with Mgm1p and previous work with Fzo1p (Fritz et al., 2001) indicate that the two proteins associate with both the outer and inner membranes. These two proteins may be located at contact sites, where the OM and IM are closely positioned. It has been suggested that OM fusion takes place at contacts sites and coordinates with IM fusion (Fritz et al., 2001). Third, the domains of Fzo1p and Ugo1p that face the intermembrane space are important for their activities (Fritz et al., 2001; H. Sesaki, unpublished observations). Nonetheless, whether the OM and IM contains the same or separate fusion machinery awaits the identification and analysis of addition fusion components.
In a previous report, Wong et al. (2000) found that although mgm1–5 mutants were defective in mitochondrial fusion, efficient mitochondrial fusion occurred after the mating of two mgm1-5 dnm1Δ double mutants. They suggested that the fusion defect seen in mgm1-5 was indirect, resulting instead from IM structural alterations caused by the lack of Mgm1p. However, using a disruption of the MGM1 open reading frame, we find that both mgm1Δ and mgm1Δ dnm1Δ mutants are completely defective in fusion. This inconsistency is not due to different strain backgrounds, because we see the same defect when MGM1 is disrupted in the W303 strain used by Wong et al. (2000). We suggest that the temperature-sensitive mgm1-5 mutant retains partial Mgm1p activity at the restrictive temperature. Although the exact role of Mgm1p in mitochondrial fusion still remained to be determined, the fusion defect seen in mgm1Δ and mgm1Δ dnm1Δ mutants is not the consequence of altered IM organization. Disruption of DNM1 in mgm1Δ cells restores the IM cristae morphology, yet mitochondrial fusion still does not occur. Moreover, mmm1 and atp21 mutants both have drastically altered IM structures (Hobbs et al., 2001; Paumard et al., 2002), but we find that mitochondrial fusion is not blocked in these mutants.
Because previous studies disagreed about the localization of Mgm1p, we reexamined its location using isolated mitochondria. Consistent with the results of Wong et al. (2000), we found that both forms of Mgm1p readily extracted from mitochondrial membranes by alkali, and protease digestion studies indicate that both forms of Mgm1p are located inside the mitochondrial outer membrane. However, our results differ from Wong et al. (2000) in that we find that the majority of Mgm1p is associated with the mitochondrial OM. Although ∼30% of the 100-kDa form of Mgm1p comigrated with IM vesicles, nearly all of the 90-kDa form and ∼70% of the 100-kDa form cosedimented with OM vesicles. Perhaps the dual location of Mgm1p explains the apparently conflicting results seen in different studies.
Mitochondrial fusion appears to be highly conserved from yeast to human, with orthologues to yeast Fzo1p present in flies (Hales and Fuller, 1997; Hwa et al., 2002) and mammals (Santel and Fuller, 2001; Rojo et al., 2002). In addition, OPA1 is an apparent human homolog of yeast Mgm1p and is defective in autosomal dominant optic atrophy, causing an early childhood visual impairment (Alexander et al., 2000; Delettre et al., 2000). We speculate that OPA1, like its yeast counterpart, is involved in mitochondrial fusion. Supporting this idea, OPA1 is located in mitochondria, and cells isolated from patients of the optic atrophy contain disorganized mitochondria (Delettre et al., 2000). If OPA1 functions like Mgm1p in mitochondrial fusion, mitochondria in mammalian cells defective in OPA1would lose their shape and mtDNA, consistent with the prediction that the optic atrophy results from defects in respiration and mitochondrial ATP production (Delettre et al., 2002). Therefore, further analysis of Mgm1p function will be important to clarify its role in mitochondrial fusion, but also to better understand mitochondrial function in general.
Acknowledgments
We thank the facilities of the Integrated Imaging Center (The Johns Hopkins University, Baltimore, MD) and Sean Jordan for his excellent technical assistance with electron microscopy. We thank N. Pfanner for antibody to Aac2p, R. Rothstein for W303, S, Michaelis for OSM373, A. Aiken Hobbs for help with gene disruption and submitochondrial fractionation, and M. Iijima for help with growth assays. We also thank J. Michael McCaffery, C. Machamer, A. Aiken Hobbs, K. Cerveny, M. Youngman, C. Dunn, H. Hoard-Fruchey, J. Holder, and M. Iijima for stimulating discussions and valuable comments on the manuscript. This work was supported by U.S. Public Health Service Grant RO1-GM54021–06 and National Science Foundation DBI Grant 009705 to R.E.J. and a postdoctoral fellowship from the Leukemia and Lymphoma Society to H.S.
Abbreviations used: OM, outer membrane; IM, inner membrane; GFP, green fluorescent protein; CFP, cyan fluorescent protein; YFP, yellow fluorescent protein; DAPI, 4′,6′-diamidino-2-phenylindole; DIC, differential interference contrast.
References
- Adams, A., Gottschling, D., Kaiser, C., and Stearns, T. (1997). Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press.
- Alexander, C. et al. (2000). OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26, 211-215. [DOI] [PubMed] [Google Scholar]
- Azpiroz, R., and Butow, R.A. (1995). Mitochondrial inheritance in yeast. Methods Enzymol. 260, 453-465. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn, J., and Voth, M. (1994). Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 27, 198-219. [DOI] [PubMed] [Google Scholar]
- Bleazard, W., McCaffery, J.M., King, E.J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J., and Shaw, J.M. (1999). The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomer, U., Pfanner, N., and Dietmeier, K. (1996). Identification of a third yeast mitochondrial Tom protein with tetratrico peptide repeats. FEBS Lett. 382, 153-158. [DOI] [PubMed] [Google Scholar]
- Brachmann, C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P., and Boeke, J.D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115-132. [DOI] [PubMed] [Google Scholar]
- Burgess, S.M., Delannoy, M., and Jensen, R.E. (1994). MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J. Cell Biol. 126, 1375-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny, K.L., McCaffery, J.M., and Jensen, R.E. (2001). Division of mitochondria requires a novel DMN1-interacting protein, Net2p. Mol. Biol. Cell 12, 309-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke, H., Binns, D.D., Ueda, H., Schmid, S.L., and Baba, T. (2001). Dynamin GTPase domain mutants block endocytic vesicle formation at morphologically distinct stages. Mol. Biol. Cell 12, 2578-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum, G., Böhni, P.C., and Schatz, G. (1982). Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Cell Biol. 257, 13028-13033. [PubMed] [Google Scholar]
- Delettre, C. et al. (2000). Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26, 207-210. [DOI] [PubMed] [Google Scholar]
- Delettre, C., Lenaers, G., Pelloquin, L., Belenguer, P., and Hamel, C.P. (2002). OPA1 (Kjer type) dominant optic atrophy: a novel mitochondrial disease. Mol. Genet. Metab. 75, 97-107. [DOI] [PubMed] [Google Scholar]
- Emtage, J.L., and Jensen, R.E. (1993). MAS6 encodes an essential inner membrane component of the yeast mitochondrial protein import pathway. J. Cell Biol. 122, 1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan, G.I., Lewis, G.K., Ramsay, G., and Bishop, J.M. (1985). Isolation of monoclonal antibodies specific for human c-myc protooncogene product. Mol Cell Biol. 5, 3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekkes, P., Shepard, K.A., and Yaffe, M.P. (2000). Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J. Cell Biol. 151, 333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz, S., Rapaport, D., Klanner, E., Neupert, W., and Westermann, B. (2001). Connection of the mitochondrial outer and inner membranes by Fzo1 is critical for organellar fusion. J. Cell Biol. 152, 683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie, A.E., Kurihara, L.J., Vallee, R.B., and Rose, M.D. (1995). DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J. Cell Biol. 130, 553-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratzer, S. et al. (1995). Mas37p, a novel receptor subunit for protein import into mitochondria. J. Cell Biol. 129, 25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote, E., Carr, C.M., and Novick, P.J. (2000). Ordering the final events in yeast exocytosis. J. Cell Biol. 151, 439-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X., and Verma, D.P. (1996). Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 15, 695-704. [PMC free article] [PubMed] [Google Scholar]
- Guan, K., Farh, L., Marshall, T.K., and Deschenes, R.J. (1993). Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr. Genet. 24, 141-148. [DOI] [PubMed] [Google Scholar]
- Hales, K.G., and Fuller, M.T. (1997). Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90, 121-129. [DOI] [PubMed] [Google Scholar]
- Hermann, G.J., and Shaw, J.M. (1998). Mitochondrial dynamics in yeast. Annu. Rev. Cell Dev. Biol. 14, 265-303. [DOI] [PubMed] [Google Scholar]
- Hermann, G.J., Thatcher, J.W., Mills, J.P., Hales, K.G., Fuller, M.T., Nunnari, J., and Shaw, J.M. (1998). Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143, 359-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs, A.E., Srinivasan, M., McCaffery, J.M., and Jensen, R.E. (2001). Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol. 152, 401-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, C.S., and Winston, F. (1987). A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation into Escherichia coli. Gene 57, 267-272. [DOI] [PubMed] [Google Scholar]
- Hwa, J.J., Hiller, M.A., Fuller, M.T., and Santel, A. (2002). Differential expression of the Drosophila mitofusin genes fuzzy onions (fzo) and dmfn. Mech. Dev. 116, 213-216. [DOI] [PubMed] [Google Scholar]
- Jensen, R.E., Hobbs, A.E., Cerveny, K.L., and Sesaki, H. (2000). Yeast mitochondrial dynamics: fusion, division, segregation, and shape. Microsc. Res. Tech. 51, 573-583. [DOI] [PubMed] [Google Scholar]
- Jones, B.A., and Fangman, W.L. (1992). Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 6, 380-389. [DOI] [PubMed] [Google Scholar]
- Labrousse, A.M., Zappaterra, M.D., Rube, D.A., and van der Bliek, A.M. (1999). C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815-826. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- Marks, B., Stowell, M.H., Vallis, Y., Mills, I.G., Gibson, A., Hopkins, C.R., and McMahon, H.T. (2001). GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature 410, 231-235. [DOI] [PubMed] [Google Scholar]
- McNiven, M.A. (1998). Dynamin: a molecular motor with pinchase action. Cell 94, 151-154. [DOI] [PubMed] [Google Scholar]
- Nigro, J.M., Sikorski, R., Reed, S.I., and Vogelstein, B. (1992). Human p53 and CDC2Hs genes combine to inhibit the proliferation of Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 1357-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niman, H.L., Houghten, R.A., Walker, L.E., Reisfeld, R.A., Wilson, I.A., Hogle, J.M., and Lerner, R.A. (1983). Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc. Natl. Acad. Sci. USA 80, 4949-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari, J., Marshall, W.F., Straight, A., Murray, A., Sedat, J.W., and Walter, P. (1997). Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell 8, 1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, K., Perlman, P.S., and Butow, R.A. (1998). The sorting of mitochondrial DNA and mitochondrial proteins in zygotes: preferential transmission of mitochondrial DNA to the medial bud. J. Cell Biol. 142, 613-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg, K.R., Vo, K.T., Michaelis, S., and Paddon, C. (1997). Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25, 451-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumard, P., Vaillier, J., Coulary, B., Schaeffer, J., Soubannier, V., Mueller, D.M., Brethes, D., di Rago, J.P., and Velours, J. (2002). The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21, 221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, D., Brunner, M., Neupert, W., and Westermann, B. (1998). Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273, 20150-20455. [DOI] [PubMed] [Google Scholar]
- Rojo, M., Legros, F., Chateau, D., and Lombes, A. (2002). Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 115, 1663-1674. [DOI] [PubMed] [Google Scholar]
- Ryan, K.R., Menold, M.M., Garrett, S., and Jensen, R.E. (1994). SMS1, a high-copy suppressor of the yeast mas6 mutant, encodes an essential inner membrane protein required for mitochondrial protein import. Mol. Biol. Cell 5, 529-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel, A., and Fuller, M.T. (2001). Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114, 867-874. [DOI] [PubMed] [Google Scholar]
- Schlossmann, J., Lill, R., Neupert, W., and Court, D.A. (1996). Tom71, a novel homologue of the mitochondrial preprotein receptor Tom70. J. Biol. Chem. 271, 17890-17895. [DOI] [PubMed] [Google Scholar]
- Schmid, S.L., McNiven, M.A., and De Camilli, P. (1998). Dynamin and its partners: a progress report. Curr. Opin. Cell Biol. 10, 504-512. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (1999). Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147, 699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (2001). UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol. 152, 1123-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever, S., Damke, H., and Schmid, S.L. (2000a). Dynamin:GTP controls the formation of constricted coated pits, the rate limiting step in clathrin-mediated endocytosis. J. Cell Biol. 150, 1137-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever, S., Damke, H., and Schmid, S.L. (2000b). Garrotes, springs, ratchets, and whips: putting dynamin models to the test. Traffic 1, 385-392. [DOI] [PubMed] [Google Scholar]
- Sever, S., Muhlberg, A.B., and Schmid, S.L. (1999). Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature 398, 481-486. [DOI] [PubMed] [Google Scholar]
- Shaw, J.M., and Nunnari, J. (2002). Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 12, 178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard, K.A., and Yaffe, M.P. (1999). The yeast dynamin-like protein, mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance [In Process Citation]. J. Cell Biol. 144, 711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R., and Hieter, P. (1989). A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweitzer, S.M., and Hinshaw, J.E. (1998). Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell 93, 1021-1029. [DOI] [PubMed] [Google Scholar]
- Takei, K., McPherson, P.S., Schmid, S.L., and De Camilli, P. (1995). Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature 374, 186-190. [DOI] [PubMed] [Google Scholar]
- Tyers, M., Tokiwa, G., Nash, R., and Futcher, B. (1992). The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 11, 1773-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, D. (1992). The Mitochondrion. New York: VCH Publishers.
- Weber, T., Parlati, F., McNew, J.A., Johnston, R.J., Westermann, B., Sollner, T.H., and Rothman, J.E. (2000). SNAREpins are functionally resistant to disruption by NSF and alphaSNAP. J. Cell Biol. 149, 1063-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, F., Dollard, C., and Ricupero-Hovasse, S.L. (1995). Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11, 53-55. [DOI] [PubMed] [Google Scholar]
- Wong, E.D., Wagner, J.A., Gorsich, S.W., McCaffery, J.M., Shaw, J.M., and Nunnari, J. (2000). The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 151, 341-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M.P. (1999). Dynamic mitochondria. Nat. Cell Biol. 1, E149-E150. [DOI] [PubMed] [Google Scholar]
- Yaffe, M.P., Jensen, R.E., and Guido, E.C. (1989). The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J. Biol. Chem. 264, 21091-21096. [PubMed] [Google Scholar]