Abstract
Binding of microRNA (miRNA) to mRNA within the RNA-induced silencing complex (RISC) leads to either translational inhibition or to destruction of the target mRNA. Both of these functions are executed by Argonaute 2 (Ago2). Using hematopoiesis in mice as a model system to study the physiological function of Ago2 in vivo, we found that Ago2 controls early development of lymphoid and erythroid cells. We show that the unique and defining feature of Ago2, the Slicer endonuclease activity, is dispensable for hematopoiesis. Instead, we identified Ago2 as a key regulator of miRNA homeostasis. Deficiency in Ago2 impairs miRNA biogenesis from precursor-miRNAs followed by a reduction in miRNA expression levels. Collectively, our data identify Ago2 as a highly specialized member of the Argonaute family with an essential nonredundant Slicer-independent function within the mammalian miRNA pathway.
Keywords: Ago2, Slicer, microRNA, hematopoeisis
MicroRNAs (miRNAs) are genome-encoded 21- to 23-base-pair (bp) RNA molecules that post-transcriptionally regulate gene expression through the inhibition of translation and/or stability of target mRNAs. MiRNA-mediated gene silencing is executed by the multiprotein RNA-induced silencing complex (RISC). At the core of RISC are Dicer and an Argonaute (Ago) protein that interact to generate miRNA and execute their function. Dicer cleaves miRNA from its precursors (Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001; Knight and Bass 2001), whereas Ago proteins (Ago1–4) bind miRNA (Lingel et al. 2003; Song et al. 2003; Yan et al. 2003) and mediate gene silencing (Hammond et al. 2001; Hutvagner and Zamore 2002). The mechanism of RISC-mediated gene silencing depends on the degree of complementarity between the miRNA and its target. Binding of miRNA–RISC to a partially complementary mRNA results in silencing through the inhibition of translation or mRNA degradation by catabolism within processing bodies (for review, see Pillai et al. 2007). In turn, direct cleavage or “slicing” of mRNA that is solely catalyzed by Ago2 (Liu et al. 2004; Meister et al. 2004; Song et al. 2004) requires perfect or near-perfect complementarity between the miRNA and the target (Hutvagner and Zamore 2002; Doench et al. 2003). To date, a single mammalian miRNA has been shown to direct Ago2-mediated slicing (Mansfield et al. 2004; Yekta et al. 2004); however, the ability of Ago2 to slice significantly mismatched targets in vitro indicates that the complementarity rules may not be as stringent as suggested (Martinez and Tuschl 2004). While the function of Ago2’s Slicer activity as the catalytic engine that powers RNA interference (RNAi) is understood, the general in vivo physiological importance of Ago2 and its endonuclease activity within the miRNA pathway remains largely unknown.
Results and Discussion
The potent regulatory function of miRNA in hematopoiesis (Chen et al. 2004; Cobb et al. 2005; Fazi et al. 2005; Muljo et al. 2005) makes this process attractive for the investigation of the physiological significance of Ago2. We therefore studied the development of the lymphoid and erythroid lineages derived from Ago2−/− bone marrow cells. The latter cells were produced by inducible inactivation of the Ago2 gene in bone marrow progenitors followed by the generation of Ago2−/− hematopoietic system in lethally irradiated wild-type mice (Supplementary Fig. S1). This approach enables the analysis of the bone marrow cell-intrinsic Ago2 function. Mice reconstituted with Ago2−/− or control Ago2fl/fl bone marrow cells are hereafter referred to as Ago2−/− or Ago2fl/fl mice, respectively.
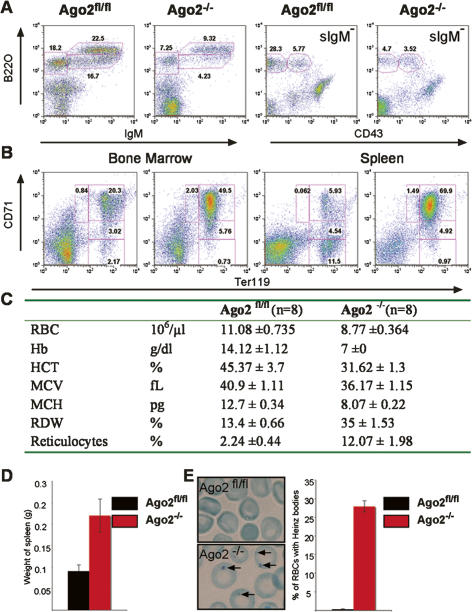
Analysis of mice 6–8 wk after reconstitution revealed that Ago2−/− bone marrow cells give rise to hematopoietic cells of various lineages (data not shown). However, the differentiation of the B lymphoid as well as erythroid lineages diverges significantly from the wild-type developmental pattern. Deficiency in Ago2 does not interfere with the generation of early pro-B (B220loCD43+IgM−) cells in the bone marrow, but affects further pre-B (B220loCD43−IgM−) cell differentiation and the subsequent generation of peripheral B cells (Fig. 1A; Table 1). Among various hematopoietic lineages in the bone marrow, erythroid cells appear to be most affected. The maturation of the Ago2−/− erythroid precursors is severely impaired. This developmental defect is characterized by a dramatic increase in the frequencies and numbers of immature Ter119hi, CD71hi basophilic erythroblasts and decrease in frequencies and numbers of Ter119hi, CD71neg mature orthochromatophilic erythroblasts in the bone marrow and spleens of Ago2−/− mice (Fig. 1B; Table 1). Ago2−/− mice display erythroid hyperplasia in both the bone marrow and spleen (Fig. 1B). The combination of ineffective erythropoiesis and erythroid hyperplasia results in splenomegaly in Ago2−/− mice (Fig. 1D). The defective erythropoiesis in the absence of Ago2 fails to generate fully functional red blood cells (RBCs). Ago2−/− mice develop severe anemia characterized by a reduction in the overall number of erythrocytes as well as the amount of hemoglobin per erythrocyte (Fig. 1C). Ago2−/− RBCs, but not control erythrocytes, contained Heinz bodies, inclusion bodies that contain Hb precipitates (Fig. 1E). The same phenotype was observed at 3 mo after reconstitution (data not shown). Therefore, these alterations do not represent a stress response to the transfer but rather reflects the long lasting consequence of Ago2 deficiency.
Figure 1.
Ago2 is required for normal hematopoiesis. Representative FACS analysis are shown of B lymphoid in the bone marrow (A) and erythroid cell populations in the bone marrow and spleen (B) of Ago2fl/fl and Ago2−/− mice. Numbers indicate the percentages of cells of the developmentally defined subpopulations. (C) Ago2 deficiency alters erythrocyte parameters in the peripheral blood. (RBC) Red blood cell; (Hb) hemoglobin; (HCT) hematocrit; (MCV) mean cell volume; (MCH) mean corpuscular hemoglobin; (RDW) red cell distribution width. (D) Splenomegaly in Ago2−/− mice. Splenomegaly was determined by the weight of spleens. Bars represent mean values (n = 8), and error bars indicate standard deviation. (E) Heinz bodies (indicated by arrows) identified by staining of blood smears (images) with methylene blue and counted by light microscopy. Bars represent mean percentages of Hz bodies in the respective genotypes (n = 4), and error bars indicate standard deviation.
Table 1.
B and erythroid precursor numbers in Ago2−/− mice
Numbers of B-lymphoid cell populations in the bone marrow and erythroid cell populations in the bone marrow and spleen of Ago2fl/fl and Ago2−/− mice. The frequency of the fraction of the nucleated cells was determined by flow cytometry and was used to calculate the total number of cells. Data shown are means in units of 106 cells with standard deviations from n = 4 mice. The significance, P value, derived from a t-test, is shown.
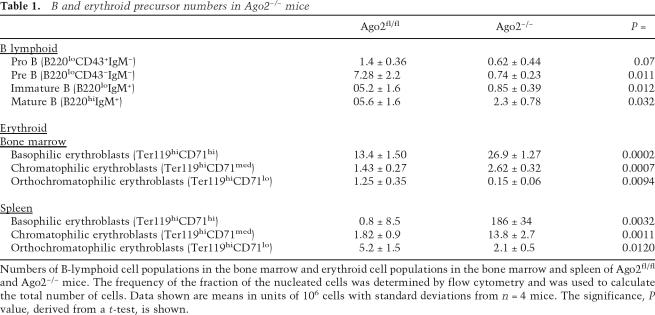
To directly test the physiological significance of “slicing,” we evaluated the ability of bone marrow cells deficient for Ago2’s “Slicer” endonuclease activity to support erythropoiesis in the bone marrow. For these purposes, we employed the Ago2D669A point mutant that is endonuclease inactive in vitro (Liu et al. 2004) and in vivo (Supplementary Fig. S2). Ago2−/− bone marrow cells were complemented with exogenous wild-type or Slicer-inactive Ago2 and used to generate bone marrow chimeras. Exogenous wild-type Ago2 and Slicer-inactive Ago2 were expressed at similar levels in both the bone marrow and bone marrow-derived splenic cells of reconstituted mice (Supplementary Fig. S3A). Most importantly, expression of either wild-type Ago2 or Slicer-inactive Ago2 could cure the impaired erythropoiesis and anemia caused by Ago2 deficiency. The expression of Slicer-inactive Ago2 restored the wild-type pattern of erythroid development in the bone marrow and spleen, rescuing ineffective erythropoiesis, erythroid hyperplasia, and splenomegaly (Fig. 2A; Supplementary Fig. S3C). In addition, expression of the Slicer-inactive Ago2 in Ago2−/− cells restored erythroid parameters and prevented Heinz body formation to the same degree as exogenous wild-type Ago2 (Fig. 2C; Supplementary Fig. S3B). Similar to the erythroid cells, expression of Slicer inactive Ago2 was able to support normal B-cell development in the bone marrow (Fig. 2B). These results show the dispensable role of Ago2’s slicer function in regulation of erythropoiesis and B-cell lymphopoiesis.
Figure 2.
Ago2 control of hematopoiesis is Slicer independent. (A,B) Slicer-inactive Ago2 supports wild-type B-cell development and erythropoiesis. Representative FACS profiles of erythroid (A) and B-lymphoid (B) populations derived from bone marrow chimeras generated with Ago2−/− cells transduced with control vector (Migr), exogenous Ago2 (Migr-Ago2), or slicer-inactive Ago2 (Migr-Ago2D669A). Transduced cells identified by the expression of GFP are shown. Numbers indicate the percentages of gated cells. (C) Slicer-inactive Ago2 normalizes erythrocyte parameters in the peripheral blood. The table displays hematological parameters of the bone marrow-derived blood cells. The frequencies (percentage of live cells) of bone marrow GFP+ cells in bone marrow chimeras used for the analysis are shown. (RBC) Red blood cell; (Hb) hemoglobin; (HCT) hematocrit; (MCV) mean cell volume; (MCH) mean corpuscular hemoglobin; (RDW) red cell distribution width.
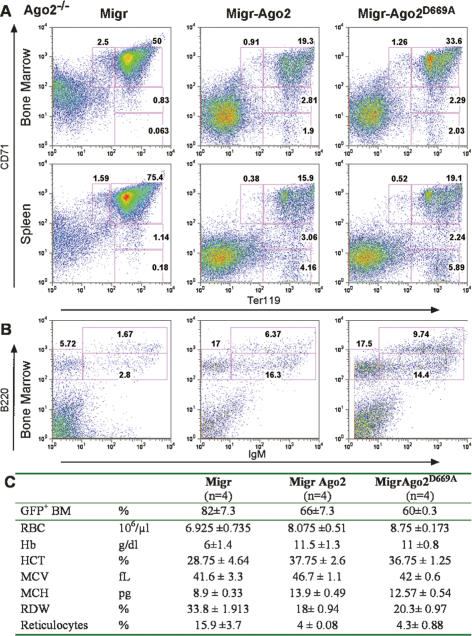
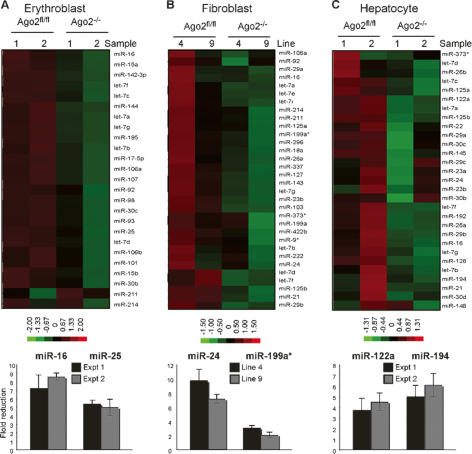
The irrelevance of Ago2-mediated slicing in hematopoiesis suggested an additional and essential role for Ago2 in the miRNA pathway. Earlier studies pointed to the potential involvement of Ago2 in miRNA homeostasis. Ago2 directly binds miRNA and is a component of the ribonucleoprotein complex that mediates miRNA biogenesis (Chendrimada et al. 2005; Gregory et al. 2005; Haase et al. 2005; Maniataki and Mourelatos 2005; Meister et al. 2005; Lee et al. 2006). To address the role of Ago2 in regulation of miRNA homeostasis, we measured the expression levels of miRNAs in Ago2−/− erythroblasts. Using miRNA microarray analysis, we observed a reduction in the levels of miRNAs in Ago2−/− erythroblasts compared with Ago2fl/fl controls (Fig. 3A). This global reduction in miRNA expression was confirmed using quantitative RT–PCR (qRT–PCR) on two representative miRNAs, and is exemplified by a five- to eightfold reduction of the respective miRNAs in Ago2−/− erythroblasts (Fig. 3A). The reduced miRNA in the absence of Ago2−/− is a general feature of Ago2 deficiency and not a result of defective erythroid differentiation per se. Similar to Ago2−/− erythroblasts both Ago2−/− fibroblasts and hepatocytes display a reduction in miRNA expression levels (Fig. 3B,C).
Figure 3.
Ago2 controls miRNA expression levels. Reduced miRNA expression levels in the absence of Ago2. The levels of miRNA expression in Ago2fl/fl and Ago2−/− erythroblasts (A), fibroblasts (B), or hepatocytes (C) were measured by miRNA array profiling (top) and qRT–PCR (bottom). (Top panels) Heat maps of miRNA expression from the Ago2fl/fl and Ago2−/− cells are shown. Two independent preparations (Sample 1 and 2) or, in the case of fibroblasts, cell lines 4 and 9, are shown. Increased or decreased amount of miRNA relative to the median are indicated in red and green, respectively; normalized fold changes are indicated in the color bar. Ranges in the color bar refer to gene standard deviations. (Bottom panels) Expression levels of two representative miRNAs were quantified by qRT–PCR. The U6 normalized expression of miRNAs in mutant cells is plotted as fold reduction over the level of miRNA expression in Ago2fl/fl cells. The mean and standard deviation of three independent measurements, each made in duplicate, are shown.
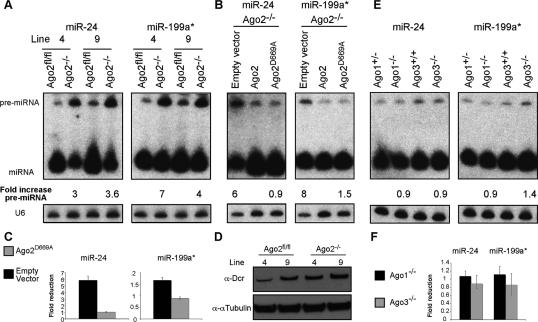
The arrested biogenesis of miRNA caused by the absence of Dicer is associated with accumulation of some but not all pre-miRNAs (Yi et al. 2006). Therefore, the accumulation of pre-miR-24 and pre-miR-199a in Ago2−/− fibroblasts, two pre-miRNAs that are readily detectible in wild-type fibroblasts, indicates a crucial role for Ago2 in miRNA biogenesis (Fig. 4A). The impaired miRNA biogenesis is independent of Ago2’s Slicer activity, as complementation of Ago2−/− cells with slicer-inacitve Ago2 restores miRNA biogenesis (Fig. 4B,C). The dispensability of Ago2’s Slicer activity in miRNA biogenesis is supported by the fact that Slicer-inactive Ago2 interacts with Dicer in vivo, and this protein complex possesses pre-miRNA processing activity in vitro (Maniataki and Mourelatos 2005). The defective miRNA biogenesis cannot be attributed to the reduced expression of Dicer that is expressed at normal levels in Ago2−/− cells (Fig. 4D). These data suggest that absence of Ago2 from RISC hampers either access of Dicer to pre-miRNA or reduces its overall enzymatic activity. The interaction of Ago2 with Dicer through the TRBP and PACT couples miRNA biogenesis with the loading of RISC. It is possible that the absence of Ago2 impairs RISC function in a fashion similar to RISC malfunction caused by reduced expression of PACT or TRBP (Chendrimada et al. 2005; Gregory et al. 2005; Haase et al. 2005; Lee et al. 2006). We demonstrate that Ago2 has an earlier function within RISC in facilitating miRNA biogenesis, this is also supported by the fact that both TRBP and PACT deficient mice, in contrast to Ago2, are viable and display minor phenotypic alterations (Zhong et al. 1999; Rowe et al. 2006). The reduced levels of miRNAs in Ago2−/− cells indicate that the other Argonautes cannot compensate for the loss of Ago2. Indeed, the negligible role of these proteins in miRNA homeostasis and hematopoiesis was confirmed in Ago1−/− or Ago3−/− cells and mice, respectively (Fig. 4E,F; Supplementary Fig. S4; U. Koenig, D. O’Carroll, and A. Tarakhovsky, in prep.)
Figure 4.
Ago2 deficiency selectively impairs miRNA biogenesis. (A) Pre-miRNA accumulation in Ago2−/− fibroblast cell lines. The expression levels of pre-miRNA and miRNA were measured by Northern blotting using probes specific for miR-24 and miR-199a*. U6 snRNA was used as a loading control. The positions of pre-miRNA and miRNA are indicated. The U6 normalized fold increase in the pre-miRNA in Ago2−/− cells is shown. (B,C) Ago2D669A restores pre-miRNA processing and miRNA expression levels in fibroblasts. (B) Pre-miR-24 and pre-miR-199a were measured and presented as in A. The U6 normalized fold increase in pre-miRNA expression levels in the indicated Ago2−/− reconstituted cell lines relative to Ago2−/− cells complemented with wild-type Ago2 are indicated. (C) Expression levels of two representative miRNAs were quantified by qRT–PCR. The U6 normalized expression of miRNAs in Ago2−/− cells complemented as indicated is plotted as fold reduction over the level of miRNA expression in Ago2−/− cells complemented with wild-type Ago2. The mean and standard deviation of three independent measurements, each made in duplicate, are shown. (D) Unaltered expression of Dicer in the absence of Ago2. The expression of Dicer in Ago2−/− fibroblasts was confirmed by immunoblotting with an α-Dcr antibody. Protein loading was controlled with α-αTubulin antiserum. (E,F) Normal expression of pre-miRNA and miRNA in Ago1- and Ago3-deficient fibroblast cell lines. (E) Total RNA was isolated from Ago1+/−, Ago1−/−, Ago3+/+, and Ago3−/− fibroblast cell lines. The expression levels of pre-miR-24 and pre-miR-199a were measured by Northern blotting and presented as above. (F) Expression levels of two representative miRNAs were quantified by qRT–PCR. The U6 normalized expression of miRNAs in the Ago1−/− and Ago3−/− cells is plotted as fold reduction over the level of miRNA expression in the respective control cell lines.
Our data identify Ago2 as a key regulator of B lymphoid and erythroid development and function. We suggest that the hemaopoieitic defects arise from a reduced threshold of miRNA-mediated gene silencing due to the overall reduction of miRNAs and the loss of Ago2-mediated translational control. The fact that the Slicer activity of Ago2 is not required for erythropoiesis as well as the development of lymphoid cells provides genetic evidence that mRNA cleavage does not play a general role in the regulation of miRNA-mediated gene silencing in vivo. Our data demonstrate that Ago2 is highly specialized member of the Argonaute family with a crucial function within RISC in the regulation of miRNA homeostasis. This may be analogous to Argonautes in Caenorhabditis. elegans, where recent studies have revealed that distinct Argonautes have specialized functions and act sequentially in the RNAi pathway (Yigit et al. 2006).
Materials and methods
Generation of an Ago2 conditional allele in mice
The Ago2 locus contains 16 exons. The targeting strategy allows Cre-mediated deletion of exons 9–11. This deletion excludes in-frame splicing of exon 8 to exons 12–16, and leads to the functional inactivation of Ago2 due to partial deletion of the PAZ domain and loss of RNase activity. In addition, according to the rules of RNA surveillance (Hentze and Kulozik 1999), the in-frame stop codons generated by frame-shift mutations introduced by the deletion of exons 9–11 is likely to destabilize the resulting primary transcript RNA and prevent production of a truncated Ago2 protein. The Ago2 gene targeting vector (pDTA–TK–Ago2) carries a neomycin resistance (neor) gene flanked with two frt sites, a loxP site (neor–frt2–loxP cassette) located 3′ of exon 11 and a loxP site (5′loxP) within the intron between exons 8 and 9. The Ago2 targeting construct was transfected into E14.1 embryonic stem (ES) cells. Southern blotting of the individual ES cell clone-derived genomic BamHI-digested DNA with probe 12i was used to identify homologous recombinants. A 4.2-kb DNA fragment corresponds to the wild-type Ago2 locus, and integration of the neor–frt2–loxP cassette 3′ of exon 11 introduces an additional BamHI site, thus increasing the size of the BamHI DNA fragment recognized by probe 12i to 5.0 kb. Targeted ES cells were used to generate mice heterozygous for the Ago2-targeted allele. These mice were then crossed to the FLP-expressing transgenic mice (FLPeR) (Farley et al. 2000) to remove the frt-flanked neor cassette, resulting in the generation of Ago2 loxP allele (Ago2 fl allele).
Generation of Ago2−/− bone marrow and analysis of bone marrow chimeras
To inactivate the Ago2 gene in the bone marrow, Ago2fl/fl mice (6–12 wk of age) that carry the MxCre transgene (Kuhn et al. 1995) (Ago2fl/fl;MxCre mice) received three i.p. injections of 250 μg per mouse of pI:pC, spaced 3 d apart. Mice were sacrificed no earlier than 10 d after the last injection and bone marrow cells were isolated and transferred into lethally irradiated (875 R) C57/Bl6 mice (6 × 106 to 7 × 106 cells per recipient). Bone marrow cells derived from Ago2fl/fl mice treated with pI:pC as described above were used as control. Bone marrow-reconstituted mice were maintained on medicated water (1000 U/mL polymixin B, 1.1 g/L neomycin sulfate) for 4 wk after reconstitution and were analyzed 6–8 wk or 3 mo after transfer. Isolation of spleen and bone marrow and subsequent FACS analysis were performed as described (Socolovsky et al. 2001; Mecklenbrauker et al. 2002). Blood parameters were analyzed using flow cytometry-based hematology (Bayer, Advia 120).
Generation of an Ago2 antibody
A synthetic peptide encompassing the first 25 amino acids of mouse Ago2 was coupled to KLH and was used as an immunogen in rabbits. Crude serum was obtained after the first boost and affinity-purified. This serum was then used in a 1:1000 dilution for Western blot analysis.
Cell extract preparation and immunodetection of Ago2
Cells were lysed in 150 mM NaCl, 20 mM Tris (pH 7.5), 1 mM EDTA, 1% Triton X-100, and Complete protease inhibitors (Roche); after 10-min centrifugation at 13,000g, the supernatant was collected and used as extract for immunoblotting, using standard protocols.
Retroviral transduction of Ago2−/− cells
The coding sequences of Ago2 and Ago2D669A were inserted into the HpaI site of MigR retroviral vector, and recombinant retroviruses were produced and used to infect Ago2−/− bone marrow or fibroblasts as described (Pear et al. 1998). Reconstitution of lethally irradiated mice was performed as above.
Derivation and manipulation of mouse embryonic fibroblast (MEF) cell lines
Two Ago2fl/fl MEF cell lines (MEF lines 4 and 9) were derived from E12.5 embryos, transformed with SV40 large T-antigen-expressing retrovirus, and propagated under standard culture conditions. To generate Ago2−/− MEFs, both Ago2fl/fl MEF clones were infected with Cre-expressing adenovirus (AdenoCre; Vector Biolabs). For the miRNA-guided cleavage assay, the same assay as described in Pillai et al. (2005) was employed.
MiRNA expression analysis
Total RNA was isolated using Trizol (Invitrogen) according to the manufacturer’s instructions. MiRNA array profiling was performed as described (Miska et al. 2004). For clustering, only miRNAs with expression levels at least five times above background were selected. Expression data were log-transformed, gene-centered (mean), and normalized, and hierarchical clustering was performed using a correlation matrix and centroid linkage using the Cluster 3.0 algorithm. qRT–PCR expression analysis of miRNAs was performed using mirVana qRT–PCR miRNA detection kit (Ambion) and Roche LightCycler 480. Northern blotting of miRNAs was performed as described (Lau et al. 2001).
Acknowledgments
We thank M. Fabry, B. Coller, A. Shet, T. Hoffmann, M. Weiss, W. Pear, O. Shestova, R. Yi, and M. Busslinger for their helpful expertise, advice, and discussions. We thank K. Saigo for the Ago2 cDNA. We acknowledge S. Buonomo for advice in generating Ago2 antiserum. D.O’C. acknowledges the support of the Irvington Institute for Immunological Research and was their National Genetics Foundation Fellow. A.T. was supported by the Irene Diamond Foundation.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1565607
References
- Chen C.Z., Li L., Lodish H.F., Bartel D.P., Li L., Lodish H.F., Bartel D.P., Lodish H.F., Bartel D.P., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R., Norman J., Cooch N., Nishikura K., Shiekhattar R., Cooch N., Nishikura K., Shiekhattar R., Nishikura K., Shiekhattar R., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb B.S., Nesterova T.B., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Nesterova T.B., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Brockdorff N., Fisher A.G., Smale S.T., Fisher A.G., Smale S.T., Smale S.T., et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Petersen C.P., Sharp P.A., Petersen C.P., Sharp P.A., Sharp P.A. siRNAs can function as miRNAs. Genes & Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley F.W., Soriano P., Steffen L.S., Dymecki S.M., Soriano P., Steffen L.S., Dymecki S.M., Steffen L.S., Dymecki S.M., Dymecki S.M. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., De Marchis M.L., Nervi C., Bozzoni I., Nervi C., Bozzoni I., Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Gregory R.I., Chendrimada T.P., Cooch N., Shiekhattar R., Chendrimada T.P., Cooch N., Shiekhattar R., Cooch N., Shiekhattar R., Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C., Baillie D.L., Fire A., Ruvkun G., Mello C.C., Fire A., Ruvkun G., Mello C.C., Ruvkun G., Mello C.C., Mello C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Haase A.D., Jaskiewicz L., Zhang H., Laine S., Sack R., Gatignol A., Filipowicz W., Jaskiewicz L., Zhang H., Laine S., Sack R., Gatignol A., Filipowicz W., Zhang H., Laine S., Sack R., Gatignol A., Filipowicz W., Laine S., Sack R., Gatignol A., Filipowicz W., Sack R., Gatignol A., Filipowicz W., Gatignol A., Filipowicz W., Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J., Caudy A.A., Kobayashi R., Hannon G.J., Kobayashi R., Hannon G.J., Hannon G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hentze M.W., Kulozik A.E., Kulozik A.E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., Zamore P.D., Zamore P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D., Balint E., Tuschl T., Zamore P.D., Tuschl T., Zamore P.D., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Ketting R.F., Fischer S.E., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H., Fischer S.E., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H., Sijen T., Hannon G.J., Plasterk R.H., Hannon G.J., Plasterk R.H., Plasterk R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.W., Bass B.L., Bass B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R., Schwenk F., Aguet M., Rajewsky K., Schwenk F., Aguet M., Rajewsky K., Aguet M., Rajewsky K., Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P., Lim L.P., Weinstein E.G., Bartel D.P., Weinstein E.G., Bartel D.P., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee Y., Hur I., Park S.Y., Kim Y.K., Suh M.R., Kim V.N., Hur I., Park S.Y., Kim Y.K., Suh M.R., Kim V.N., Park S.Y., Kim Y.K., Suh M.R., Kim V.N., Kim Y.K., Suh M.R., Kim V.N., Suh M.R., Kim V.N., Kim V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A., Simon B., Izaurralde E., Sattler M., Simon B., Izaurralde E., Sattler M., Izaurralde E., Sattler M., Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Joshua-Tor L., Hannon G.J., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Maniataki E., Mourelatos Z., Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes & Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield J.H., Harfe B.D., Nissen R., Obenauer J., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Harfe B.D., Nissen R., Obenauer J., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Nissen R., Obenauer J., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Obenauer J., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Zuker M., Pasquinelli A.E., Ruvkun G., Pasquinelli A.E., Ruvkun G., Ruvkun G., et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- Martinez J., Tuschl T., Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes & Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecklenbrauker I., Saijo K., Zheng N.Y., Leitges M., Tarakhovsky A., Saijo K., Zheng N.Y., Leitges M., Tarakhovsky A., Zheng N.Y., Leitges M., Tarakhovsky A., Leitges M., Tarakhovsky A., Tarakhovsky A. Protein kinase Cδ controls self-antigen-induced B-cell tolerance. Nature. 2002;416:860–865. doi: 10.1038/416860a. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T., Patkaniowska A., Dorsett Y., Teng G., Tuschl T., Dorsett Y., Teng G., Tuschl T., Teng G., Tuschl T., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T., Landthaler M., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T., Urlaub H., Luhrmann R., Tuschl T., Luhrmann R., Tuschl T., Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Miska E.A., Alvarez-Saavedra E., Townsend M., Yoshii A., Sestan N., Rakic P., Constantine-Paton M., Horvitz H.R., Alvarez-Saavedra E., Townsend M., Yoshii A., Sestan N., Rakic P., Constantine-Paton M., Horvitz H.R., Townsend M., Yoshii A., Sestan N., Rakic P., Constantine-Paton M., Horvitz H.R., Yoshii A., Sestan N., Rakic P., Constantine-Paton M., Horvitz H.R., Sestan N., Rakic P., Constantine-Paton M., Horvitz H.R., Rakic P., Constantine-Paton M., Horvitz H.R., Constantine-Paton M., Horvitz H.R., Horvitz H.R. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo S.A., Ansel K.M., Kanellopoulou C., Livingston D.M., Rao A., Rajewsky K., Ansel K.M., Kanellopoulou C., Livingston D.M., Rao A., Rajewsky K., Kanellopoulou C., Livingston D.M., Rao A., Rajewsky K., Livingston D.M., Rao A., Rajewsky K., Rao A., Rajewsky K., Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear W.S., Miller J.P., Xu L., Pui J.C., Soffer B., Quackenbush R.C., Pendergast A.M., Bronson R., Aster J.C., Scott M.L., Miller J.P., Xu L., Pui J.C., Soffer B., Quackenbush R.C., Pendergast A.M., Bronson R., Aster J.C., Scott M.L., Xu L., Pui J.C., Soffer B., Quackenbush R.C., Pendergast A.M., Bronson R., Aster J.C., Scott M.L., Pui J.C., Soffer B., Quackenbush R.C., Pendergast A.M., Bronson R., Aster J.C., Scott M.L., Soffer B., Quackenbush R.C., Pendergast A.M., Bronson R., Aster J.C., Scott M.L., Quackenbush R.C., Pendergast A.M., Bronson R., Aster J.C., Scott M.L., Pendergast A.M., Bronson R., Aster J.C., Scott M.L., Bronson R., Aster J.C., Scott M.L., Aster J.C., Scott M.L., Scott M.L., et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Basyuk E., Bertrand E., Filipowicz W., Bertrand E., Filipowicz W., Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Filipowicz W., Bhattacharyya S.N., Filipowicz W., Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Rowe T.M., Rizzi M., Hirose K., Peters G.A., Sen G.C., Rizzi M., Hirose K., Peters G.A., Sen G.C., Hirose K., Peters G.A., Sen G.C., Peters G.A., Sen G.C., Sen G.C. A role of the double-stranded RNA-binding protein PACT in mouse ear development and hearing. Proc. Natl. Acad. Sci. 2006;103:5823–5828. doi: 10.1073/pnas.0601287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolovsky M., Nam H., Fleming M.D., Haase V.H., Brugnara C., Lodish H.F., Nam H., Fleming M.D., Haase V.H., Brugnara C., Lodish H.F., Fleming M.D., Haase V.H., Brugnara C., Lodish H.F., Haase V.H., Brugnara C., Lodish H.F., Brugnara C., Lodish H.F., Lodish H.F. Ineffective erythropoiesis in Stat5a−/−5b−/− mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- Song J.J., Liu J., Tolia N.H., Schneiderman J., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L., Liu J., Tolia N.H., Schneiderman J., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L., Tolia N.H., Schneiderman J., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L., Schneiderman J., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L., Martienssen R.A., Hannon G.J., Joshua-Tor L., Hannon G.J., Joshua-Tor L., Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L., Smith S.K., Hannon G.J., Joshua-Tor L., Hannon G.J., Joshua-Tor L., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Yan K.S., Yan S., Farooq A., Han A., Zeng L., Zhou M.M., Yan S., Farooq A., Han A., Zeng L., Zhou M.M., Farooq A., Han A., Zeng L., Zhou M.M., Han A., Zeng L., Zhou M.M., Zeng L., Zhou M.M., Zhou M.M. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Yekta S., Shih I.H., Bartel D.P., Shih I.H., Bartel D.P., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yi R., O’Carroll D., Pasolli H.A., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E., O’Carroll D., Pasolli H.A., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E., Pasolli H.A., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E., Dietrich F.S., Tarakhovsky A., Fuchs E., Tarakhovsky A., Fuchs E., Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- Yigit E., Batista P.J., Bei Y., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Batista P.J., Bei Y., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Bei Y., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C., Mitani S., Simard M.J., Mello C.C., Simard M.J., Mello C.C., Mello C.C. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Zhong J., Peters A.H., Lee K., Braun R.E., Peters A.H., Lee K., Braun R.E., Lee K., Braun R.E., Braun R.E. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat. Genet. 1999;22:171–174. doi: 10.1038/9684. [DOI] [PubMed] [Google Scholar]