Abstract
Pre-mRNA splicing specifically deposits the exon junction complex (EJC) onto spliced mRNA, which is important for downstream events. Here, we show that EJC components are primarily recruited to the spliceosome by association with the intron via the intron-binding protein, IBP160. This initial association of EJC components occurs in the absence of the final EJC-binding site on the exon. RNA interference (RNAi) knockdown of IBP160 arrested EJC association with cytoplasmic RNAs following nonsense-mediated decay. We propose that the intron has a crucial role in the early steps of EJC formation and is indispensable for the subsequent formation of a functional EJC.
Keywords: Exon junction complex, pre-mRNA splicing, intron, nonsense-mediated decay, spliceosome
The removal of introns from pre-mRNAs is conducted by a large ribonucleprotein complex called the spliceosome. During the splicing reaction, it has been recognized that the spliceosome deposits a set of protein factors on the nascent mRNA that shape subsequent events in gene expression (Maniatis and Reed 2002; Maquat 2004). The exon junction complex (EJC) is a multiprotein complex that is deposited 20–24 nucleotides (nt) upstream of an exon–exon junction upon completion of the splicing reaction (Le Hir et al. 2000; Maquat 2004; Tange et al. 2004). To date, 16 proteins including transiently interacting factors have been identified as EJC components (Tange et al. 2005). The tetrameric core of this complex is composed of eIF4AIII, Y14, Magoh, and MNL51; the EJC is anchored to the upstream exon of the mature mRNA by eIF4AIII (Ballut et al. 2005; Tange et al. 2005). The EJC is a multifunctional complex involved in mRNA export, nonsense-mediated mRNA decay (NMD), translation, and mRNA localization in metazoans (Maquat 2004; Tange et al. 2004). Among the EJC components, REF, Y14, and/or Magoh have been observed to bind to the mRNA export receptor TAP (Kataoka et al. 2001; Le Hir et al. 2001), whereas RNPS1 and Y14 interact with hUpf3, a factor required for NMD (Kim et al. 2001; Lykke-Andersen et al. 2001). The EJC is only assembled onto mature mRNA that has been produced as a result of pre-mRNA splicing; however, the pathway of assembly remains to be described in molecular detail. Proteomic analyses have revealed that the majority of EJC components are recruited to the spliceosome prior to the second catalytic step of the splicing reaction (Jurica et al. 2002; Makarov et al. 2002; Reichert et al. 2002; Merz et al. 2007). Immunoprecipitation experiments conducted in a simple in vitro splicing system indicated stepwise binding of successive EJC components to the mRNA (Kataoka and Dreyfuss 2004). The composition of the complex that interacts with the sequence upstream of the exon junction, which corresponds to the EJC-binding site, is dynamically altered during the splicing reaction (Reichert et al. 2002). However, the spliceosomal protein(s) responsible for recruiting EJC components remained to be identified. We recently demonstrated that IBP160, a helicase-like spliceosomal protein, specifically binds ∼40 nt upstream of the intron branch site in the spliceosomal C1 complex. IBP160 binding is not sequence dependent, but is position dependent (Hirose et al. 2006). IBP160 is critical for assembly of the intron-encoded box C/D snoRNP. However, it also binds introns that do not encode a snoRNA, suggesting that the function of IBP160 during pre-mRNA processing is more general than snoRNP biogenesis.
In this paper, we provide evidence that IBP160 is responsible for the initial binding of EJC components to the spliceosome by linking them to the intron in the C1 complex. The association of EJC components with the spliceosome can occur in the absence of the final EJC-binding region in the upstream exon. For two noncoding RNAs (ncRNAs), depletion of IBP160 by RNA interference (RNAi) reduced EJC deposition and resulted in NMD arrest, which is dependent on the integrity of the EJC. These data suggest that a significant role of the intron and the intron-binding protein IBP160 is the initial binding of EJC components to the spliceosome, which is a prerequisite for subsequent assembly of the functional EJC.
Results and Discussion
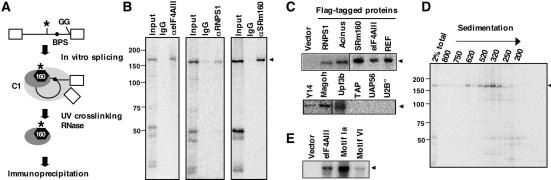
We recently reported that SRm160, an EJC component, is associated with intron-bound IBP160 in the C1 complex (Hirose et al. 2006). This association raises the interesting possibility that the function of IBP160 may be correlated with that of SRm160. Site-specific UV cross-linking enabled us to visualize intron-bound IBP160 in the C1 complex (Fig. 1A) This technique was applied here to examine whether intron-bound IBP160 associates with EJC components other than SRm160. For these experiments, Adenovirus (Adv) pre-mRNA was prepared as a substrate for an in vitro splicing assay. A GG 3′ splice mutation was introduced to arrest the second catalytic step of splicing, resulting in the capture of the C1 complex. Next, the mutated substrate pre-mRNA was specifically 32P-labeled at the −40 nt position relative to the branch site. The 32P-label was then transferred by UV cross-linking to the protein bound to this specific site, IBP160. Thus, we could easily distinguish intron-bound IBP160 from unbound IBP160 or other proteins. Extensive RNase treatment after UV cross-linking releases 32P-IBP160, but maintains its interactions with associated proteins. In this assay, therefore, the association of EJC components with IBP160 can be assessed simply by examining whether the cross-linked (32P-labeled) IBP160 coimmunoprecipitates with a given EJC component. Figure 1B shows that antibodies against SRm160, as well as two other EJC components, eIF4AIII or RNPS1, coimmunoprecipitated the cross-linked IBP160. Further immunoprecipitation experiments using nuclear extracts from HEK293 cells, each expressing a different Flag-tagged EJC component (see Supplementary Fig. 1A), revealed that all the EJC components efficiently coimmunoprecipitated the cross-linked IBP160 (Fig. 1C). These data indicate that the majority of EJC components indirectly bind the intron by interacting with IBP160 in the C1 complex. Similar results were obtained with a β-globin pre-mRNA with a GG 3′ splice site mutation (Supplementary Fig. 2), indicating the generality of this interaction.
Figure 1.
Interaction of EJC components with intron-bound IBP160 in the spliceosomal C1 complex. (A) The experimental design for coimmunoprecipitation of UV-cross-linked IBP160. The 32P-labeled site, located 40 nt upstream of the branchpoint site (BPS) in Adv pre-mRNA, is indicated by an asterisk. The 3′ splice site substituted with GG is shown. The hexagon (black), the smaller oval (dark gray), and the larger oval (light gray) represent IBP160, the IBP160 subcomplex, and the whole spliceosome, respectively. (B,C) Coimmunoprecipitation of the cross-linked IBP160 with antibodies against eIF4AIII, RNPS1, or SRm160 (B), or Flag tag (C). Aliquots of the total samples (10%) were loaded in the input lanes. The EJC component expressed in each HEK293 nuclear extract used is indicated above each lane in C. (D) Size fractionation of the IBP160 subcomplex by gel filtration chromatography (Sephacryl S500HR column). The numbers to the left of the panel represent the molecular weight markers. (E) Coimmunoprecipitation with Flag-eIF4AIII mutants. The mutated motifs are shown above each lane. The arrowheads show the 32P-labeled cross-linked IBP160 in B–E.
In contrast, the association of TAP, an mRNA export receptor that interacts with the EJC (Kataoka et al. 2001; Le Hir et al. 2001), was not detected (Fig. 1C). These results suggest that the entire EJC is not formed in the C1 complex, and that further RNP remodeling upon exon ligation may be required for the formation of the complete EJC. Similarly, UAP56, a component of other complexes that assemble onto mature mRNA (Cheng et al. 2006; Nojima et al. 2007), was not detected (Fig. 1C), indicating that IBP160 interacts specifically with EJC components rather than with other mRNA-binding factors. Most importantly, the association of U2B″, a core protein component of the U2 snRNP, was not detected (Fig. 1C). U2 snRNP interacts with an intronic sequence located up to 25 nt upstream of the branchpoint; therefore, it is likely to be positioned close to IBP160 in the C1 complex. We found that a component of the U2 snRNP-associated SF3b complex could interact with IBP160 (Hirose et al. 2006). This result indicated that the U2 snRNP was disrupted into smaller subcomplexes upon RNase A/T1 treatment after UV cross-linking. Thus, our immunoprecipitations did not merely precipitate the entire spliceosome. This hypothesis was confirmed by our experiment showing that U2, U5, and U6 snRNAs could no longer be detected in the RNase A/T1-treated nuclear extracts that were used for immunoprecipitation (data not shown). Furthermore, size fractionation by gel filtration chromatography of the subcomplex containing cross-linked IBP160 reproducibly indicated the presence of two populations of IBP160 subcomplexes with peaks at 620 kDa and 340 kDa (Fig. 1D). These data support the above results, and indicate that the spliceosomes are disrupted and divided into smaller subcomplexes, and that a subcomplex containing the cross-linked IBP160 transiently associates with EJC components. The other components of the IBP160 subcomplex remain to be identified. The previous proteome analysis suggested that IBP160 is closely linked to the PRP19-associated complex (NTC) (Makarov et al. 2002; Makarova et al. 2004). We also observed that Flag-hPRP19 coprecipitates with the cross-linked IBP160 (data not shown), suggesting that the IBP160 subcomplex may overlap at least in part with the NTC. These results suggest that the intron-bound IBP160 may play an instructive role in the initial association of EJC components with the spliceosome. Alternatively, the recruited components may already be assembled into the intact EJC on the upstream exon EJC-binding site in the C1 complex and secondarily associate with IBP160.
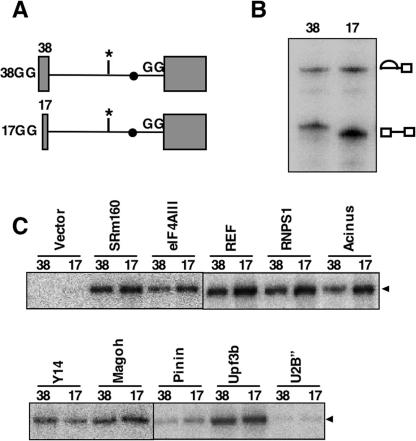
To examine the importance of the upstream exon in the association of EJC components with the C1 complex, two modified Adv-GG substrates with short upstream exons (17 nt: 17GG; 38 nt: 38GG) were employed for in vitro splicing (Fig. 2A). A previous study showed that shortening the upstream exon to 17 nt, but not 38 nt, results in defective EJC formation on the spliced mRNA because of the lack of an EJC-binding site (Le Hir et al. 2001). Consistent with previous reports, we confirmed by RNase H protection and immunoprecipitation that correct EJC assembly took place in our in vitro splicing system (Supplementary Fig. 3). If the EJC were fully assembled and anchored to the upstream exon in the C1 complex, the association of EJC components with the spliceosome would be abolished only with the 17GG construct. We found that the splicing of both 17GG and 38GG proceeded efficiently to the C1 complex (Fig. 2B). Immunoprecipitation of cross-linked IBP160 with each Flag-tagged EJC component clearly showed that shortening the upstream exon to 17 nt did not affect the association of EJC components with IBP160 in the C1 complex (Fig. 2C), indicating that the initial association of EJC components with the spliceosome does not require the upstream exon that eventually serves for the assembly of the intact EJC. A recent proteomic analysis reached the same conclusion (Merz et al. 2007). Furthermore, two mutants of Flag-eIF4AIII (mutations in motif Ia or motif VI) (see Supplementary Fig. 1B), which have lost the ability to induce NMD or EJC formation but retain the ability to associate with the spliceosome (Shibuya et al. 2006), were found to associate with intron-bound IBP160 (Fig. 1E). This supports our hypothesis that the initial binding of EJC components to the spliceosome is tightly linked to their ability to interact with intron-bound IBP160. These observations raise the intriguing possibility that the intron bound to the IBP160 subcomplex is required for subsequent EJC assembly.
Figure 2.
Association of EJC components with the C1 complex in the absence of the final EJC-binding site in the upstream exon. (A) The two shortened pre-mRNA splicing substrates are shown schematically. The lengths of the upstream exons are shown on the left. The labeled nucleotide and the 3′ splice site mutation are shown as in Figure 1A. (B) In vitro splicing of 17GG and 38GG pre-mRNAs. The identities of the RNA species are shown on the right. (C) Coimmunoprecipitation of IBP160 cross-linked either to 38GG or to 17GG, indicated as 38 or 17 above each lane, with the αFlag antibody as in Figure 1C.
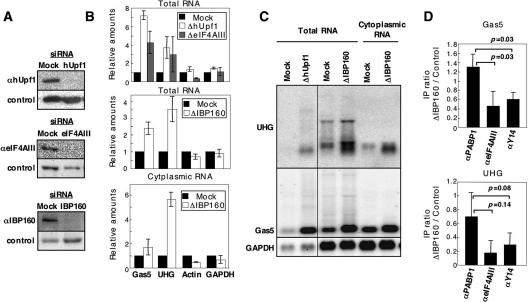
To demonstrate the significance of IBP160 in EJC function, we used small interfering RNA (siRNA) to knock down IBP160 in HeLa cells. Since our recent results indicated that depletion of IBP160 from nuclear extracts destabilizes in vitro spliced exons (Hirose et al. 2006), EJC assembly during in vitro splicing could not be assessed in extracts from IBP160-depleted cells. Instead, we investigated the effect of IBP160 depletion by examining the ability of ΔIBP160 cells to induce NMD. siRNAs against hUPF1, eIF4AIII, or IBP160 were introduced into HeLa cells and were found to efficiently deplete each protein (Fig. 3A). Subsequent quantitative RT–PCR measurement of the levels of two endogenous NMD target ncRNAs (Gas5 and UHG) (Tycowski et al. 1996; Smith and Steitz 1998) indicated that both the Gas5 and UHG ncRNAs were markedly stabilized in ΔhUPF1 (350%∼700%) and ΔeIF4AIII cells (300%∼400%) while the levels of two control mRNAs (β-actin and GAPDH) were unaltered (Fig. 3B). These data confirm that these ncRNAs are genuine NMD targets, and that NMD arrest was induced by defective EJC formation. Importantly, knockdown of IBP160 increased the levels of the two ncRNAs (250%∼350%) as much as those in ΔeIF4AIII cells, while the levels of the control mRNAs were almost unaltered (Fig. 3B). The increase in the levels of the ncRNAs was observed in the cytoplasmic fraction (Fig. 3B), indicating that the increase is not caused by defects in nuclear export. Recently, it has been reported that a subset of shuttling SR proteins (e.g., 9G8 and SRp20) is able to recruit TAP onto the mature mRNA and to enhance mRNA export (Huang et al. 2003). It is possible that these mRNA export adaptors might compensate for any defects in EJC function during nuclear export. IBP160 is a general intron-binding protein, but was previously shown not to be essential for splicing in vitro (Hirose et al. 2006). Northern blot analysis also showed that aberrant accumulation of unspliced RNA species of all four transcripts were not observed in ΔIBP160 cells (Fig. 3C). Taken together, these data strongly suggest that EJC assembly failed in the absence of IBP160, leading to NMD arrest in the ΔIBP160 cells.
Figure 3.
IBP160 is required for efficient EJC association and NMD. (A) Western blot to examine the effects of RNAi. The siRNAs used are indicated above the panels. The antibodies used are shown on the left. (B) Quantitative RT–PCR to measure the levels of two ncRNAs (Gas5 and UHG) and two control mRNAs (Actin and GAPDH) in siRNA-treated cells. The bottom panel shows quantitation of the cytoplasmic RNAs. (C) Northern blot analysis with the RNAs used in B. The slowly migrating bands in the UHG panel are likely to be the splicing intermediates containing certain intron(s). (D) Immunoprecipitation of two ncRNAs (Gas5 and UHG) associated with EJC in the cytoplasm of HeLa cells. The RNA levels immunoprecipitated with αPABP1, αeIF4AIII, and αY14 were measured by quantitative RT–PCR. The ratios of immunoprecipitated RNA efficiencies between control and ΔIBP160 cells were calculated from three independent experiments and graphed. The immunoprecipitation (IP) efficiency is represented as the ratio (percentage) between RNA amounts in immunoprecipitates and input (IP efficiency = RNA amounts in immunoprecipitates/input × 100). The immunoprecipitation efficiency of each antibody from the control cells is as follows: αPABP1, 1.4% for Gas5 and 1.6% for UHG; αeIF4AIII, 0.02% for Gas5 and 0.15% for UHG; αY14, 0.75% for Gas5 and 4.2% for UHG. The statistical analysis of the results between αPABP1 and either αeIF4AIII or αY14 were examined by t-test; each P-value is indicated above the bar.
To demonstrate the role of IBP160 more directly, the levels of EJC-bound cellular RNAs in control and ΔIBP160 cells were compared. EJC-associated cellular RNAs were obtained by coimmunoprecipitating the cytoplasmic fraction of siRNA-treated HeLa cells experiments with αeIF4AIII or αY14 antibodies. Since EJC is obliterated by the translating ribosome upon mRNA export to the cytoplasm (Dostie and Dreyfuss 2002), it is difficult to obtain substantial amounts of EJC-associated cytoplasmic mRNAs. We chose UHG and Gas5 ncRNAs to capture the cytoplasmic ribonucleoprotein complexes associated with EJC, because we expected that EJC would remain associated with these ncRNAs, which possess translatable small ORFs close to their 5′ termini (see Supplementary Fig. 4). Coimmunoprecipitation of Gas5 and UHG ncRNAs with eIF4AIII and Y14 revealed that the EJC association was remarkably reduced in ΔIBP160 cells, while that of PABP1 remained relatively constant (Fig. 3D). These data additionally suggest that the intron-mediated primary association of EJC with the spliceosome is a prerequisite for EJC assembly onto the ligated exon.
Our immunoprecipitation experiments indicated that IBP160 remains associated with the lariat intron but may dissociate prior to debranching of the lariat structure, since lariats, but not linearlized intron fragments, were detected in the IBP160 precipitates (Supplementary Fig. 5A). In contrast, association of SRm160 with the lariat intron was poorly detected. Instead, it was found to stably associate with the ligated exon (Supplementary Fig. 5B). These data suggest that remodeling of the IBP160 subcomplex takes place upon exon ligation, resulting in the production of distinct post-splicing complexes containing the ligated exon and the lariat intron (Fig. 4). It has been reported that hPRP22 plays a critical role in the release of the ligated exon from the spliceosome (Ohno and Shimura 1996). We observed that Flag-hPRP22 efficiently coprecipitated the cross-linked IBP160 (Supplementary Fig. 6). This interaction requires the intact RS domain of hPRP22, which was previously reported to be necessary for association with the spliceosome (Ohno and Shimura 1996). This result suggests that hPRP22 is another component of the IBP160 subcomplex that may be involved in remodeling of the IBP160 subcomplex for the release of ligated exons. It would be intriguing to try similar coimmunoprecipitation experiments using cross-linked IBP160 with EJC components in the presence of previously reported dominant-negative hPRP22 mutants (Ohno and Shimura 1996). We reported previously that the functional EJC was deposited by the U12-type (minor) spliceosome (Hirose et al. 2004). Our immunoprecipitation of RNA species produced by in vitro splicing of P120 pre-mRNA containing a U12-type intron shows that IBP160 is associated with the lariat U12-type intron (Supplementary Fig. 5C). This suggests that the role played by IBP160 in EJC formation is a common feature of splicing by both the U2- and U12-type spliceosomes. In summary, we have demonstrated an interaction of EJC components with the intron-bound spliceosomal subcomplex containing IBP160 in the C1 stage prior to exon ligation. This initial binding of EJC components to the intron is likely to be essential for the assembly of the functional EJC that induces NMD (Fig. 4). Our data provide a novel mechanistic insight into the role of introns during mRNA biogenesis.
Figure 4.
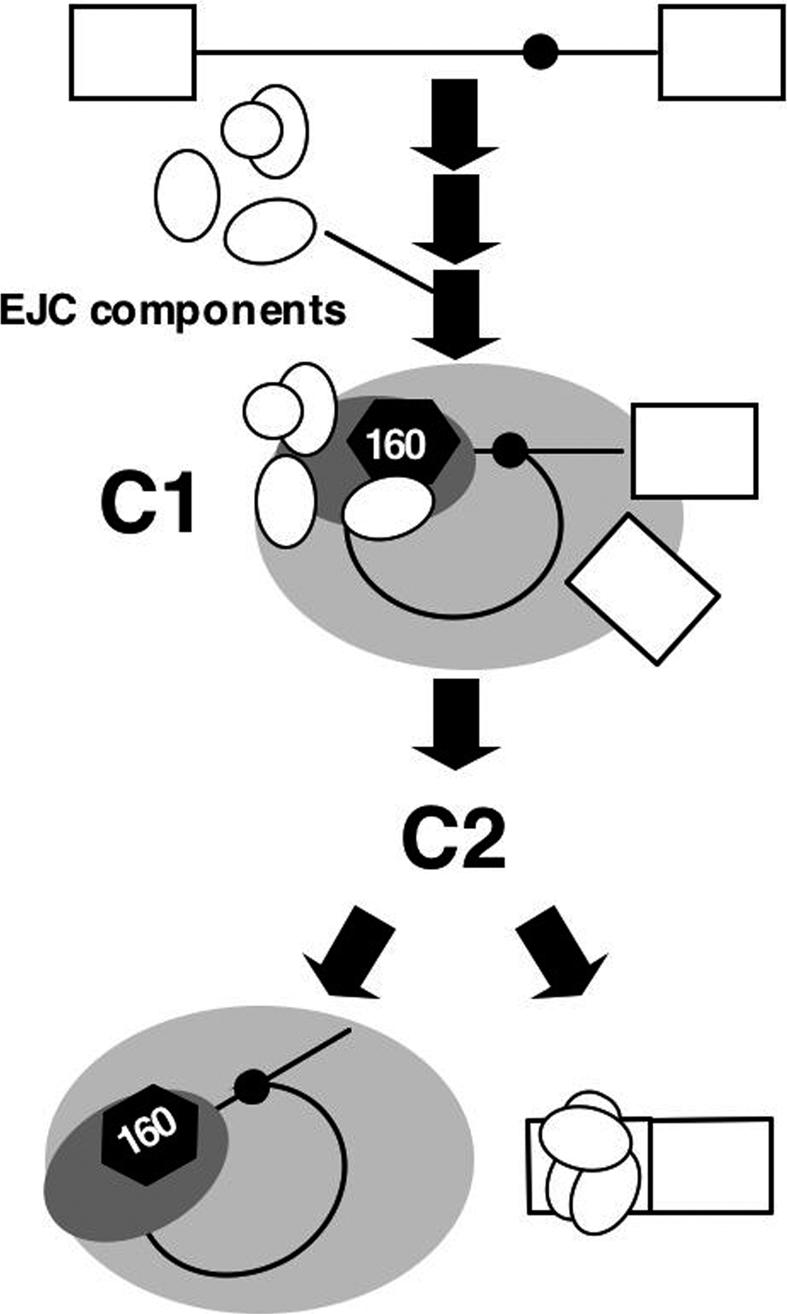
Model of EJC formation during pre-mRNA splicing. The white ovals represent EJC components. Other symbols are as shown in Figure 1A.
Materials and methods
Plasmid construction
The Adv, β-globin, Gas5, and P120 splicing substrates have been described previously (Hirose et al. 2004, 2006). The 38GG and 17GG Adv mutants were constructed by PCR cloning. Flag-RNPS1, Flag-Y14, Flag-REF, and Flag-Magoh were constructed by Jens Lykke-Andersen (Lykke-Andersen et al. 2001). Other Flag-tagged protein constructs were obtained by cloning PCR-amplified cDNA fragments into the pcDNA-Flag vector. The eIF4AIII mutants were constructed using the QuikChange Site-Directed mutagenesis kit (Stratagene).
Site-specific labeling of RNA
Site-specific labeling of splicing substrates has been described previously (Hirose et al. 2006). In brief, for labeling at −40 nt relative to the branch site of the Adv intron or β-globin intron, PCR was used to obtain the template DNA for either the upstream RNA or the downstream RNA relative to the site to be labeled. These PCR fragments were used as templates for in vitro transcription with T7 RNA polymerase. For the upstream RNA, transcription was performed in the presence of an RNA cap analog (Invitrogen). For the downstream RNA, GMP was added into the transcription reaction. The 5′ terminus of the purified downstream RNA was dephosphorylated with CIP, followed by 32P-labeling with T4 polynucleotide kinase. The capped upstream RNA (10 pmol) was ligated to the 32P-5′-end-labeled downstream RNA (15 pmol) with T4 DNA ligase (50 U; Roche) in the presence of a bridging DNA oligonucleotide (30 mer, 15 pmol) for 3 h at 25°C. The ligated RNA was separated from the unligated RNAs and purified by PAGE.
Preparation of nuclear extracts, in vitro splicing, and UV cross-linking
Nuclear extracts from HeLa cells were prepared by standard procedures (Dignam et al. 1983). Nuclear extracts containing Flag-tagged proteins were prepared from HEK293 cells transfected with a pcDNA3-Flag plasmid as described (Hirose et al. 2004). The expression of each Flag-tagged protein was confirmed by the appearance of a single band in Western blots using αFlag antibody (M2; Sigma). In vitro splicing reactions and UV cross-linking were carried out as described previously (Hirose et al. 2006). In brief, pre-mRNAs with site-specific 32P labeling (100 fmol) were incubated in 20 μL of reaction mixtures containing 2.4 mM MgCl2, 0.5 mM ATP, 20 mM creatine phosphate, 2% polyvinyl alcohol, and 12 μL of nuclear extract. For splicing in the presence of Flag-tagged proteins, equal volumes of HeLa nuclear extract (6 μL) and Flag-tagged protein-containing HEK293 nuclear extract (6 μL) were mixed. In vitro splicing of the P120 substrate with an U12-type intron was carried out as described (Tarn and Steitz 1996). UV cross-linking was carried out after incubating under conditions of in vitro splicing. UV light (1.8 J/cm2) was applied to the open-top reaction tube on ice using a UV cross-linker CL-1000 (UVP). RNase A and RNase T1 were added and incubated for 15 min at 37°C, followed by precipitation with 50% acetone. Precipitated proteins were dissolved in the SDS loading buffer and fractionated by SDS-PAGE. An autoradiographic image was captured and quantified using a Fuji BAS2500 Bio-imaging analyzer. For gel filtration analysis, the UV-cross-linked sample was loaded onto a Sephacryl S500HR column (Amersham) equilibrated with NET2 buffer (20 mM Tris-HCl at pH 7.9, 150 mM KCl, 0.5% NP40) containing 2 mM MgCl2. Molecular weight standards were analyzed in parallel under the same conditions.
Antibodies and immunoprecipitation
Antibodies were purchased or provided as follows: anti-Flag M2 and anti-PABP1 from Sigma, anti-SRm160 from B. Blencowe, anti-eIF4AIII from G. Dreyfuss, and anti-RNPS1 from A. Mayeda. Anti-IBP160 was obtained as antiserum against the synthetic peptide as described (Hirose et al. 2006). For coimmunoprecipitation experiments, the anti-SRm160 monoclonal antibodies were bound to protein A-Sepharose (PAS) beads via rabbit anti-mouse IgG + IgM (Pierce). All other antibodies were bound directly to Dynabeads protein G (Invitrogen). For immunoprecipitation, the in vitro splicing reaction was fivefold larger (100 μL). The reaction mixture was diluted 10-fold with NET2 buffer immediately after RNase treatment and mixed with the antibody-Dynabeads conjugates for 3 h at 4°C. The beads were washed four times with NET2 buffer using an automatic magnetic bead washer (Thermo). Bound proteins were eluted by directly adding SDS loading buffer to the beads. For immunoprecipitation of the EJC-associated cellular RNAs, the cytoplasmic fraction of HeLa cells (∼2 × 107 cells) was prepared as described (Dostie and Dreyfuss 2002) and mixed with the antibody-Dynabeads conjugates for 3 h at 4°C. The beads were washed four times with HNT buffer (Dostie and Dreyfuss 2002) using an automatic magnetic bead washer (Thermo). Precipitated RNAs were recovered using Sepasol-RNAI (Nacalai).
RNAi, Northern blot, and quantitative RT–PCR
HeLa cells were grown to 30%–50% confluency in six-well tissue culture dishes. The siRNAs for IBP160 (Hirose et al. 2006), hUPF1 (Mendell et al. 2002), or eIF4AIII (Shibuya et al. 2004) were synthesized by Sigma Genosys, Inc. The control siRNA was purchased from Ambion. The siRNA duplex (50 nM) was administered to the HeLa cells using Lipofectamine 2000 reagent according to the manufacturer’s instructions (Invitrogen). After 48 h (68 h for the ΔIBP160 cells), total RNA and cytoplasmic RNA were prepared using TRIzol reagent or a PARIS kit, respectively. Quantitative RT–PCR and Northern blotting were carried out as described previously (Sasaki et al. 2007).
Acknowledgments
We thank J.A. Steitz, K. Watanabe, T. Kin, and N. Kataoka for useful discussions and encouragement. We thank J. Lykke-Andersen, B. Blencowe, A. Mayeda, and G. Dreyfuss for providing antibodies or plasmid constructs. We also thank M. Nagai for technical assistance. This research was supported by a PRESTO program grant from the Japan Science and Technology agency (JST) (to T.H.), a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) (to T.H.), a grant from the National Institute of Biomedical Innovation (to T.I. and M.H.), and a grant from the New Energy and Industrial Technology Development Organization (NEDO) (to T.H.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1557907
References
- Ballut L., Marchadier B., Baguet A., Tomasetto C., Seraphin B., Le Hir H., Marchadier B., Baguet A., Tomasetto C., Seraphin B., Le Hir H., Baguet A., Tomasetto C., Seraphin B., Le Hir H., Tomasetto C., Seraphin B., Le Hir H., Seraphin B., Le Hir H., Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Cheng H., Dufu K., Lee C.S., Hsu J.L., Dias A., Reed R., Dufu K., Lee C.S., Hsu J.L., Dias A., Reed R., Lee C.S., Hsu J.L., Dias A., Reed R., Hsu J.L., Dias A., Reed R., Dias A., Reed R., Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz R.M., Roeder R.G., Lebovitz R.M., Roeder R.G., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Dreyfuss G., Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- Hirose T., Shu M.-D., Steitz J.A., Shu M.-D., Steitz J.A., Steitz J.A. Splicing of U12-type introns deposits an exon junction complex competent to induce nonsense-mediated mRNA decay. Proc. Natl. Acad. Sci. 2004;101:17976–17981. doi: 10.1073/pnas.0408435102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Ideue T., Nagai M., Hagiwara M., Shu M.-D., Steitz J.A., Ideue T., Nagai M., Hagiwara M., Shu M.-D., Steitz J.A., Nagai M., Hagiwara M., Shu M.-D., Steitz J.A., Hagiwara M., Shu M.-D., Steitz J.A., Shu M.-D., Steitz J.A., Steitz J.A. A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Mol. Cell. 2006;23:673–684. doi: 10.1016/j.molcel.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Huang Y., Gattoni R., Stevenin J., Steitz J.A., Gattoni R., Stevenin J., Steitz J.A., Stevenin J., Steitz J.A., Steitz J.A. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Jurica M.S., Licklider L.J., Gygi S.R., Grigorieff N., Moore M.J., Licklider L.J., Gygi S.R., Grigorieff N., Moore M.J., Gygi S.R., Grigorieff N., Moore M.J., Grigorieff N., Moore M.J., Moore M.J. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N., Dreyfuss G., Dreyfuss G. A simple whole cell lysate system for in vitro splicing reveals a stepwise assembly of the exon–exon junction complex. J. Biol. Chem. 2004;279:7009–7013. doi: 10.1074/jbc.M307692200. [DOI] [PubMed] [Google Scholar]
- Kataoka N., Diem M.D., Kim V.N., Yong J., Dreyfuss G., Diem M.D., Kim V.N., Yong J., Dreyfuss G., Kim V.N., Yong J., Dreyfuss G., Yong J., Dreyfuss G., Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon–exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Kataoka N., Dreyfuss G., Kataoka N., Dreyfuss G., Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde E., Maquat L.E., Moore M.J., Izaurralde E., Maquat L.E., Moore M.J., Maquat L.E., Moore M.J., Moore M.J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield D., Izaurralde E., Moore M.J., Gatfield D., Izaurralde E., Moore M.J., Izaurralde E., Moore M.J., Moore M.J. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu M.-D., Steitz J.A., Shu M.-D., Steitz J.A., Steitz J.A. Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Makarov E.M., Makarova O.V., Urlaub H., Gentzel M., Will C.L., Wilm M., Luhrmann R., Makarova O.V., Urlaub H., Gentzel M., Will C.L., Wilm M., Luhrmann R., Urlaub H., Gentzel M., Will C.L., Wilm M., Luhrmann R., Gentzel M., Will C.L., Wilm M., Luhrmann R., Will C.L., Wilm M., Luhrmann R., Wilm M., Luhrmann R., Luhrmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- Makarova O.V., Makarov E.M., Urlaub H., Will C.L., Gentzel M., Wilm M., Luhrmann R., Makarov E.M., Urlaub H., Will C.L., Gentzel M., Wilm M., Luhrmann R., Urlaub H., Will C.L., Gentzel M., Wilm M., Luhrmann R., Will C.L., Gentzel M., Wilm M., Luhrmann R., Gentzel M., Wilm M., Luhrmann R., Wilm M., Luhrmann R., Luhrmann R. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 2004;23:2381–2391. doi: 10.1038/sj.emboj.7600241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Reed R., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., ap Rhys C.M., Dietz H.C., ap Rhys C.M., Dietz H.C., Dietz H.C. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- Merz C., Urlaub H., Will C.L., Luhrmann R., Urlaub H., Will C.L., Luhrmann R., Will C.L., Luhrmann R., Luhrmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13:116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T., Hirose T., Kimura H., Hagiwara M., Hirose T., Kimura H., Hagiwara M., Kimura H., Hagiwara M., Hagiwara M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 2007;282:15645–15651. doi: 10.1074/jbc.M700629200. [DOI] [PubMed] [Google Scholar]
- Ohno M., Shimura Y., Shimura Y. A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes & Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- Reichert V.L., Le Hir H., Jurica M.S., Moore M.J., Le Hir H., Jurica M.S., Moore M.J., Jurica M.S., Moore M.J., Moore M.J. 5′ Exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes & Dev. 2002;16:2778–2791. doi: 10.1101/gad.1030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y.T.F., Sano M., Kin T., Asai K., Hirose T., Sano M., Kin T., Asai K., Hirose T., Kin T., Asai K., Hirose T., Asai K., Hirose T., Hirose T. Identification and characterization of human non-coding RNAs with tissue-specific expression. Biochem. Biophys. Res. Commun. 2007;357:991–996. doi: 10.1016/j.bbrc.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Shibuya T., Tange T.O., Sonenberg N., Moore M.J., Tange T.O., Sonenberg N., Moore M.J., Sonenberg N., Moore M.J., Moore M.J. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- Shibuya T., Tange T.O., Stroupe M.E., Moore M.J., Tange T.O., Stroupe M.E., Moore M.J., Stroupe M.E., Moore M.J., Moore M.J. Mutational analysis of human eIF4AIII identifies regions necessary for exon junction complex formation and nonsense-mediated mRNA decay. RNA. 2006;12:360–374. doi: 10.1261/rna.2190706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.M., Steitz J.A., Steitz J.A. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange T.O., Nott A., Moore M.J., Nott A., Moore M.J., Moore M.J. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Tange T.O., Shibuya T., Jurica S., Moore M.J., Shibuya T., Jurica S., Moore M.J., Jurica S., Moore M.J., Moore M.J. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn W.Y., Steitz J.A., Steitz J.A. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT–AC) intron in vitro. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- Tycowski K.T., Shu M.-D., Steitz J.A., Shu M.-D., Steitz J.A., Steitz J.A. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]