Abstract
Dosage compensation is a process required to balance the expression of X-linked genes between males and females. In Drosophila this is achieved by targeting the dosage compensation complex or the male-specific lethal (MSL) complex to the male X chromosome. In order to study the mechanism of targeting, we have studied two X-chromosomal genes, mof and CG3016, using chromatin immunoprecipitation as well as immuno-FISH analysis on transgenic flies. We show that MSL complex recruitment requires the genes to be in a transcriptionally active state. MSL complex recruitment is reversible because blocking transcription severely reduces MSL binding to its target genes. Furthermore, targeting cues are found toward the 3′ end of the gene and depend on the passage of the transcription machinery through the gene, whereby the type of promoter and the direction of transcription are dispensable. We propose a model of dynamic MSL complex binding to active genes based on exposed DNA target elements.
Keywords: MSL, MOF, roX, transcription, dosage compensation, X chromosome
Sex determination often involves differentiation of a pair of chromosomes such that one sex retains the original two copies (homogametic), whereas in the other sex one of the homologous chromosomes degenerates almost completely with only the exception of a few sex-specific genes (heterogametic). The deleterious effects of aneuploidy creates the need for a compensatory process that equalizes the gene products between the two sexes (Charlesworth and Charlesworth 2005). In Drosophila, dosage compensation occurs by increasing transcription of most genes on the single male X chromosome to compensate for transcription of both X chromosomes in females. Genetic studies have identified five male-specific lethal (MSL) genes—msl1, msl2, msl3, males absent on the first (mof), and maleless (mle)—that are important for the regulation of dosage compensation. The products of these genes (collectively referred to as MSL genes), as well as two noncoding RNAs (roX1 and roX2), assemble in a large complex called the dosage compensation complex (DCC), or the MSL complex, which is targeted to hundreds of sites on the male X chromosome (Lucchesi et al. 2005; Mendjan and Akhtar 2006; Straub and Becker 2007).
One of the most intriguing features of the MSL complex is its ability to recognize its targets on the male X chromosome. Studies that involved P-element-mediated insertions of X-chromosomal genes on autosomes and X–autosome translocations have shown that most X-linked genes were still targeted when inserted on an autosome. In contrast, autosomal genes translocated to the X remained untargeted (Fagegaltier and Baker 2004; Oh et al. 2004). Targeting therefore most likely happens on genes individually and may not require the presence of 35–40 so-called “entry sites” (Kelley et al. 1999).
Recent chromatin profiling studies have confirmed the preference of the MSL complex for individual genes. The MSLs were found to bind locally to genes with MSL occupancy predominantly at the 3′ of genes (Alekseyenko et al. 2006; Gilfillan et al. 2006; Legube et al. 2006). Moreover, the affinity of MSL1 for its targets correlates with its dosage-compensated state (Gilfillan et al. 2006; Legube et al. 2006). Therefore, the affinity of the MSL complex might have evolved concurrent with the need for dosage compensation of its targets. This is in agreement with the findings that those genes strongly bound by MSL1 are more generally essential genes (Gilfillan et al. 2006).
Despite the large number of MSL targets recently identified by chromatin profiling studies, a universal targeting sequence motif has not been identified for the MSL complex. Also, the proposal that the MSL complex simply targets those genes on the X that are transcriptionally active does not seem to apply universally since there are many genes on the X that are transcriptionally active but not bound by the MSLs (Alekseyenko et al. 2006; Gilfillan et al. 2006; Legube et al. 2006). It seems conceivable that transcription is a prerequisite for target recognition but not sufficient by itself, because most target genes are transcribed, and those few genes that show differential expression are only bound when the gene is active (Alekseyenko et al. 2006). Although no universal targeting sequence has been found, several short degenerative sequences with some predictive power have been identified (Dahlsveen et al. 2006; Gilfillan et al. 2006). It is therefore possible that these degenerative sequences are only exposed when the gene is active and the chromatin is present in an open conformation.
An assumption of the above hypothesis would be the ability of the MSL complex to associate with genes only while transcription is ongoing, presumably at the stage of transcriptional elongation. Therefore, targeting cues may reside in the body of the gene as opposed to the original reports of upstream enhancer sequences present in dosage-compensated genes (for review, see Baker et al. 1994), but in agreement with recent MSL profiling studies (Smith et al. 2001; Alekseyenko et al. 2006; Gilfillan et al. 2006; Legube et al. 2006).
In order to gain better insights into the targeting mechanism, we studied two neighboring genes, CG3016 and mof, for their targeting properties as they recently have shown to be targeted by the MSL complex in genome-wide analysis (Gilfillan et al. 2006; Legube et al. 2006). The mof gene encodes the histone acetyltransferase in the MSL complex that specifically acetylates Lys 16 of histone H4 (H4K16). Apart from the two roX RNA-encoding genes, mof is the only other known MSL member whose gene is located on the X chromosome. Interestingly, we find that transcriptional activation is required for targeting of the MSL complex and that polymerase passage through the gene is a prerequisite for target recognition. We also show that this is true for most genes on the X chromosome, as blocking transcription by α-amanitin treatment greatly reduced binding of the MSL complex to X-chromosomal genes. Finally, we show that in addition to transcription, targeting to the mof and CG3016 genes most likely is a combination of degenerative DNA sequences toward the 3′ end of these genes only to be recognized when the gene is transcriptionally active.
Results
MSL proteins are enriched on the region spanning the mof and CG3016 genes
In order to precisely map the binding pattern of MSLs on the mof and CG3016 loci, we performed chromatin immunoprecipitation (ChIP) experiments in Drosophila Schneider cells (SL-2) using specific antibodies directed against MSL1, MSL3, and MOF. SL-2 cells are a male 16-h embryonic cell line with an MSL-binding pattern closely reflecting that of late-stage embryos (Alekseyenko et al. 2006). The immunoprecipitated material was analyzed by quantitative real-time PCR (Q-PCR) and presented as the percentage recovery compared with input DNA (Fig. 1A). The roX2 high-affinity site, which served as a positive control, showed highly enriched MSL binding (Fig. 1A, lane 15) as opposed to gapdh (located on chromosome 2R) (Fig. 1A, lane 14) and the runt gene (located on the X chromosome) (Fig. 1A, lane 13), but is dosage-compensated in an MSL-independent fashion (Gergen 1987; Smith et al. 2001). Binding was enriched on the body of the gene for both CG3016 and mof for all three MSL proteins tested (Fig. 1, lanes 1–12). Unlike MSL1 and MSL3, there is also considerable binding of MOF to the promoter regions (see Fig. 1, lanes 1,5). Whether the binding of MOF to promoters reflects its general binding pattern on a genome-wide scale remains to be tested.
Figure 1.
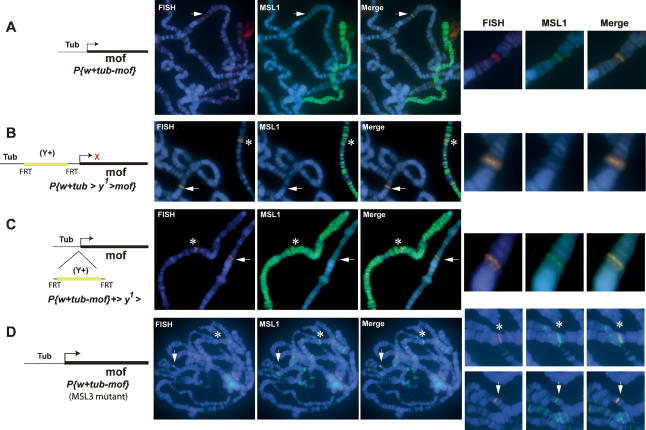
Transcriptional-dependent recruitment of MSL1 to the mof gene. (A) ChIP in SL-2 cells on the CG3016 and mof genes with antibodies directed against MSL1, MSL3, and MOF. Levels of enrichment are determined by Q-PCR and depicted as the percentage recovery of input DNA. The position of the primer pairs is schematically shown by green arrows. (B–D) MSL-1 immunostaining (green) in combination with DNA-FISH (red) directed against the mini-white gene of a polytene chromosome carrying an autosomal 6.7-kb P-element insertion encompassing mof and the coding region of CG3016 (B), or an autosomal P-element insertion of mof with its endogenous promoter (C) or without its endogenous promoter (D). Recruitment of MSL1 to the transgenes is indicated by an arrow. The position of the endogenous mof gene is shown by an asterisk. DNA was stained with Hoechst 322 (blue).
We next wished to test if a large genomic fragment including mof and most of CG3016 was able to target the MSL complex ectopically when inserted on an autosomal location by means of P-element-mediated transformation. As is shown in Figure 1B, this 6.7-kb fragment (mof 6.7) showed robust MSL1 recruitment visualized by staining with an MSL1 antibody in combination with fluorescence in situ hybridization (immuno-FISH) to visualize the location of the transgene (see Materials and Methods). The presence of MSL1 and the direct visualization of targeting on polytene chromosomes enabled further detailed analysis of the nature of targeting to this locus.
Taken together, these results show that MSL1 protein is enriched on mof and CG3016 genes and that this region is able to recruit the MSL complex autonomously on an autosomal location.
MSL1 is recruited to the mof gene in a transcription-dependent manner
In order to study targeting to individual genes, we first generated transgenic flies carrying the mof gene with its endogenous promoter sequences (mof3.5). The mof gene showed ectopic MSL1 recruitment in almost all independent lines that were tested (Fig. 1C; Supplementary Table 1). Recently, the MSL-binding pattern to polytene chromosomes was suggested to reflect the distribution of active genes (Sass et al. 2003). Although the vast majority of genes bound by MSL1 are indeed active, several active genes on the X chromosome escape MSL1 binding (Alekseyenko et al. 2006; Gilfillan et al. 2006; Legube et al. 2006). Therefore, in order to address directly whether or not transcription is a prerequisite for targeting, we analyzed targeting of the MSL complex to the mof gene in the absence of the endogenous promoter sequences (mof 2.4Δ prom). Clearly, targeting to these lines was lost as we did not observe MSL1 binding on the transgene in any of the lines studied (Fig. 1D; Supplementary Table 1).
Although the results above suggest that the absence of transcription leads to the loss of MSL1 binding, it is possible that targeting sequences are present in the promoters of the respective genes as it was originally suggested for several genes (Baker et al. 1994). In order to address if the promoter requirement is due to a targeting signal embedded in the promoter sequence or solely due to transcriptional activation, mof was fused downstream from the tubulin promoter. The tubulin gene is located on chromosome 3, and therefore its promoter should not contain any X-chromosome-specific sequences. Interestingly, a tubulin promoter-driven mof transgene (tub-mof) also showed ectopic recruitment of MSL1 (Fig. 2A), as well as MSL3, MLE, and MOF (Supplementary Fig. 1). In contrast, tubulin-driven expression of the pho gene (Klymenko et al. 2006), encoded on the fourth chromosome, did not show any MSL1 recruitment, further confirming the specificity of MSL1 binding to the mof gene (Supplementary Fig. 2A; Supplementary Table 1).
Figure 2.
The mof transgene driven by a tubulin promoter is a target for MSL1 and overcomes the requirement for MSL3. (A) Tubulin-driven mof gene. (B,C) Tubulin-driven mof gene (tub > y+ > mof) containing a yellow gene cassette between the promoter and the gene such that mof expression becomes inducible upon excision of the yellow gene cassette by expression of flippase. Recruitment of MSL1 before (B) and after (C) excision is shown. (D) Same as in A, however, in this case the transgenic line carrying tub-mof was crossed into the msl-3083 mutant background by using females mutant for msl-3083 expressing low levels of MSL2 from an Hsp83 promoter. Recruitment of MSL1 to the tub-mof transgene is indicated by an arrow. The position of the endogenous mof gene is shown by an asterisk. Red cross (×) represents no transcription. DNA was stained with Hoechst 322 (blue).
In order to study transcription-dependent targeting at the same genomic position and thereby addressing any possible position effect variegation (PEV)-related effects, we also created a transcription on/off assay by using a previously developed system that makes use of FRT-flip recombinase-mediated excision of a yellow gene inserted between the tubulin promoter and the gene of interest, in our case the mof gene (Struhl and Basler 1993). When present, the yellow gene blocks transcription from the tubulin promoter. This assay can therefore be used to study recruitment of MSL complex to the same location in the inactive and active states. Using this assay, we observed that mof was only recognized as a MSL1 target when transcription was allowed by excision of the yellow gene (Fig. 2B,C; Supplementary Table1). We confirmed activation of transcription after excision of the yellow gene by rescuing the male-specific lethality caused by the absence of MOF (data not shown). Since mof driven by a tandem repeat of five GAL4-binding sites (UAS-mof) also showed targeting of MSL1 (Supplementary Fig. 2B; Supplementary Table 1), we conclude that targeting solely requires transcriptional activity irrespective of the promoter used. The determinant of targeting, therefore, most likely is to be found in the gene region and not in the promoter sequence. Furthermore, the observation that UAS-driven expression of mof without GAL4 activation also displayed MSL1 recruitment suggests that transcription is required but the levels of transcription are not important.
A limited number of sites on the X chromosome are defined as high-affinity sites because they have the ability to recruit partial complexes of MSL1 and MSL2 in the absence of the other MSLs (Kelley et al. 1999). Since mof is located in a cytological location of the previously mapped high-affinity site at position 5c (Lyman et al. 1997; Demakova et al. 2003), we tested by DNA-FISH if this site coincides with the location of mof. Since homozygous msl-3083 male third instar larvae are rarely recovered, we used female flies expressing MSL2 in an msl-3083 mutant background. These flies assemble partial complexes on the X chromosome and are routinely used to study high-affinity sites (Kelley et al. 1999; Kageyama et al. 2001; Dahlsveen et al. 2006). As shown in Figure 2D, the mof endogenous gene is found in very close proximity to the high-affinity site, but upon close examination appears to be not the site itself but just beside it (Fig. 2D, see asterisks). We verified this result by crossing the genomic construct containing the mof locus (mof3.5) in an msl-3083 background, which also showed no targeting (data not shown). Surprisingly, however, when the tubulin-driven mof transgene was crossed in an msl-3083 background, we created what appeared to be a high-affinity site, as we could observe recruitment of MSL1 in the absence of MSL3 (Fig. 2D, arrow). We obtained similar results with this line in an mle1 mutant background, albeit with a substantially weaker MSL1 recruitment (data not shown).
Taken together, the results above show that the MSL complex is recruited to the mof gene in a transcription-dependent manner. Our results suggest that also on other regions of the X chromosomes, otherwise known as lower-affinity sites, partial MSL complexes are able to recognize their targets autonomously without MSL3 or MLE. We therefore propose that low-affinity sites may override the requirement for MSL3 or MLE if their expression is driven from a strong promoter such as tubulin. This is presumably due to the combinatorial effect of chromatin modifications associated with high levels of transcription that may cooperate to expose certain DNA target sequences to MSL1 and MSL2, which may not be exposed otherwise.
MSL complex targeting is compromised in α-amanitin-treated cells
Knowing that transcription is a prerequisite for binding of the MSL complex (Fig. 2B,C), we wished to test for the plasticity of the bound state. Somewhat conflicting hypotheses on the subject of timing and maintenance of targeting the MSL complex have been put forward recently. On one hand, the MSL complex is thought to be targeted in early development, presumably following the present transcriptional pattern, and maintained in the course of development irrespective of transcriptional changes (Alekseyenko et al. 2006; Legube et al. 2006). On the other hand, subtle developmental changes in the binding pattern of the MSL complex on polytene chromosomes and the presence of genes in early development (6-h embryos) that are bound by MSLs but do not correlate with transcriptional status, as judged by RNA Pol II chromatin-binding profiles (Gilfillan et al. 2006), suggest that differential binding takes place.
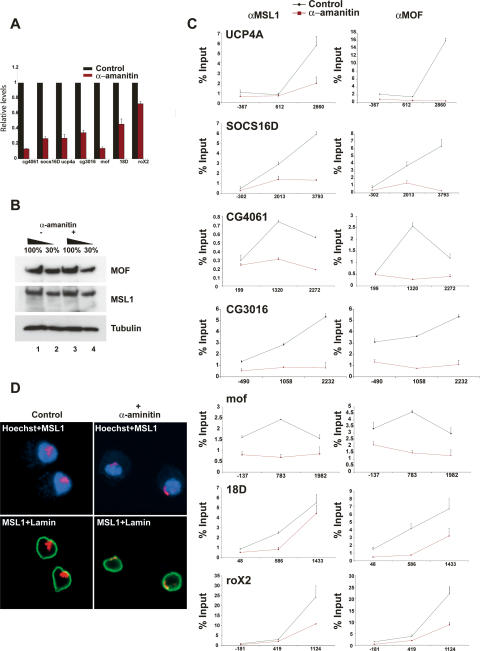
Since many MSL target genes are housekeeping genes that are continuously transcribed (Gilfillan et al. 2006; Legube et al. 2006), stable binding throughout development could be the rule with some differentially transcribed genes being the exception. In order to test this hypothesis, we blocked transcription by means of α-amanitin treatment in Schneider (SL-2) cells. We argued that if binding is maintained irrespective of transcriptional changes, genes should preserve their normal MSL-binding pattern when transcription is blocked. As expected, after a 20-h α-amanitin (Pol II inhibitor) treatment, transcript levels of most genes tested were significantly reduced compared with untreated control levels (Fig. 3A). Visually the cells showed normal morphology (data not shown), and the protein levels of MSL1 and MOF were comparable with the control situation (Fig. 3B). Furthermore, roX2 RNA levels in treated cells were mildly affected in comparison with the control situation, very likely reflecting a higher stability of this RNA (Fig. 3A). In addition to the mof gene, we tested four additional X-chromosomal genes (UCP4A, SOCS16D, CG4061, and CG3016) by ChIP. Interestingly, all the tested genes show a dramatic reduction of MSL binding following α-amanitin treatment (Fig. 3C). Intriguingly, MSL binding to the high-affinity sites such as roX2 and 18D was significantly less susceptible to α-amanitin treatment, presumably reflecting their ability to recruit the complex ectopically independently of transcription (Kelley et al. 1999; Kageyama et al. 2001; Oh et al. 2003, 2004). Consistent with these observations, we also found severely reduced staining of MSL1 on the X-chromosomal territory in α-amanitin-treated SL-2 cells in comparison with the control cells (Fig. 3D). These results suggest that MSL binding is reversible and depends directly on the transcriptional state of the targets. The rigid binding of MSLs therefore is not an embryonic irreversible preset condition, but most probably reflects the continuous transcriptional activity of its targets.
Figure 3.
Recruitment of MSL1 and MOF on X-chromosomal genes is severely affected upon blocking transcription by α-amanitin. (A) Q-RT–PCR analysis of expression levels normalized to mock (100%) after a 20-h α-amanitin treatment. (B) Western blot analysis from whole-cell extracts prepared from mock- or α-amanitin-treated cells. Western blots were probed with MSL1, MOF, and Tubulin antibodies as indicated. (C) ChIP in SL-2 cells with antibodies directed against MSL1, and MOF after 20-h mock or α-amanitin treatment. Levels of enrichment are determined by Q-PCR and depicted as the percentage recovery of input DNA. Enrichment on each gene was systematically tested using primer pairs located at the beginning, middle, and end of genes. Position of the PCR probes with respect to the transcription start site (+1) is indicated on the X-axis; for example, for UCP4A, the first primer pair is located 367 bp upstream of the transcription start site, and the second and third primer pairs are located 612 bp and 2860 bp downstream, respectively. (D) Immunofluorescence with MSL1 (red) and Lamin (green) in Schneider (SL-2) cells of mock- or α-amanitin-treated cells. DNA was stained with Hoechst 322 (blue).
Genic sequences contribute to MSL complex recruitment on the mof gene
Although it is conceivable that targeting requires the transcription machinery to pass through the gene for a certain targeting signal to become exposed, it remains plausible that MSL binding is achieved by the recruitment of transcriptional activators to the promoter; e.g., in the scenario in which the MSL complex slides along the DNA pushed forward by transcribing polymerase (Gilfillan et al. 2006). For this purpose, we created a mof construct that has the mof endogenous promoter but in a reverse orientation (mof 3.5prom AS), such that transcription does not pass through the mof gene. As is shown in Figure 4A, this transgene is unable to recruit MSL1. Thus, transcription activation is not sufficient for targeting MSL1 to mof, instead transcription needs to run through the gene to reveal mof as an MSL target.
Figure 4.
Targeting by MSL1 to the mof gene involves DNA sequences embedded in the coding region. MSL1 immunostaining (green) in combination with DNA-FISH (red) for the location of the transgene for insertions containing mof with an antisense promoter (A), tubulin-driven antisense mof gene (B), endogenous promoter-driven mof derivative lacking 670 bp of the 3′ end (C), or endogenous promoter-driven 3′ end of mof (1863–2533 bp) (D). Recruitment of MSL1 to the transgenes is indicated by an arrow. DNA was stained with Hoechst 322 (blue).
To date, it remains unknown what the targeting signal embodies. Several groups over the past years have attempted to identify a universal targeting sequence, but despite these attempts, beside several short degenerative sequences with limited predictive power, no general binding sequence has been identified (Alekseyenko et al. 2006; Dahlsveen et al. 2006; Gilfillan et al. 2006; Legube et al. 2006). It is possible that the predictive power of these sequences is limited because they need to be exposed in order to be recognized. It is conceivable, therefore, that for a target to be exposed, transcription needs to run through the gene whereby, if the signal involves degenerative DNA sequences, the orientation of transcription should not make a difference. Therefore, in order to test this hypothesis we placed a tubulin promoter at the 3′ end of the mof gene such that it would make a mof antisense transcript (tub-mof2.4AS). If targeting involves a DNA element only to be recognized when the chromatin is in an open conformation, this transgene should retain its binding capacities for MSL1. This construct is, indeed, able to recruit MSL1 (Fig. 4B, top panel; Supplementary Table 1). Interestingly, occasional spreading was observed on one of the lines (<5% of the nuclei) (Fig. 4B, bottom panel). This is possibly caused by the presence of targeting cues toward the 3′ end that are now more exposed due to the close proximity of the tubulin promoter. In order to test if the 3′ end of mof is essential for targeting, we created a mof construct similar to mof 3.5 but now lacking 600 base pairs (bp) of the 3′ end of the mof gene (mof2.9Δ 3prime). This construct, despite being transcriptionally active (Supplementary Fig. 3), failed to recruit MSL1 in all lines observed (Fig. 4C; Supplementary Table 1), which is in agreement with the overall binding preference of MSLs to the 3′ end of genes (Smith et al. 2001; Alekseyenko et al. 2006; Gilfillan et al. 2006). We next tested whether the 3′ end of the mof gene alone is sufficient to recruit MSL1 in the absence of the surrounding sequences. However, we were unable to observe recruitment of MSL1 on this fragment (Fig. 4D).
Our data suggest that it is most likely that the targeting signal for the MSL complex is a DNA sequence embedded in the coding region of mof, only to be exposed when the gene is transcribed, and that the sequences toward the 3′ end of mof are necessary but not sufficient for MSL1 recruitment as a small DNA element, out of context of the entire gene.
Ectopic recruitment of MSL1 to CG3016 also involves transcriptional activity and sequences toward the 3′ end of the gene
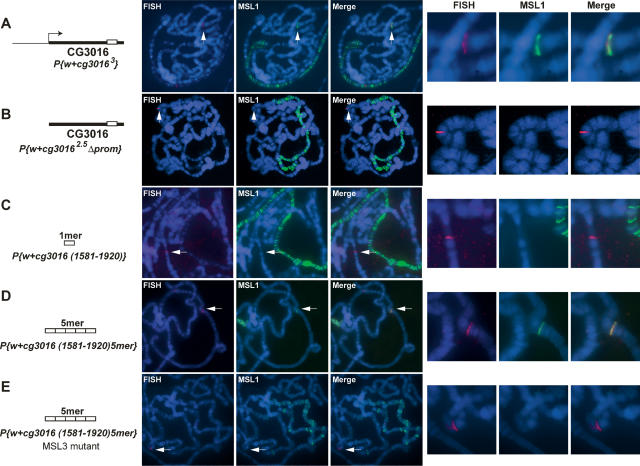
In order to test whether transcription-dependent recruitment of the MSL complex was not unique for the mof gene but represented a more general feature of other X-chromosomal genes, we next generated independent transgenes carrying CG3016 with or without its endogenous promoter sequences (CG30163, CG3016 2.5Δ prom) (Supplementary Table 1). Similar to the mof gene, we also observed ectopic recruitment of MSL1 to CG3016 in the presence of its promoter but not in its absence (Fig. 5A,B). We were next interested in studying the contribution of the sequences toward the 3′ end of the CG3016 gene in MSL1 targeting. For this purpose, we mapped a 339-bp region near the 3′ end of CG3016 that shows maximal MSL enrichment by ChIP and sequence similarity to the previously characterized conserved consensus sequence present on both roX1/2 high-affinity sites (Supplementary Fig. 4; Park et al. 2003). We therefore created transgenes containing 339 bp of this sequence as a monomer [P{w+ CG3016(1581–1920)}]. Interestingly, we observed that similar to the mof 3′ end, this sequence alone was unable to recruit MSL1 ectopically. Intriguingly however, multimerization [containing five tandem repeats, P{w+ CG3016(1581–1920) 5mer}] of this sequence restored binding of MSL1, indicating that different from the high-affinity sites that recruit MSLs as an ∼300-bp monomer (Kageyama et al. 2001; Park et al. 2003; Oh et al. 2004) but similar to the mof situation, the affinity of MSL1 for the CG3016 sequences is too low to be recognized when placed out of context of the gene itself, but can be revealed upon multimerization (Fig. 5D).
Figure 5.
Transcription-dependent targeting of MSL1 to CG3016 and the identification of a targeting sequence toward the 3′ end of the gene. MSL1 immunostaining (green) in combination with DNA-FISH (red) on an autosomal P-element insertion carrying CG3016 with (A) or without (B) its endogenous promoter. (C) A 339-bp fragment (represented by a white box) corresponding to nucleotide positions 1581–1920 bp from the transcription start site of CG3016. Multimerization of the 339-bp sequence (five tandem repeats) in a wild-type (D) and an msl-3083 mutant (E) background. Recruitment of MSL1 to the transgenes is indicated by an arrow. DNA was stained with Hoechst 322 (blue).
The three previously mapped high-affinity sites (roX1, roX2, and 18D) have been shown to maintain their ability to recruit MSL1 in an msl-3083 mutant background as a small ∼300-bp monomer for roX1/2 and as a multimer in the case of the 18D high-affinity site (Kageyama et al. 2001; Park et al. 2003; Oh et al. 2004). Although CG3016 is not a high-affinity site, we were interested to test if multimerization of a low-affinity site could overcome the requirement for MSL3. Therefore, to address this issue (as described above in Fig. 2D), we crossed the transgenic flies carrying the CG3016 multimer with flies expressing MSL2 in an msl-3083 mutant background and analyzed MSL1 binding to female polytene chromosomes. Interestingly, we observed that the CG3016 multimer failed to recruit MSL1 in an msl-3083 mutant background (Fig. 5E).
These results show that similar to the mof gene, MSL complex recruitment on the CG3016 gene requires transcriptional activity and sequences embedded within the body of the gene. We also show that even though multimerization of 3′ of CG3016 restores the binding of MSL1 in the wild-type context, the affinity of MSL1 for this CG3016 sequence is too weak to be maintained in the absence of MSL3.
Discussion
In this study, we provide direct evidence for transcription-dependent recruitment of the MSL complex to X-linked genes. We show that passage of the transcription machinery through the gene is important, whereas the type of promoter and the direction of transcription are not. Our data demonstrate that the recruitment signal lies within the transcribed portion of the gene and that targeting occurs independently of neighboring nucleation sites as we can successfully assess recruitment of X-linked genes on autosomal locations in a transcription-dependent manner. Our data support a model for MSL complex targeting to transcriptionally active X-linked genes that encode a certain combination of small degenerative target sequences in their transcribed region.
It has been proposed previously that one can create MSL complex-binding sites on X-chromosomal regions, normally devoid of the MSL complex, by driving strong transcription via 14 tandem repeats of GAL4-binding sequences (Sass et al. 2003). However, because integration of the EP lines used was random and the sites of insertion were not characterized, the nature of the transcribed unit was unclear, and therefore any assumptions made about the importance of certain sequences, direction, or strength of transcription of the targeted unit remained speculative. Furthermore, since the autosomal EP lines failed to recruit MSL complex, it remained unclear to what extent the X chromosomal context played a role in MSL recruitment on these insertion sites. It may therefore be important to study MSL targeting autonomously, out of the X-chromosomal context, to avoid the complication that may be caused by high local concentration of MSLs on the X chromosome in combination with strong transcriptional activation as previously reported (Sass et al. 2003). Furthermore, in contrast to what has been proposed previously (Sass et al. 2003), our data suggest that transcription alone is unlikely to be the sole targeting signal— first, because many genes on the X chromosome are transcribed but not targeted by MSLs (Alekseyenko et al. 2006; Gilfillan et al. 2006; Legube et al. 2006), and second, as shown in this study, the mof gene loses its targeting properties without sequences toward the 3′ end while still being transcribed (Fig. 4; Supplementary Fig. 3).
Our data show that most likely the targeting signals for the MSL complex include DNA sequences embedded in the coding regions of mof and CG3016 only to be exposed when the gene is transcribed. These target sequences are found toward the 3′ end and are necessary but not sufficient for MSL1 recruitment, out of context of the entire gene. It remains possible, however, that additional elements also contribute to MSL1 target recognition. Such elements could either be DNA sequences or simply other chromatin-associated factors/modifications that facilitate MSL1 target recognition in a chromatin context. Alternatively, an RNA secondary structure might be involved in MSL recruitment. Such a RNA structure would be a feature of the mRNA sequence. Since the secondary RNA structure formed is independent of the orientation of transcription, the situation created in the case in which mof is transcribed in an antisense direction would be similar if not identical to the situation in which mof is transcribed in a sense orientation. It is unlikely, however, that a natural antisense transcript is involved in MSL recruitment because a mof transgene without a promoter failed to recruit MSL1 where it would still have the potential to produce an antisense RNA molecule (Fig. 1D).
We failed to find any predicted target sequences in the mof gene, and although the CG3016 gene shows some similarity to the roX1/2 high-affinity consensus sequences, CG3016 does not behave as a high-affinity site, as judged by MSL3-dependent recruitment. This once again emphasizes the difficulty of predicting MSL target sequences. In agreement with previous studies, we conclude that most probably the nature of these sequences determines the affinity of MSL for its targets and not the level of transcription, as any type of promoter reveals mof as a target (Dahlsveen et al. 2006; Gilfillan et al. 2006). Our data may also explain in part the limited predictive power of MSL binding, since targeting only occurs when the gene is transcribed and thus the sequence is exposed. Therefore, MSL-binding predictions should also take into account the transcription state of genes or possibly the chromatin state, for example, by creating a genome-wide DNase I map (Sabo et al. 2004) of the X chromosome.
Unlike the full-length mof or CG3016 genes, the affinity of MSL1 for the 3′ end sequences of the mof and CG3016 genes is not sufficient for visualization on polytene chromosomes. Furthermore, even though multimerization of sequences toward the 3′ of CG3016 restores the binding of MSL1 in the wild-type context, the affinity of MSL1 for this CG3016 sequence is too weak to be maintained in the absence of MSL3. These results show that, on the one hand, there are “high-affinity sites” such as roX genes that display transcriptional-independent MSL-targeting sites that are able to recruit MSLs as relatively small DNA sequences (Kageyama et al. 2001; Oh et al. 2004). And, on the other hand, the majority of sites present on the X-chromosome are “low- or moderate-affinity sites” for MSL1 that require exposure of their target sequences presumably by transcriptional activity or artificially by multimerization in order to be recognized as an MSL target sequence. We therefore propose a model of MSL recruitment to the majority of X-linked target genes based on a combination of active transcription and the presence of DNA target elements.
Future bioinformatics studies encompassing several MSL target genes should reveal whether the 3′ ends of X-linked genes harbor a particular consensus sequence, binding sites for other factors, or even a particular secondary structure that together with transcriptional activity contributes to MSL target recognition of X-linked genes.
Materials and methods
ChIP
All ChIP experiments were performed at least three times using independent chromatin preparations as described in Orlando and Paro (1993). Briefly, SL-2 cells were grown in Schneider medium (GIBCO) containing 10% FCS. Cells (1 × 108) were cross-linked with formaldehyde for 8 min. Sonication was performed (26 × 30 sec) at maximum power (Bioruptor; CosmoBio) in lysis buffer (50 mM HEPES/KOH at pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% DOC, 0.1% SDS + Complete protease inhibitors [Roche]). One-hundred micrograms of chromatin and 3 μL of polyclonal antibody were used per immunoprecipitation (MSL1, MSL3 raised in rats, and MOF raised in rabbit as described previously; Mendjan et al. 2006). Immunoprecipitated complexes were isolated by adding protein A/G-Sepharose (Roche) followed by four washing steps: twice in lysis buffer, once in DOC buffer (10 mM Tris at pH 8, 0.25 M LiCl, 0.5% NP-40, 0.5% DOC, 1 mM EDTA), and once in TE (pH 8). DNA was eluted in 1× elution buffer (1% SDS, 0.1 M NaHCO3) for 20 min at room temperature, followed by reversal of cross-links overnight at 65°C. DNA was purified by a 30-min incubation in 37°C RNase A (0.2 mg/mL) and 2 h in Proteinase K (0.05 mg/mL), followed by phenol/chloroform extraction and DNA precipitation. Each ChIP was resuspended in 100 μL of TE.
Q-PCR
For the spanning of the cytological location 5c5, primers were designed to space approximately every 500 bp, which is the average size of the chromatin used in this study (primers available on request). The roX2, runt, and gapdh primer pairs are as described in Legube et al. (2006). Q-PCR analysis of the ChIP was performed using the SYBR Green PCR master mix (Applied Biosystem), 100 ng of each primer, and 1 μL of the immunoprecipitated DNA, in an ABI7500 Real-time PCR Instrument (Applied Biosystem). The formula [percentage ChIP/input] = [E(Ctinput − CtChIP) * 100%] (where E represents the primer efficiency) was used to calculate the percentage recovery after ChIP as compared with input. For the analysis of the RNA levels, RNA was first reverse-transcribed using the SuperScript RT (Invitrogen) and 500 ng of random hexamer. One microliter of the cDNA was then subjected to real-time PCR using the SYBR Green PCR master mix (Applied Biosystem) and 10 pmol/μL each primer. The primers designed in the middle of the genes in the ChIP experiment were used for the analysis of the transcript levels. For the α-amanitin ChIP experiment, primer pairs were designed to amplify 100- to 200-bp fragments in the beginning, middle, and end of the genes. For the Q-RT–PCR analysis of the transcript levels, the RT reaction was performed as described in the RT–PCR section below. RNA levels were normalized to mitochondrial RNA and depicted as the percentage recovery of input DNA after α-amanitin treatment compared with mock-treated cells.
RT–PCR
For the RT–PCR in Supplementary Figure 3, third instar larvae were snap-frozen in liquid nitrogen and crushed with a mortar and pestle. RNA was then collected following instructions from the Qiagen RNeasy kit. RT reactions were performed with random hexamers using Invitrogen SuperSciptII. RT reactions were loaded as a dilution series (1, 0.1, and 0.001 μL of RT mix). For RT–PCR on the transgenes, a primer specific for the CaSpeR vector was used in combination with a mof-specific primer. In contrast, for the expression of the endogenous mof gene, both primers were encoded within the coding sequence of mof.
α-Amanitin treatment
SL-2 cells were grown to a density of ∼4 × 106 and treated for 20 h with either α-amanitin (15 μg/μL; Sigma) or plain insect medium. A total of 1 × 108 cells were used for ChIP, 1 × 106 cells were used to make a nuclear extract as described in Akhtar et al. (2000), and 1 × 106 cells were used for RNA purification. The samples were analyzed by SDS-PAGE followed by Western blot analysis.
Immuno-FISH on polytene staining
Preparation of polytene chromosomes and immuno-FISH were performed as described (http://www.igh.cnrs.fr/equip/cavalli/Lab%20Protocols/Immunostaining.pdf). The location of the target genes was detected with specific probes made against the mini-white gene present in our transgenic cassettes. The probes for FISH were generated using random primed dioxygen-dUTP labeling (Roche) of the mini-white gene. MSL1, MSL3, MOF, and MLE antibodies were used at 1:500 dilution (Mendjan et al. 2006). Images were captured with an AxioCamHR CCD camera on a Zeiss Axiovert200M microscope using a 100× PlanApochomat NA 1.4 oil immersion objective.
Fly genetics
Flies were raised on standard cornmeal–agar–yeast medium at 18°C or 25°C. To generate transgenic flies carrying different insertions, all fragments were cloned using PCR-based strategy (primers available on request) into the p[PCaSpeR]4 vector (Pirrotta 1988), except for w-;P{w+ UAS-mof} and w-;P{w+ tub-mof}, where the mof cDNA was cloned downstream from multiple GAL4-binding sites, pP[UAST] (Brand and Perrimon 1993), and downstream from a 2.4-kb tubulin promoter sequence, respectively. For the line y− w−;P{w+ tub < yellow+ > mof}, the yellow cassette was subcloned from p[PJ35] (Struhl and Basler 1993) into the tub-mof vector. Transgenic flies were made by P-element-mediated transformation of the w1118 (also referred to as w−) recipient stock (Rubin and Spradling 1982). Lines were selected for second or third chromosome insertions. To generate msl-3083 mutant larvae that carry P{w+ tub-mof} or P{w+ cg3016 1581–1920 5mer}, y− w−; msl-3083 {hsp83-msl-2} females were crossed to w−; msl-3083 P{w+ tub-mof}/TM6C, Sb Tb or P{w+ cg3016 1581–1920 5mer}/TM6C, Sb Tb males. msl-3083 female larvae, w−; msl-3083 P{hsp83-msl-2}/msl-3083 P{w+ tub-mof} were distinguished by the absence of the Tb phenotype. To create y− w−; P{w+ tub-mof} from y− w−; P{w+ tub < y+ < mof}, male flies were crossed to female y w P{70FLP}3F and progeny was subjected to heat shock at various times during development. Since the stocks were mutant for the endogenous y gene, progeny was selected for a loss of the y+ marker.
Acknowledgments
We thank members of the laboratory for helpful discussions. We thank Ann-Mari Voie for fly injections. We are grateful to Ritsuko Suyama for her excellent technical expertise for generating transgenic flies. We thank Juerg Mueller for providing the tubulin-driven pho flies. We are grateful to Andreas Ladurner, Eileen Furlong, and Francois Spitz for critical reading of the manuscript. This work was supported by DFG SPP1129 “Epigenetics” and the EU funded NoE “Epigenome” to A.A.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.430807
References
- Akhtar A., Zink D., Becker P.B., Zink D., Becker P.B., Becker P.B. Chromodomains as RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- Alekseyenko A.A., Larschan E., Lai W.R., Park P.J., Kuroda M.I., Larschan E., Lai W.R., Park P.J., Kuroda M.I., Lai W.R., Park P.J., Kuroda M.I., Park P.J., Kuroda M.I., Kuroda M.I. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes & Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B.S., Gorman M., Marin I., Gorman M., Marin I., Marin I. Dosage compensation in Drosophila. Annu. Rev. Genet. 1994;28:491–521. doi: 10.1146/annurev.ge.28.120194.002423. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Charlesworth B. Sex chromosomes: Evolution of the weird and wonderful. Curr. Biol. 2005;15:R129–R131. doi: 10.1016/j.cub.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Dahlsveen I.K., Gilfillan G.D., Shelest V.I., Lamm R., Becker P.B., Gilfillan G.D., Shelest V.I., Lamm R., Becker P.B., Shelest V.I., Lamm R., Becker P.B., Lamm R., Becker P.B., Becker P.B. Targeting determinants of dosage compensation in Drosophila. PLoS Genet. 2006;2:e5. doi: 10.1371/journal.pgen.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demakova O.V., Kotlikova I.V., Gordadze P.R., Alekseyenko A.A., Kuroda M.I., Zhimulev I.F., Kotlikova I.V., Gordadze P.R., Alekseyenko A.A., Kuroda M.I., Zhimulev I.F., Gordadze P.R., Alekseyenko A.A., Kuroda M.I., Zhimulev I.F., Alekseyenko A.A., Kuroda M.I., Zhimulev I.F., Kuroda M.I., Zhimulev I.F., Zhimulev I.F. The MSL complex levels are critical for its correct targeting to the chromosomes in Drosophila melanogaster. Chromosoma. 2003;112:103–115. doi: 10.1007/s00412-003-0249-1. [DOI] [PubMed] [Google Scholar]
- Fagegaltier D., Baker B.S., Baker B.S. X chromosome sites autonomously recruit the dosage compensation complex in Drosophila males. PLoS Biol. 2004;2:e341. doi: 10.1371/journal.pbio.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen J.P. Dosage compensation in Drosophila: Evidence that daughterless and Sex-lethal control X chromosome activity at the blastoderm stage of embryogenesis. Genetics. 1987;117:477–485. doi: 10.1093/genetics/117.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan G.D., Straub T., de Wit E., Greil F., Lamm R., van Steensel B., Becker P.B., Straub T., de Wit E., Greil F., Lamm R., van Steensel B., Becker P.B., de Wit E., Greil F., Lamm R., van Steensel B., Becker P.B., Greil F., Lamm R., van Steensel B., Becker P.B., Lamm R., van Steensel B., Becker P.B., van Steensel B., Becker P.B., Becker P.B. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes & Dev. 2006;20:858–870. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y., Mengus G., Gilfillan G., Kennedy H.G., Stuckenholz C., Kelley R.L., Becker P.B., Kuroda M.I., Mengus G., Gilfillan G., Kennedy H.G., Stuckenholz C., Kelley R.L., Becker P.B., Kuroda M.I., Gilfillan G., Kennedy H.G., Stuckenholz C., Kelley R.L., Becker P.B., Kuroda M.I., Kennedy H.G., Stuckenholz C., Kelley R.L., Becker P.B., Kuroda M.I., Stuckenholz C., Kelley R.L., Becker P.B., Kuroda M.I., Kelley R.L., Becker P.B., Kuroda M.I., Becker P.B., Kuroda M.I., Kuroda M.I. Association and spreading of the Drosophila dosage compensation complex from a discrete roX1 chromatin entry site. EMBO J. 2001;20:2236–2245. doi: 10.1093/emboj/20.9.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R.L., Meller V.H., Gordadze P.R., Roman G., Davis R.L., Kuroda M.I., Meller V.H., Gordadze P.R., Roman G., Davis R.L., Kuroda M.I., Gordadze P.R., Roman G., Davis R.L., Kuroda M.I., Roman G., Davis R.L., Kuroda M.I., Davis R.L., Kuroda M.I., Kuroda M.I. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Klymenko T., Papp B., Fischle W., Kocher T., Schelder M., Fritsch C., Wild B., Wilm M., Muller J., Papp B., Fischle W., Kocher T., Schelder M., Fritsch C., Wild B., Wilm M., Muller J., Fischle W., Kocher T., Schelder M., Fritsch C., Wild B., Wilm M., Muller J., Kocher T., Schelder M., Fritsch C., Wild B., Wilm M., Muller J., Schelder M., Fritsch C., Wild B., Wilm M., Muller J., Fritsch C., Wild B., Wilm M., Muller J., Wild B., Wilm M., Muller J., Wilm M., Muller J., Muller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes & Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legube G., McWeeney S.K., Lercher M.J., Akhtar A., McWeeney S.K., Lercher M.J., Akhtar A., Lercher M.J., Akhtar A., Akhtar A. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes & Dev. 2006;20:871–883. doi: 10.1101/gad.377506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi J.C., Kelly W.G., Panning B., Kelly W.G., Panning B., Panning B. Chromatin remodeling in dosage compensation. Annu. Rev. Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Lyman L.M., Copps K., Rastelli L., Kelley R.L., Kuroda M.I., Copps K., Rastelli L., Kelley R.L., Kuroda M.I., Rastelli L., Kelley R.L., Kuroda M.I., Kelley R.L., Kuroda M.I., Kuroda M.I. Drosophila male-specific lethal-2 protein: Structure/function analysis and dependence on MSL-1 for chromosome association. Genetics. 1997;147:1743–1753. doi: 10.1093/genetics/147.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S., Akhtar A., Akhtar A. The right dose for every sex. Chromosoma. 2006;116:95–106. doi: 10.1007/s00412-006-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S., Taipale M., Kind J., Holz H., Gebhardt P., Schelder M., Vermeulen M., Buscaino A., Duncan K., Mueller J., Taipale M., Kind J., Holz H., Gebhardt P., Schelder M., Vermeulen M., Buscaino A., Duncan K., Mueller J., Kind J., Holz H., Gebhardt P., Schelder M., Vermeulen M., Buscaino A., Duncan K., Mueller J., Holz H., Gebhardt P., Schelder M., Vermeulen M., Buscaino A., Duncan K., Mueller J., Gebhardt P., Schelder M., Vermeulen M., Buscaino A., Duncan K., Mueller J., Schelder M., Vermeulen M., Buscaino A., Duncan K., Mueller J., Vermeulen M., Buscaino A., Duncan K., Mueller J., Buscaino A., Duncan K., Mueller J., Duncan K., Mueller J., Mueller J., et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Oh H., Park Y., Kuroda M.I., Park Y., Kuroda M.I., Kuroda M.I. Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes & Dev. 2003;17:1334–1339. doi: 10.1101/gad.1082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Bone J.R., Kuroda M.I., Bone J.R., Kuroda M.I., Kuroda M.I. Multiple classes of MSL binding sites target dosage compensation to the X chromosome of Drosophila. Curr. Biol. 2004;14:481–487. doi: 10.1016/j.cub.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Orlando V., Paro R., Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Park Y., Mengus G., Bai X., Kageyama Y., Meller V.H., Becker P.B., Kuroda M.I., Mengus G., Bai X., Kageyama Y., Meller V.H., Becker P.B., Kuroda M.I., Bai X., Kageyama Y., Meller V.H., Becker P.B., Kuroda M.I., Kageyama Y., Meller V.H., Becker P.B., Kuroda M.I., Meller V.H., Becker P.B., Kuroda M.I., Becker P.B., Kuroda M.I., Kuroda M.I. Sequence-specific targeting of Drosophila roX genes by the MSL dosage compensation complex. Mol. Cell. 2003;11:977–986. doi: 10.1016/s1097-2765(03)00147-3. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnology (N. Y.) 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- Rubin G.M., Spradling A.C., Spradling A.C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Sabo P.J., Humbert R., Hawrylycz M., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., Humbert R., Hawrylycz M., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., Hawrylycz M., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., McArthur M., Stamatoyannopoulos J.A., Stamatoyannopoulos J.A. Genome-wide identification of DNaseI hypersensitive sites using active chromatin sequence libraries. Proc. Natl. Acad. Sci. 2004;101:4537–4542. doi: 10.1073/pnas.0400678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass G.L., Pannuti A., Lucchesi J.C., Pannuti A., Lucchesi J.C., Lucchesi J.C. Male-specific lethal complex of Drosophila targets activated regions of the X chromosome for chromatin remodeling. Proc. Natl. Acad. Sci. 2003;100:8287–8291. doi: 10.1073/pnas.1332749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.R., Allis C.D., Lucchesi J.C., Allis C.D., Lucchesi J.C., Lucchesi J.C. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem. 2001;276:31483–31486. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- Straub T., Becker P.B., Becker P.B. Dosage compensation: The beginning and end of generalization. Nat. Rev. Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- Struhl G., Basler K., Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]