Abstract
Efficient ribosome biogenesis requires coordination of a highly complex series of events. Early events include pre-RNA transcription, processing, and modification. Analysis in yeast has demonstrated that t-UTPs, components of the U3 snoRNA-containing pre-rRNA processing complex, are required for efficient transcription of ribosomal genes (rDNA) by RNA polymerase I (pol I). Here, we characterize human t-UTPs and establish that their ability to link transcription and pre-rRNA processing is evolutionarily conserved. The pol I transcription factor UBF binds extensively across rDNA throughout the cell cycle, resulting in a specialized form of chromatin that is the hallmark of active nucleolar organizer regions (NORs). Transcriptionally silent pseudo-NORs are ectopic, chromosomally integrated, artificial arrays that mimic this specialized chromatin structure. Pseudo-NORs sequester t-UTPs and factors linking transcription with pre-rRNA modification (Nopp140 and Treacle). Recruitment is independent of transcription, the underlying DNA sequence, and location within the nucleolus. Previously, we have demonstrated that pseudo-NORs sequester every component of the pol I transcription machinery. Taken together, these results highlight the importance of the specialized chromatin structure at active NORs in coordinating early events in ribosome biogenesis and nucleolar formation.
Keywords: Nucleolus, rDNA transcription, pre-rRNA processing, nucleolar organizer region (NOR), t-UTPs, UBF
The production of 18S, 5.8S, and 28S ribosomal RNAs (rRNAs) is a highly elaborate and evolutionarily conserved process that occurs in a discrete subnuclear compartment, the nucleolus. The human genome contains 300–400 copies of a 43-kb ribosomal gene (rDNA) repeat distributed among five nucleolar organizer regions (NORs). Transcription of rDNA by RNA polymerase I (pol I) yields a 47S pre-rRNA that is matured by a complex series of endonucleolytic and exonucleolytic cleavages as well as rRNA methylations and pseudouridylations prior to assembly with ribosomal proteins to yield ribosomal subunits. Ribosome synthesis is a major metabolic undertaking in all cells and as such is a highly coordinated exercise (Warner 1999). This is highlighted by the finding that perturbations in ribosome biogenesis provide a major cellular stress-sensing mechanism (Olson 2004).
Although transcription is silenced during mitosis, rDNA chromatin on competent NORs remains undercondensed and forms a visible structure termed the secondary constriction. Many factors involved in ribosome biogenesis remain associated with competent NORs during mitosis (Jordan et al. 1996; Roussel et al. 1996), facilitating the rapid onset of rDNA transcription and nucleolar reformation as cells exit mitosis. The nucleolus can be divided into three morphological domains. Current evidence suggests that the fibrillar center (FC) represents pools of factors required for transcription and pre-rRNA processing together with nontranscribed rDNA (Raska et al. 2006). Transcription occurs either in the dense fibrillar component (DFC) or at the FC/DFC interface with early pre-rRNA processing occurring in the DFC and ribosomal subunit assembly in the granular component. The colocalization of many factors involved in transcription and pre-rRNA processing, observed by light microscopy, and the close juxtaposition of these processes, observed by electron microscopy, provide further evidence of coordination or indeed coupling between these steps of ribosome biogenesis. But perhaps the most convincing demonstration of this point comes from electron micrographs of actively transcribing ribosomal genes that reveal the presence of highly condensed terminal knobs at the 5′ end of nascent pre-RNA transcripts (Miller and Beatty 1969), now known to be an assembly of pre-rRNA processing complexes (Mougey et al. 1993).
While a number of the early events in rRNA production have been studied in isolation, how these processes are coordinated is less well understood. The pol I transcription machinery has been characterized in detail (Russell and Zomerdijk 2005). It is now well established that one component of the pol I machinery, upstream binding factor (UBF), binds extensively across the ribosomal gene repeat including both the intergenic spacer (IGS) and 47S coding sequences, performing both a structural role (O’Sullivan et al. 2002; Mais et al. 2005) and a role in transcriptional elongation (Stefanovsky et al. 2006). In human cells it appears that <10% of pol I is engaged in elongation complexes at any given time (Dundr et al. 2002). This excess appears to colocalize with UBF and ribosomal genes within nucleoli. The form of chromatin specified by extensive UBF binding can recruit the pol I machinery independent of transcription (Mais et al. 2005). This point was most convincingly demonstrated by the finding that arrays of a heterologous UBF binding site inserted at ectopic sites formed novel secondary constrictions during metaphase. During interphase these so-called pseudo-NORs sequester the pol I machinery independent of both transcription and the nucleolus. Presumably, UBF-mediated recruitment maintains a localized high concentration of the pol I machinery to support the rates of transcription observed on ribosomal genes. An extension of this logic is that components of the pre-rRNA processing machinery could be recruited to ribosomal gene chromatin prior to their engagement in complexes on the pre-rRNA. This would provide a means of coordinating these two key steps in ribosome biogenesis.
Cleavages and modifications of rRNA are directed by two classes of small nucleolar RNAs (snoRNAs) (Decatur and Fournier 2003). Several box C/D snoRNAs, including U3, U8, and U14, are essential for pre-rRNA cleavage events. The remaining box C/D snoRNAs and H/ACA snoRNAs direct methylation and pseudouridine formation, respectively. Box C/D snoRNAs associate with four core proteins, 15.5K, NOP56, NOP58, and the methyltransferase fibrillarin. H/ACA snoRNAs associate with Gar1, Nhp2, Nop10, and the pseudouridylase Nap57 (also known as dyskerin) (for review, see Meier 2005). U3 snoRNA is required for the cleavage events that result in 18S rRNA production and base pairs with sequences in the 5′ ETS and 18S rRNA (Kass et al. 1990). It is found in both a small 12S monoparticle as well as a larger (80S–90S) complex. The 12S particle comprises the four core box C/D snoRNP proteins and an additional U3-specific protein, hU3–55K (Lubben et al. 1993). The 80S particle was known to comprise the components of the 12S monoparticle and additional proteins including MPP10, Imp3, and Imp4 (Granneman et al. 2003). Proteomic analysis of this larger complex in yeast identified previously known U3 snoRNA-associated proteins as well as an array of proteins derived from open reading frames whose function had not previously been characterized, but many of which were conserved in humans (Dragon et al. 2002). These proteins were collectively termed UTPs (U Three Proteins), and the complex was termed the SSU (small ribosomal subunit) processome. Now it is generally agreed that the SSU processome/90S pre-ribosome is the terminal knob. Depletion of SSU processome components results in loss of terminal knobs on the 5′ end of nascent pre-rRNA. Grandi et al. (2002) purified a 90S complex similar in composition to the SSU processome and showed that it contains pre-rRNA sequences including the 5′ ETS. Depletion of a subset of UTPs (transcriptional or t-UTPs) results in up to a fourfold drop in the number of nascent transcripts on each rDNA repeat (Gallagher et al. 2004). Further evidence indicated that t-UTPs function as a distinct subcomplex that is recruited to rDNA independently of the remaining SSU components. A distinct t-UTP complex was also purified by the Greenblatt laboratory and termed UTPA (Krogan et al. 2004).
Here, we characterize the human orthologs of these t-UTPs and demonstrate their involvement in both transcription and U3 snoRNA-dependent pre-rRNA processing. We show that t-UTPs but not other components of the mature processome are recruited directly to the form of ribosomal gene chromatin specified by extensive UBF binding. Recruitment occurs independent of transcription, the underlying DNA sequence, and nucleolar context, but is strictly UBF-dependent. We also show that TCOF1, a nucleolar protein that has links with both transcription and methylation of rRNA (Hayano et al. 2003; Valdez et al. 2004; Gonzales et al. 2005), and Nopp140, a chaperone for box H/ACA snoRNPs (Meier 2005), are both also recruited to ribosomal gene chromatin independent of transcription. These results highlight the importance of the specialized chromatin structure associated with extensive UBF binding in coordinating ribosome biogenesis.
Results
Identification of human t-UTPs
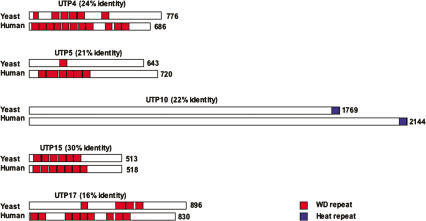
In yeast, t-UTPs link rDNA transcription and the pre-rRNA processing steps mediated by U3 snoRNP (Gallagher et al. 2004). To investigate if such links occur in human cells, we first sought to identify the human counterparts of yeast t-UTPs. TBLASTN searches of cDNA sequences with yeast peptide sequences identified open reading frames that are probable human orthologs of yeast UTP4, UTP5, UTP10, UTP15, and UTP17. BLAST searches failed to identify orthologs of yeast UTP8 and UTP9. This is unsurprising as they are also absent in fission yeast. Sequence conservation between yeast and human (h) t-UTPs ranges between 16% identity in the case of UTP17 and 30% identity for UTP15 (Fig. 1). Human UTP4, UTP5, UTP15, and UTP17 each contain multiple WD repeats. UTP10 is devoid of recognizable motifs except for the presence of a conserved Heat repeat at its extreme C terminus. A further indication that these open reading frames represent the true orthologs of yeast t-UTPs is that peptides derived from each of them have been identified in a proteomic analysis of human nucleoli (Andersen et al. 2002). Coding sequences for each human t-UTP were introduced into the Gateway recombination-based cloning system (Invitrogen) to facilitate production of recombinant proteins and generation of expression constructs encoding epitope-tagged fusion proteins (see Materials and Methods for the source of full-length cDNA clones).
Figure 1.
Yeast and human t-UTPs. Alignments between yeast and human t-UTPs were performed with ClustalW (EMBL–EBI). Yeast and human t-UTPs are drawn in cartoon form with percentage amino acid sequence identity shown above each pair. Numbers on the right indicate the length of each open reading frame. (Red blocks) WD repeats; (blue blocks) Heat repeats.
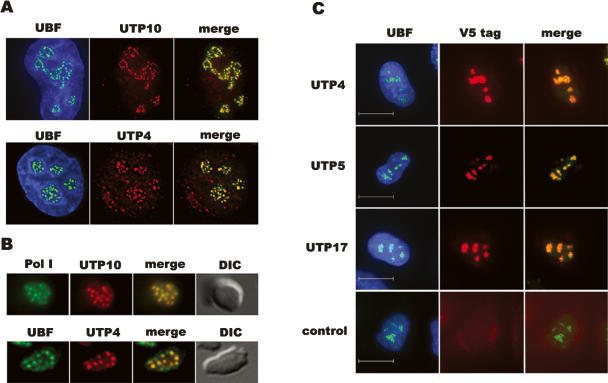
Endogenous human UTP10 and UTP4 are clearly enriched in nucleloli as revealed by staining of HeLa cells with affinity-purified antibodies raised against recombinant protein (Fig. 2A). Staining of nucleoli isolated from HeLa cells with these antibodies further reveals that UTP4 and UTP10 precisely colocalize with foci (FC/DFC) of pol I and UBF within nucleoli, as would be expected for proteins that function in both transcription and early pre-rRNA processing (Fig. 2B). To confirm this result and address the localization of the remaining t-UTPs, HeLa cells were transfected with plasmids encoding V5 epitope–UTP fusion proteins. Staining of transfected cells with antibodies against UBF to visualize nucleoli and a mAb that recognizes the V5 epitope reveal that in addition to UTP4 and UTP10, UTP5 and UTP17 also localize to nucleoli (Fig. 2C). For reasons that are not clear to us, expression from UTP15 constructs is not detectable in transfected cells. Nevertheless, we are confident that it is the ortholog of the yeast protein due to its presence in the nucleolar proteome (Andersen et al. 2002) and the effects observed on rRNA synthesis upon depletion of its mRNA (see below).
Figure 2.
T-UTPs localize to nucleoli in HeLa cells. (A) HeLa cells were stained with rhodamine-conjugated affinity-purified antibodies against UTP10 (upper panels) and UTP4 (lower panels). Nuclei were visualized with DAPI, and nucleoli were identified by co-staining with FITC-conjugated affinity-purified antibodies against UBF. (B) Nucleoli purified from HeLa cells were stained with UTP10 or UTP4 antibodies as above. FC/DFC structures within nucleoli were identified by co-staining with FITC-conjugated antibodies against either the pol I subunit RPA43 or UBF. Nucleoli were revealed using DIC optics. (C) HeLa cells were transfected with plasmids encoding V5 epitope-tagged UTP4, UTP5, and UTP17. Tagged proteins were identified with a monoclonal antibody against the V5 tag combined with a rhodamine-labeled secondary antibody. Nucleoli were visualized with UBF antibodies as above. Nontransfected cells provided a control. Scale bars, 10 μm.
Human t-UTPs are required for both efficient transcription and pre-rRNA processing
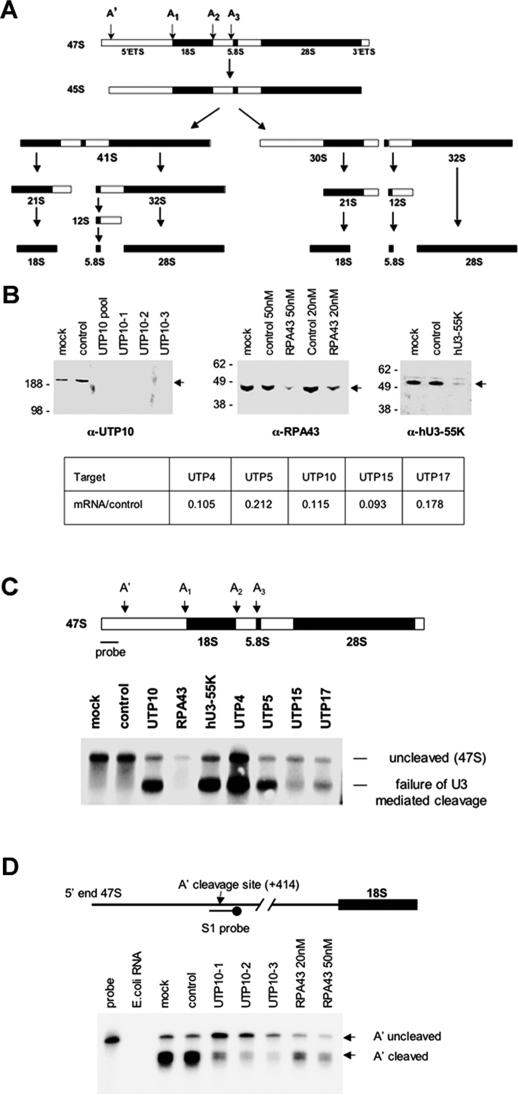
To address their role in rRNA biogenesis, we undertook siRNA-mediated depletion of individual t-UTPs. HeLa cells were transfected with pools or individual siRNA duplexes (Dharmacon) directed against t-UTPs, the pol I subunit RPA43, the U3 snoRNP protein hU3–55K, or a control duplex. SiRNA directed against RPA43 was expected to exhibit only transcriptional effects and U3–55K only pre-rRNA processing defects. Due to the abundance and stability of these targets, HeLa cells were initially transfected at low cell density. A second round of transfection was performed after 48 h and cells analyzed after a further 48 h. Where appropriate antibodies were available, the effectiveness of depletion was monitored by Western blotting (Fig. 3B). Both the pool and individual duplexes directed against UTP10 result in its depletion to undetectable levels. Whereas the standard concentration of siRNA (20 nM) was partially effective in RPA43 depletion, a higher concentration (50 nM) was more effective. hU3–55K was efficiently depleted with the standard concentration of duplex. As antibodies against UTP4 do not work sufficiently well in Western blotting, and antibodies against UTP5, UTP15, and UTP17 are unavailable, the efficiency of depletion was monitored by real-time RT-PCR (Fig. 3B; Supplemental Fig. 1). This demonstrated that t-UTP mRNA levels were reduced by between five- and 10-fold.
Figure 3.
SiRNA depletion of t-UTPs impairs pre-rRNA processing. (A) A schematic of the alternative pre-rRNA processing pathways that occur in HeLa cells (adapted with permission from Hadjiolova et al. 1993); © 1993 Blackwell Publishing. The position of U3 snoRNP-dependent cleavage sites (A′–A3) is shown above the 47S pre-rRNA. (B) HeLa cells were transfected with Smartpool or individual siRNA duplexes at a standard concentration of 20 nM. Duplexes targeting RPA43 were also used at a higher concentration (50 nM). Western blots were performed on equal amounts of total protein form mock, control, and siRNA-transfected cells. The identity of the siRNA duplex employed is shown above each sample, the antibody used is indicated below each panel, molecular weight markers (kDa) are on the left, and the position of the relevant antigen is shown (arrow) on the right of each panel. To assess the efficiency of siRNA-mediated knockdown of t-UTP mRNAs, quantitative real-time PCR was performed on reverse-transcribed RNA. (Table) mRNA level for each targeted UTP relative to the control (see Supplemental Fig. 1 for details). (C) A Northern blot was performed on RNA extracted from cells transfected with each of the t-UTPs, RPA43 (50 nM), and hU3–55K duplexes. RNA from control duplex and mock-transfected cells provided controls. (Arrows) Bands representing the intact 47S and the aberrant processing intermediate resulting from failure of U3-dependent cleavages. The location of the probe and sites of U3-dependent cleavage on the 47S are indicated in the cartoon above. (D) S1 nuclease protection assays were performed on RNA extracted from cells transfected with control, UTP10 (three separate duplexes), and RPA43 duplexes (20 nM and 50 nM). RNA from mock-transfected cells provided an additional control. E. coli RNA was used to demonstrate complete digestion of nonhybridizing probe under the conditions used. The positions of A′ uncleaved (full-length protected probe) and A′ cleaved signals are indicated (arrows on the right) (see Supplemental Fig. 3 for quantitation of this gel). The location of the A′ cleavage site and the S1 probe is shown in cartoon form above.
U3 snoRNP is required for cleavage at four sites within the pre-rRNA. The initial cleavage site, A′, is at nucleotide (nt) +414 of the pre-rRNA (Kass et al. 1987). Sites A1 and A2 border the 18S coding sequence, and site A3 is located on the 5′ side of the 5.8S coding sequence (Fig. 3A). To assess the impact of factor depletion on pre-rRNA processing, Northern blots were performed on RNA extracted from siRNA-treated cells. The probe utilized (nt +1 to +339) is derived from sequences on the 5′ side of the primary cleavage site and recognizes only the uncleaved 47S pre-rRNA in RNA from mock-transfected and control duplex-transfected cells. Depletion of hU3–55K severely compromises U3-dependent cleavage as evidenced by the accumulation of an aberrant processing intermediate comprising 5′ ETS, 18S, ITS1, and 5.8S sequences. The identity of this intermediate was confirmed using a number of other probes (Supplemental Fig. 2). No processing defects are observed upon depletion of RPA43, but as expected the level of 47S pre-rRNA is markedly reduced due to decreased levels of transcription. Depletion of each t-UTP clearly affects U3-dependent cleavage as revealed by accumulation of the same novel intermediate observed upon hU3–55k depletion, thus confirming their role in U3-dependent processing. As failure of U3-dependent cleavage results in stabilization of RNA sequences detected by this probe, we cannot assess transcriptional effects of t-UTP depletion using this assay. In any case, Northern blotting is not a reliable measure of ongoing transcription as nascent transcripts are not readily visualized due to their undefined length. Furthermore, due to the rapidity with which pre-rRNA is processed, it is likely that the majority of RNA species containing 5′ ETS sequences are nascent transcripts (Warner 2001). Nevertheless, it would appear that depletion of individual t-UTPs has differing effects on RNA levels. We reproducibly observe higher levels of the aberrant processing intermediate in UTP4-depleted cells than cells depleted in the other t-UTPs.
The earliest U3-snoRNP-dependent cleavage event in pre-rRNA processing is cleavage at the A′ site. In order to specifically address defects in this cleavage, we designed a 60-nt probe for use in S1 nuclease protection assays (Fig. 3D). Levels of full-length protected probe and truncated probe provide a quantitative measure of uncleaved and A′ cleaved transcripts, respectively. The ratio of these signals provides a measure of the efficiency of cleavage, and the combined signals provide an improved measure of the level of ongoing transcription. In mock and control duplex-transfected cells, we observe that the majority of transcripts detected have already been cleaved at the A′ site. Quantitation of this gel is shown in Supplemental Figure 3. In RPA43-depleted cells, we observe a similar ratio of cleaved to uncleaved but the total signal is reduced, indicative of lower levels of transcription. In cells treated with duplexes directed against UTP10, we observe a shift toward the majority of transcripts being uncleaved as well as a drop in total signal, especially evident with duplex 3. S1 nuclease protection assays, using short oligonucleotide probes, detect both released and nascent transcripts, thus providing a more quantitative measure of ongoing transcription than Northern blots. It would appear that depletion of UTP10 results in lower levels of transcription and that those transcripts that are produced are less likely to be cleaved by the U3 snoRNP.
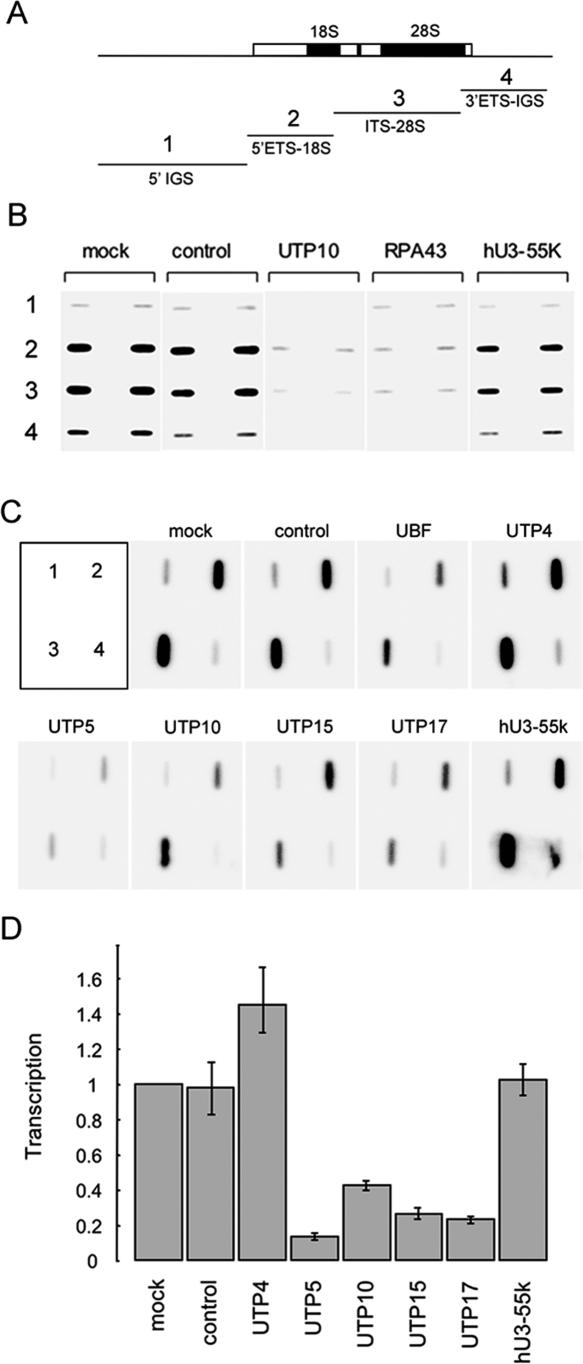
Nuclear run-on transcription assays provide a direct measure of ongoing transcription and were utilized to demonstrate that t-UTP-depleted cells exhibit reduced levels of ribosomal gene transcription. Run-on reactions were performed in the presence of αP32-GTP using equal numbers of nuclei isolated from siRNA-treated cells. Labeled RNA was then hybridized to slot blots of cloned ribosomal DNA (Fig. 4A). Figure 4B shows the results of nuclear run-on experiments comparing the effects of depleting UTP10, RPA43, and hU3–55k. This experiment clearly demonstrates that depletion of UTP10 and RPA43 have similarly reduced levels of ongoing transcription (fourfold drop) in comparison with controls. Note that depletion of hU3–55K has little effect.
Figure 4.
SiRNA depletion of tUTPs impairs transcription of pre-rRNA. (A) Labeled RNAs recovered from run-on transcription reactions were used to probe slot blots loaded with plasmids containing subcloned rDNA fragments. (1) (5′ IGS) An 11.9-kb EcoRI restriction fragment positioned immediately upstream of the promoter. (2 and 3) (5′ ETS-18S and ITS-28S) 5.3-kb and 7.1-kb EcoRI restriction fragments, respectively, both from the transcribed region. (4) (3′ ETS-IGS) A 5.7-kb EcoRI/BamHI restriction fragment that includes the 3′ ETS and 4.7 kb of IGS. (B) Run-on transcription reactions were performed with nuclei isolated from cells transfected with control, UTP10, RPA43, and hU3–55K siRNA duplexes. Nuclei from mock-transfected cells provided an additional control. Labeled RNAs were used to probe slot blots loaded in duplicate as indicated. (C) Run-on transcription reactions were performed in triplicate with nuclei from cells transfected with the indicated duplexes. Labeled RNAs were used to probe slot blots, loaded as indicated in the upper left panel. One set of experiments is shown. (D) Signals from the above triplicate sets of run-on transcription reactions were quantified using a phosphorimager and expressed relative to signals obtained from mock-transfected cells.
Using nuclei from cells treated with duplexes targeting each of the t-UTPs, run-on transcription reactions were performed in triplicate to provide a statistically more accurate measure of the transcriptional consequences. The results from one of the triplicate experiments are shown in Figure 4C and the quantitation from all three experiments in Figure 4D. These results confirm roles for t-UTPs in transcription by pol I. Depletion of UTP5, UTP10, UTP15, and UTP17 results in markedly reduced levels of transcription. Depletion of UTP4 has no negative effect on transcription, consistent with the results from Northern blotting (Fig. 3C). The differential effects of t-UTP depletion will be discussed below.
Human t-UTPs are recruited to ribosomal gene chromatin independent of transcription
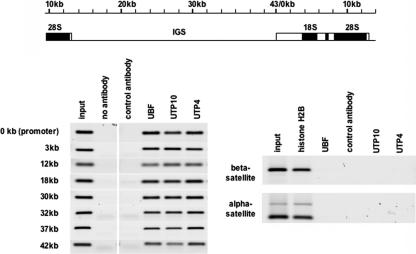
The observation that t-UTPs are required for efficient transcription of ribosomal genes suggests that they are recruited to rDNA prior to their inclusion into the terminal knob structure at the 5′ end of the pre-rRNA. Recruitment could be mediated by direct interaction with promoter DNA or indirectly through interactions with the transcription machinery. Previously, we have demonstrated that the nonengaged fraction of the pol I transcription machinery is associated with rDNA chromatin, predominantly with the IGS (Mais et al. 2005). This prompted us to look at the distribution of t-UTPs on the rDNA repeat. Nucleolar chromatin was prepared from HeLa cells fixed with 0.25% formaldehyde as previously described (Mais et al. 2005). Chromatin immunoprecipitation (ChIP) was then performed with affinity-purified antibodies against UTP4, UTP10, and UBF. DNA recovered from these and control ChIPs were analyzed by PCR with primer pairs from across the 43-kb human ribosomal gene repeat (Fig. 5). As previously published, UBF was found to interact with sequences across the rDNA repeat (O’Sullivan et al. 2002). It is also abundantly clear that both UTP4 and UTP10 bind to chromatin throughout the rDNA repeat. The specificity of ChIP is revealed by a number of controls. Firstly, rDNA sequences are not detected in either control ChIP. Secondly, PCR with primers against pericentromeric alpha-satellite DNA and beta-satellite DNA, located on NOR-bearing chromosome arms, reveals that these sequences are present in nucleoli and are bound by histone H2B but not by UBF, UTP4, or UTP10. As t-UTPs bind to sequences across the IGS, we can surmise that they are recruited to rDNA independent of transcription. Given the diversity of DNA sequences across the rDNA repeat, we can also infer that t-UTP recruitment is not specified by the underlying DNA sequence; rather, it is the consequence of protein–protein interactions arising from the specialized nature of ribosomal gene chromatin.
Figure 5.
T-UTPs associate with chromatin across the rDNA repeat. ChIP was performed on nucleolar chromatin with affinity-purified UBF, UTP10, and UTP4 (anti-peptide) antibodies. Control ChIPs used either a control antibody or no antibody. PCR was performed with aliquots of each ChIP and the input chromatin, using primer pairs that are distributed across the repeat (left). In order to position the primer pairs utilized (left), a cartoon drawing of the rDNA repeat is shown above. As a control, PCR was performed on ChIPs with antibodies against histone H2B, UBF, UTP4, UTP10, and a control antibody using beta-satellite- and alpha-satellite-specific primer pairs (right).
Recently, we have established that extensive UBF binding underlies the specialized form of chromatin associated with rDNA (Mais et al. 2005). Transcription by pol I can be specifically inhibited in vivo by incubating cells in a low concentration of Actinomycin D (Act D). This results in segregation of the nucleolus and formation of associated cap-like structures that contain UBF. In Supplemental Figure 4, we demonstrate that UTP10 precisely colocalizes with UBF in these structures in Act D-treated HeLa cells. This provides further support for a model in which t-UTPs are recruited to ribosomal gene chromatin independent of transcription.
t-UTPs but not other components of the terminal knob associate with pseudoNORs
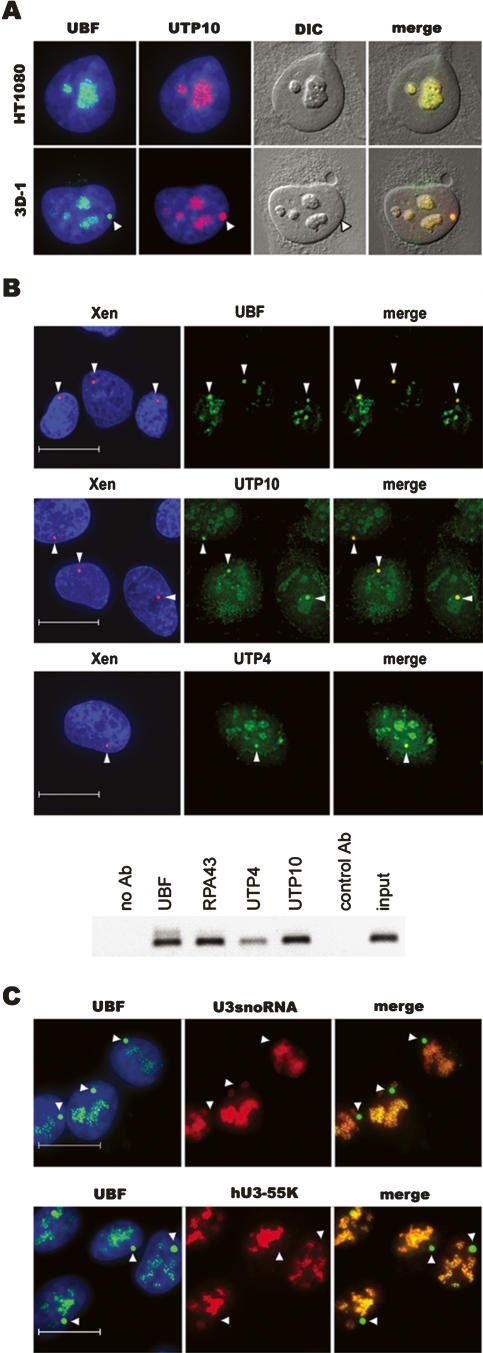
Large arrays of a heterologous UBF binding sequence (XEn, enhancer elements from Xenopus laevis rDNA) integrated at ectopic sites on human chromosomes result in formation of structures we have termed pseudo-NORs (Mais et al. 2005). During interphase, pseudo-NORs sequester not only UBF, but also every component of the pol I transcription machinery so far analyzed. Furthermore, pseudo-NORs can sequester these factors even when present on non-NOR-bearing chromosomes and localized outside of nucleoli. Pseudo-NORs offer a unique opportunity to study recruitment of proteins directly to rDNA chromatin. Furthermore, we can be more certain that they are devoid of transcription compared with nucleoli in Act D-treated cells. The pseudo-NOR-containing cell line 3D-1 was derived from the human cell line HT1080 and contains a 1.5-Mb array of XEn sequences on the long arm of chromosome 10. To confirm transcription independent recruitment of human t-UTPs, 3D-1 cells were stained with antibodies recognizing endogenous t-UTPs. Staining of 3D-1 cells with affinity-purified antibodies reveals that UTP10 is highly enriched within pseudo-NORs (Fig. 6A). To further demonstrate colocalization of t-UTPs with pseudo-NORs, we performed combined immuno-FISH (Fig. 6B). Staining of 3D-1 with antibodies against UBF, UTP10, or UTP4 reveals that these proteins colocalize with pseudo-NORs identified using a spectrum red-labeled XEn DNA probe. Localization of the remaining t-UTPs to pseudo-NORs was demonstrated by transfection of 3D-1 cells with plasmids encoding epitope-tagged proteins (data not shown). Targeting of UBF, pol I, and t-UTPs to pseudo-NORs was also confirmed by ChIP (Fig. 6B). These experiments clearly establish that t-UTPs target to pseudo-NORs, providing the most convincing evidence for transcription-independent recruitment.
Figure 6.
T-UTPs but not components of the U3 snoRNP associate with pseudo-NORs. (A) HT1080 and pseudo-NOR containing 3D-1 cells were co-stained with affinity-purified UTP10 (red) and UBF (green) antibodies. (B) Combined immuno-FISH was performed on 3D-1 cells. Cells were stained with affinity-purified UBF (green), UTP10 (green), and UTP4 (green) antibodies. Pseudo-NORs were revealed by hybridization with XEn DNA (red). ChIP was performed on nuclear chromatin with affinity-purified UBF, RPA43, UTP10, and UTP4 (anti-peptide) antibodies. Control ChIPs used either a control antibody or no antibody. PCR was performed with aliquots of each ChIP and the input chromatin, using a XEn-specific primer pair. (C) (Upper panels) U3 snoRNA was detected in 3D-1 cells by FISH with a Cy3.5-labeled oligonucleotide probe (red). Nucleoli and pseudo-NORs were visualized by staining with affinity-purified UBF (green) antibodies. (Lower panels) 3D-1 cells were co-stained with affinity-purified UBF (green) antibodies and hU3–55K antisera (red). (Arrowheads, A–C) Pseudo-NORs. Note that pseudo-NORs are visible under DIC optics. Scale bars, 10 μm.
Terminal knobs comprise many components including t-UTPs (Dragon et al. 2002; Grandi et al. 2002; Gallagher et al. 2004). Our finding that t-UTPs are recruited to ribosomal gene chromatin independent of transcription and consequently prior to terminal knob formation prompts the obvious question: When are U3 snoRNA and the remaining protein components of the terminal knob recruited? Once again, pseudo-NORs provide an opportunity to address this issue. In human cells, U3 snoRNA is found in two distinct ribonucleprotein (RNP) complexes: the 12S monoparticle and the terminal knob (Grandi et al. 2002; Granneman et al. 2003). The 12S monoparticle also contains 15.5K, Nop56p, Nop58p, fibrillarin, and hU3–55K proteins. Previously, we have demonstrated that fibrillarin does not associate with pseudo-NORs (Mais et al. 2005). Fibrillarin is, however, not a specific marker for the U3 snoRNP, as it is found associated with all box C/D snoRNAs. To specifically address the U3 snoRNP, we have determined if U3 snoRNA and the U3 snoRNP-specific protein hU3–55K associate with pseudo-NORs (Fig. 6C). RNA FISH with a U3 snoRNA-specific probe and staining with hU3–55K antibodies clearly demonstrates that these components do not associate with pseudo-NORs, implying a transcriptional dependence on recruitment. These results agree with a recent report that targeting of U3 snoRNP components to transfected rDNA plasmids is dependent on pol I transcription (Kopp et al. 2007). Staining of 3D-1 cells with an antibody against UTP12 (a member of the b-UTP subcomplex) (Krogan et al. 2004) and transfection of cells with GFP-tagged hImp4 and hMpp10 reveal that these terminal knob components do not associate with pseudo-NORs (data not shown). Thus, it would appear that as terminal knob constituents, t-UTPs are probably unique in being recruited to ribosomal gene chromatin independent of transcription.
TCOF-1 and Nopp140 associate with pseudo-NORs
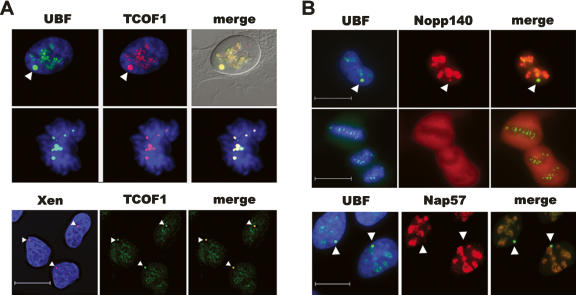
Pseudouridylation and 2′-O methylation of pre-rRNA carried out by box H/ACA and box C/D snoRNPs, respectively, occur cotranscriptionally (Warner 2001; Decatur and Fournier 2003). Are transcription and establishment of these modifications linked? Intriguingly, evidence for such links already exists. Nucleolar phosphoprotein Nopp140 associates with box H/ACA snoRNPs (Meier and Blobel 1994) and pol I (Yang et al. 2000). A related nucleolar phosphoprotein, TCOF1/Treacle, interacts with box C/D snoRNPs (Hayano et al. 2003) and binds to UBF (Valdez et al. 2004). If TCOF1 and Nopp140 perform a similar role to t-UTPs, we might predict that they would both be recruited to pseudo-NORs whereas components of box C/D and box H/ACA snoRNPs would not. Antibody staining and combined immuno-FISH of 3D-1 cells reveals that TCOF1 is highly enriched at pseudo-NORs. To reiterate, fibrillarin, a component of all box C/D sno-RNPs, does not associate with pseudo-NORs (Mais et al. 2005). We note also that TCOF1 remains associated with both NORs and pseudo-NORs through mitosis (Fig. 7A). Furthermore, TCOF1 colocalizes with UBF in Act D-treated HeLa cells (Supplemental Fig. 3). These observations provide further proof of its ability to interact with ribosomal gene chromatin independent of transcription. Antibody staining of 3D-1 cells with Nopp140 antibodies reveals that it also associates with pseudo-NORs, although apparently not to the same degree as observed with TCOF1 (Fig. 7B). Nap57 (dyskerin), a component of box H/ACA snoRNPs, clearly does not target to pseudo-NORs (Fig. 7B). These results are in full agreement with a role for t-UTPs, TCOF1, and Nopp140 in linking transcription with pre-rRNA processing and modification.
Figure 7.
TCOF1 and Nopp140 associate with pseudo-NORs. (A) 3D-1 cells stained with affinity-purified UBF (green) antibodies and TCOF1 antibodies (red). (Upper panels) An interphase cell; (middle panels) a mitotic cell; (lower panels) combined immuno-FISH performed on 3D-1 cells, with TCOF1 antibodies (green) and pseudo-NORs revealed by hybridization with XEn DNA (red). (B) 3D-1 cells stained with affinity-purified UBF (green) antibodies and Nopp140 antibodies (red). (Upper panels) An interphase cell; (middle panels) a mitotic cell; (lower panels) 3D-1 cells stained with affinity-purified UBF (green) antibodies and Nap57 antibodies (red). (Arrowhead, A,B) Pseudo-NORs. Scale bars, 10 μm.
UBF is required for transcription-independent recruitment of Pol I, t-UTPs, and TCOF1
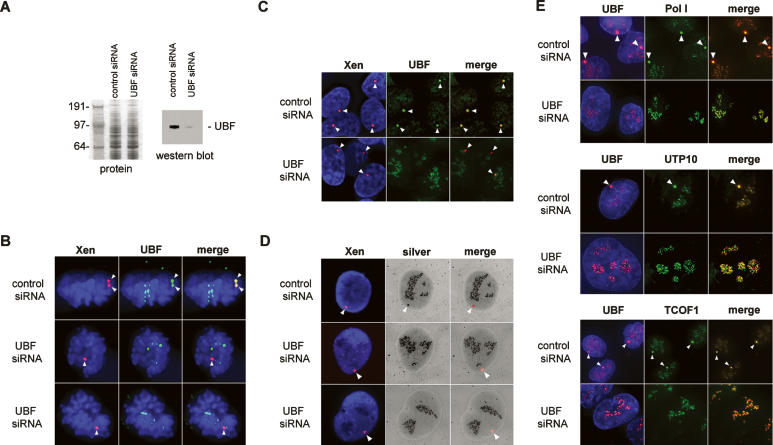
The inference from this and previous work (Mais et al. 2005) is that UBF is required for transcription-independent recruitment of the pol I machinery and factors linking transcription with pre-rRNA processing. To test this hypothesis, we depleted UBF in the pseudo-NOR-containing cell line. 3D-1 cells were repeatedly transfected with either control or UBF siRNA duplexes. The ability of the siRNA duplexes to reduce cellular levels of UBF was demonstrated by Western blotting (Fig. 8A). Combined immuno-FISH shows that transient depletion of UBF preferentially effect pseudo-NORs. In cells treated with a control siRNA duplex, every pseudo-NOR shows strong UBF antibody staining throughout the cell cycle. Metaphase and interphase cells are shown in Figure 8,B and C, respectively. In cells treated with UBF siRNA duplexes, the majority of pseudo-NORs are negative for UBF. During mitosis, pseudo-NORs lack UBF staining whereas NORs are positive (Fig. 8B). In interphase cells, pseudo-NORs also lack UBF staining but nucleoli are positive (Fig. 8C).
Figure 8.
UBF is required for recruitment of pol I, t-UTPs, and TCOF1 to pseudo-NORs. (A) 3D-1 cells were repeatedly transfected with either control or UBF siRNA duplexes. Depletion of UBF was monitored by Western blot (right panel) loaded with equal amounts of protein (left panel). The sizes of molecular weight markers (kDa) are indicated on the left. (B) Combined immuno-FISH was performed on 3D-1 cells transfected with control or UBF duplexes. Cells were stained with affinity-purified UBF (green), and pseudo-NORs were revealed by hybridization with XEn DNA (red). (Upper panels) A control siRNA transfected cell in metaphase, (middle and lower panels) UBF siRNA-transfected cells in telophase. (C) Combined immuno-FISH performed as above on control and UBF siRNA-transfected interphase cells (upper and lower panels, respectively). (D) Combined FISH and silver staining was performed on 3D-1 cells transfected with control (upper panels) or UBF (middle and lower panels) duplexes. Pseudo-NORs were revealed by hybridization with XEn DNA (red) prior to silver staining. (E) 3D-1 cells transfected with control (upper panels in each set) or UBF (lower panels in each set) duplexes were stained with UBF (red) and either Pol I (RPA43), UTP10, or TCOF1 antibodies (green). (Arrowheads, B–E) Pseudo-NORs.
Interphase nucleoli strongly stain with silver due to the presence of many argyophilic proteins. Previously, we have observed that pseudo-NORs are also strongly positive for silver staining during interphase (Prieto and McStay 2005). This presumably reflects the wide variety of nucleolar proteins recruited to pseudo-NORs. To determine if the ability of pseudo-NOR to recruit these argyophilic nucleolar proteins is UBF-dependent, we first visualized pseudo-NORs by FISH using a spectrum red-labeled XEn DNA probe, then silver staining was performed. Strikingly, we observe complete loss of silver staining at pseudo-NORs in cells transfected with UBF, but not control siRNA duplexes (Fig. 8D), confirming a role for UBF. To address the recruitment of specific factors, we co-stained both control and UBF siRNA-transfected cells with UBF and either pol I, UTP10, or TCOF1 antibodies (Fig. 8E). These experiments reveal a strict UBF requirement for recruitment of the pol I transcription machinery and factors linking transcription with pre-rRNA processing.
Discussion
Gallagher et al. (2004) identified seven so-called t-UTPs—UTP4, UTP5, UTP8, UTP9, UTP10, UTP15, and UTP17—that link transcription and pre-rRNA processing in yeast. A number of lines of evidence suggest that t-UTPs function as a complex. Firstly, Krogan et al. (2004) identified the same t-UTPs in a complex termed UTPA. Secondly, t-UTPs form a complex in the absence of U3 snoRNA (Gallagher et al. 2004). Finally, ChIP demonstrated that at least six of the seven t-UTPs associate with rDNA chromatin (Gallagher et al. 2004). At least two of these t-UTPs, UTP8 and 9, apparently associate with chromatin independent of transcription. Although, the finding in the same experiment that Nop1, the yeast homolog of fibrillarin and a core component of box C/D snoRNPs, also associated with chromatin suggests that transcription may not have been fully repressed in this experiment.
Here, we have identified and characterized the human orthologs of t-UTPs. Antibodies against two of them (UTP4 and UTP10) demonstrate that the endogenous proteins colocalize within the FC/DFC, and tagged versions of these and remaining t-UTPs localize to nucleoli. Northern blots clearly demonstrate that depletion of each t-UTP results in accumulation of an aberrant processing intermediate, consistent with failure of U3 snoRNA-dependent cleavage events. This result was further confirmed by analyzing cleavage at the A′ site in the 5′ ETS by S1 nuclease protection. A transcriptional role was unequivocally demonstrated by nuclear run-on analysis. We conclude that a function for t-UTPs in linking transcription and pre-rRNA processing has been conserved throughout evolution.
For the t-UTP complex to link transcription with processing, one could imagine that it must make protein–protein contacts with both machineries. Since depletion of UTP4 has consequences only on processing, we can infer that, unlike yeast, its primary role in human cells is to link the t-UTP complex with the remaining components of the terminal knob.
Yeast UTP8 and UTP9 lack common protein–protein interaction motifs such as WD and Heat repeats. They also appear to be the most divergent components of the t-UTP complex. Homologs of these proteins are present in related yeast strains such as Candida albicans but are missing in the more distantly related fission yeast and in all higher eukaryotes. We presume that as yet unidentified components of the human t-UTP/UTPA complex perform the role of yUTP8 and 9. In addition to the seven t-UTPs, the UTPA complex contains the protein Pol5, so named because it bears sequence similarity with DNA polymerases (Krogan et al. 2004). Apparently, this protein is also required for optimum transcription and binds to rDNA chromatin (Gallagher et al. 2004). Interestingly, a highly related human protein (>15% identity) termed p160 or Myb-binding protein 1A is localized predominantly in the nucleolus (Tavner et al. 1998; Andersen et al. 2002).
Having established that the human t-UTP complex is required for optimal rDNA transcription in human cells, we next sought to determine if it associated with rDNA chromatin. In previous work, we had made the somewhat surprising finding that a significant fraction of the pol I machinery associates with IGS chromatin (Mais et al. 2005). Our interpretation was that this represents the pools of unengaged transcription machinery known to be present within nucleoli (Dundr et al. 2002). Here, we demonstrate that t-UTPs likewise associate with the IGS. As there is no evidence for transcription in the human IGS, we can reasonably assume that t-UTPs can be recruited to rDNA independent of transcription. Further support comes from the colocalization of UBF and UTP10 observed in Act D-treated cells. However, the demonstration that pseudo-NORs sequester t-UTPs provides incontrovertible proof of transcription-independent recruitment to rDNA chromatin. Furthermore, the location of pseudo-NORs outside of nucleoli allows us to clearly separate recruitment of t-UTPs from recruitment of other terminal knob components including U3 snoRNA, hU3–55K, and fibrillarin. At the moment, we do not know precisely how the human t-UTP complex is recruited to rDNA chromatin, but we can say with certainty that it is UBF-dependent.
We believe that t-UTPs directly influence transcription, rather than inducing a feedback mechanism that inhibits pol I activity. Firstly, depletion of hU3–55K has a profound effect on U3 snoRNA-dependent processing but no appreciable effect on transcription. Secondly, the recruitment of t-UTPs to chromatin positions them in the right place at the right time to directly influence transcription. Finally, the rapidity with which depletion of t-UTPs in yeast results in inhibition of pre-rRNA synthesis also indicates a direct role in transcription (Gallagher et al. 2004). In the longer term, however, one would clearly expect that disruption of processing by targeting non-tUTP components of the processome (such as hU3–55k) would induce nucleolar stress and result in transcription inhibition. There is now ample evidence that disruption of ribosome biogenesis induces a p53-dependent nucleolar stress pathway (Olson 2004). Note that HeLa cells are functionally negative for p53. If the same experiments were performed in cells that support a p53 response, we may have observed more pronounced effects on transcription upon depletion of hU3–55K. In fact, we chose to work on HeLa cells so we could discriminate between direct and indirect effects of factor depletion. Next, we have to discriminate between effects on transcriptional initiation and elongation. While our data do not speak directly to this issue, the work in yeast clearly favors a model in which t-UTPs influence rates of transcription initiation or the early phase of transcriptional elongation. This is the simplest interpretation of the chromosomal spread data in which the number of nascent transcripts over each repeat drops fivefold upon t-UTP depletion (Gallagher et al. 2004).
A protein here described as UTP4 has also been identified as cirhin, the product of the CIRH1A gene. North American Indian childhood cirrhosis (NAIC), a recessive genetic disorder, arises as a consequence of a missense mutation (R565W) in CIRH1A (Chagnon et al. 2002). More recently, it was demonstrated that cirhin is localized principally in the nucleolus, although the similarity to yeast UTP4 was not commented on (Yu et al. 2005). It remains to be seen if defects in ribosome biogenesis are responsible for NAIC pathology.
Human UTP10 had previously been identified as an uncharacterized protein termed BAP28. Zebrafish embryos homozygous for a mutant bap28 allele display excess apoptosis primarily in the central nervous system (Azuma et al. 2006). As predicted, rRNA synthesis is impaired in this mutant. This increased apoptotic cell death appears to be mediated by p53, as inhibition of p53 expression with an antisense morpholino leads to a significant reduction of cell death. These results are in keeping with the role of the nucleolus as a major cellular stress sensor (Olson 2004).
Modification of rRNA by 2′-O-methylation and pseudouridylation are thought to occur cotranscriptionally (Warner 2001; Decatur and Fournier 2003). Thus, snoRNAs are likely to associate with nascent transcripts. As t-UTPs apparently facilitate loading of the U3 snoRNA onto nascent pre-rRNA, we might expect therefore that factors that are not core components of box C/D and H/ACA snoRNAs would facilitate their recruitment. By analogy with t-UTPs, we might also expect that such factors would be recruited to rDNA chromatin independent of transcription. Two such candidate proteins have been described: TCOF1/Treacle and Nopp140. Treacher Collins syndrome (TCS) is an autosomal dominant disorder of craniofacial development. Treacle, the protein product of TCOF1, the gene mutated in TCS, is a nucleolar phosphoprotein that interacts directly with both UBF (Valdez et al. 2004) and Nopp56/58 present in box C/D snoRNPs (Hayano et al. 2003). Furthermore, it has been reported that knockdown of TCOF1 expression results in reduction of both rDNA transcription (Valdez et al. 2004) and pre-rRNA methylation (Gonzales et al. 2005). A related nucleolar phosphoprotein, Nopp140, associates with NAP57 (dyskerin), the pseudouridylation catalytic activity of box H/ACA snoRNPs (Meier and Blobel 1994), and is not required for pseudouridylation in vitro (Wang et al. 2002) but may link this process with transcription in vivo. Nopp140 is found associated with pol I (Chen et al. 1999) and mutant derivatives can act as dominant inhibitors of transcription by pol I in vivo (Yang et al. 2000).
Here, we have demonstrated that like t-UTPs, TCOF1 and Nopp140 are recruited to ribosomal gene chromatin independent of both their partner snoRNPs and transcription. It is likely that TCOF1 is recruited through direct interaction with UBF. Nopp140 may be recruited through interaction with pol I. Interestingly, the majority of pol I dissociates from mitotic NORs during metaphase, as does Nopp140 (Leung et al. 2004). In contrast, both UBF and TCOF1 remain associated with NORs through mitosis.
In summary, electron microscopy has told us that rDNA transcription and pre-rRNA processing are intimately linked both in space and time. The multiple levels at which these processes are now thought to be linked illustrates the fundamental importance of coordinating steps in ribosome biogenesis. Recently, transcription elongation by pol I has been linked to both processing and ribosome assembly in yeast (Schneider et al. 2007). It is unclear at the moment if the yeast t-UTP complex appears forms part of this mechanism (Gallagher et al. 2004). Our identification and characterization of human t-UTPs illustrates that links between transcription and processing are conserved throughout evolution. Importantly, we have shown that the context in which transcribed sequences are located is likely to be a major contributing factor in vivo. Extensive UBF binding on human NORs specifies a distinct form of chromatin that facilitates recruitment of the pol I transcription machinery, t-UTPs, TCOF1, and Nopp140. These results highlight the importance of NOR structure in recruiting factors required for coordinating early steps in ribosome biogenesis.
Materials and methods
Plasmids
Image clones encoding UTP4 (Accession number: BC009348), UTP10 (Accession number: CD643675), UTP15 (Accession number: CX784161), and UTP17 (Accession number: BC040567) were obtained from www.geneservice.co.uk. A cDNA clone encoding UTP5, KIAA0007 (Accession number: D26488), was obtained from www.kazusa.or.jp/huge/. UTP4, 10, and 17 ORFs were amplified by PCR and introduced into pENTR/D-TOPO (Invitrogen). An NcoI/NotI fragment encoding the full-length UTP5 open reading frame was subcloned from KIAA0007 into pENTR4 (Invitrogen). Note that sequences surrounding the initiation codon were converted to an NcoI restriction site prior to subcloning. Sequence alignment of KIAA with putative UTP5 clones from other mammalian species indicates that the open reading frame in KIAA0007 most likely initiates at nucleotide position 30. All Gateway entry clones were fully sequenced prior to transfer into destination vectors. An open reading frame encoding UTP15 was also introduced into pENTR4, but we were unable to detect expression from this construct using a number of destination vectors. UTP4, UTP5, and UTP17 were transferred into the destination vector pcDNA6.2/nLumio-DEST (Invitrogen) in order to generate mammalian expression constructs that introduce an N-terminal V5 tag.
Cell culture
HT1080 and HeLa cells were grown in Dulbeco’s MEM (+Glutamax, sodium pyruvate, and 4.5 g/L glucose; GIBCO) supplemented with 10% fetal bovine serum and 50 U/mL of penicillin/streptomycin. To maintain the clone 3D-1, the medium was supplemented with 5 μg/mL blasticidin (GIBCO). HeLa cells were transfected using TransIT reagent (Mirus Bio Corp.) following the manufacturer’s protocol. HT1080 and 3D-1 were transfected using a standard calcium phosphate protocol.
Antibodies
A PCR product encoding the N-terminal 120 amino acids of human UTP10 including a novel stop codon was introduced into the Gateway entry vector pENTR/D-TOPO. A KpnI/XhoI restriction fragment encoding the C-terminal 184 amino acids of UTP4 isolated from its image clone (see above) was subcloned into the Gateway entry vector pENTR3C (Invitrogen). Each insert was then transferred into the Gateway destination vector pDEST17 (Invitrogen) for production of N-terminal 6-His-tagged fusion proteins in Escherichia coli. Purified fusion proteins were used to immunize sheep. Antibodies were affinity purified as described previously (Mais et al. 2005). Anti-peptide antibodies were raised by immunizing rabbits with a peptide encoding amino acids 133–147 inclusive of UTP4. This antibody was raised and affinity purified by Eurogentec. Rabbit antibodies against TCOF1/Treacle were provided by Michael Dixon (University of Manchester). Rabbit antibodies against Nopp140 (serum RS8) and NAP57 (serum RU8) were provided by Tom Meier (Albert Einstein College of Medicine, NY). Rabbit antibodies against hU3–55K were provided by Nicholas Watkins (University of Newcastle upon Tyne). Mouse monoclonal antibodies against the V5 tag were from Serotec. Affinity-purified UBF and RPA43 antibodies have been previously described (Mais et al. 2005). Anti-sheep and rabbit horseradish peroxidase-conjugated secondary antibodies were purchased from Sigma and New England Biolabs, respectively. Dye-conjugated secondary antibodies used in immunofluorescent staining were purchased from Jackson ImmunoResearch. The histone H2B polyclonal antibody used in ChIP was purchased from Upstate.
Cell staining and image capture
Immunofluorescent staining, silver staining, and combined immuno-FISH were performed as described previously (Mais et al. 2005) except that after staining, slides were mounted in Vectashield plus DAPI (Vector Laboratories). DIC and Z-stacks of fluorescent images were captured using a Photometric Coolsnap HQ camera and Volocity 4 imaging software (Improvision) with a 63× Plan Apochromat Zeiss objective mounted on a Zeiss Axioplan2 imaging microscope. Flattened images of deconvolved Z-stacks (iterative restoration) are used. In the experiments shown in Figure 8B–D, where we demonstrate lack of staining at pseudo-NORs, special care was taken to include the entire nuclear volume in the Z-stack series.
Nucleoli were isolated from HeLa cells as previously described (Muramatsu et al. 1963; see http://www.lamondlab.com/f7nucleolarprotocol.htm). Purified nucleoli were spread onto polylysine-coated slides, air dried, and fixed in PBS containing 1.0% formaldehyde. Antibody staining was performed as above.
SiRNA
SiRNA duplexes were purchased as Smartpools from Dharmacon. In some cases, individual duplexes from the Smartpool were used in transfections. The control duplex has been described previously (Valdez et al. 2004). 5 × 105 HeLa cells were seeded per 6-cm-diameter dish. The initial transfection with 80 μmol (20 nM final concentration) of Smartpool duplex oligos using 2 μL of Dharmafect 1 transfection reagent was performed 24 h later. A second round of transfection was performed after a further 48 h, and cells were harvested 48 h after that. The efficiency of depletion was monitored by Western blotting. When antibodies were unavailable, the efficiency of mRNA knockdown was monitored by RT-PCR using an Opticon 2 (MJ Research) real-time PCR machine (see Supplemental Fig. 1 for details).
For UBF depletion in 3D-1 cells, four rounds of transfection with UBF Smartpool duplexes were performed at 24-h intervals. Twelve hours after the second round of transfection, cells were split and seeded onto fresh plates. Twelve hours after the final transfection, cells were trypsinized and seeded onto polylysine-coated slides. After a further 12 h, cells were fixed in 4% paraformaldehyde and prepared for combined immuno-FISH as described previously (Mais et al. 2005).
Northern blots
RNA was isolated from cells using TRI reagent (Ambion) and following the protocol provided by the manufacturer. Northern blots were performed by electrophoresis of 10-μg samples of RNA in standard formaldehyde–agarose gels followed by transfer in 10× SSC buffer to Hybond-N+ membranes (Amersham Biosciences) and UV cross-linking (UV-Stratalinker 2400, Stratagene). To probe Northern blots, a PCR product encoding nt 1–339 of the 47S pre-rRNA was labeled using random-prime labeling with α-32PdCTP. Membranes were probed and washed as described previously (O’Sullivan et al. 2002).
S1 nuclease protection assays
The probe used in S1 nuclease protection assays was a HYPUR gel-purified 60-mer oligonucleotide (5′-CGGACCCGGCCCGG GAGAGCACGACGTCACCACATCGATCACGAAGAGCCCC CCGGGAGC-3′) supplied by MWG. The probe oligonucleotide (200 ng) was 5′ end-labeled with γ-32P ATP and T4 polynucleotide kinase. Due to secondary structure within the probe, it was denatured in a boiling water bath prior to labeling. For each S1 nuclease protection assay, a 10-μg sample of RNA is resuspended in 26 μL of H2O together with 3 μL of 10× Hybridization buffer (3 M NaCl, 0.1 M Tris pH 7.9, 10 mM EDTA) and 1 μL (2 ng) of labeled probe. Hybridizations were incubated for 3 h at 65°C and then placed on ice. Two hundred seventy microliters of chilled S1 nuclease buffer (1 mM ZnSO4, 30 mM Na acetate pH 5.4, 50 mM NaCl) containing 50 units of S1 nuclease (AP Biotech) was added and the reaction incubated for 30 min at 37°C. Nuclease digestion was stopped by the addition of 50 μL of 5 M ammonium acetate, 5 μL of 0.5 M EDTA, and 10 μL of 10% SDS. Following ethanol precipitation and washing with 70% ethanol, pellets were resuspended in 6 μL of loading buffer (80% deionized formamide, 0.01% xylene cyanol and bromophenol blue dyes in 1× TBE). Denatured samples were electrophoresed on 9% denaturing (7 M urea) polyacrylamide gels run in 1× TBE, and signals were visualized and quantified using a Molecular Imager (BioRad).
Nuclear run-on transcription reactions
Nuclei were prepared from control and siRNA-transfected HeLa cells grown on 6-cm plates as follows. Cells were rinsed twice with ice cold PBS, scraped into 1-mL ice-cold PBS, and transferred to a chilled Eppendorf tube. Cells were pelleted by centrifugation at 500g for 5 min in a refrigerated microcentrifuge. The cell pellet was resuspended in 500 μL of ice-cold NP40 lysis buffer A (10 mM Tris pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.5% NP40) and incubated on ice for 5 min. Cell lysis was monitored by microscopy. Nuclei were pelleted by centrifugation at 500g for 5 min. The pellet was resuspended in a further 500 μL of ice-cold NP40 lysis buffer A and repelleted. The nuclear pellet was then resuspended in 50 μL of ice-cold glycerol storage buffer (50 mM Tris pH 8.3, 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA), flash frozen, and stored at −80°C.
Nuclear run-on reactions were performed as follows. Normalized numbers of nuclei in 50 μL of glycerol storage buffer were combined with 50 μL of 2× reaction mix (10 mM Tris pH 8.0, 5 mM MgCl2, 150 mM KCl, 5 mM dithiothreitol, and 0.1 mM CTP, UTP, and ATP) and 2 μL of α-32P GTP, then incubated for 15 min at 30°C with shaking. RNA was isolated from run-on reactions using TRI reagent (Ambion) and was then used to probe slot blots of rDNA subclones (see Fig. 4A for details). Hybridization and washes were performed as described previously (O’Sullivan et al. 2002).
ChIP
Nucleolar ChIP was performed as described previously (Mais et al. 2005) except that complexes were eluted from beads by boiling in a total volume of 200 μL of a 10% slurry of Chelex 100 (BioRad) in water (Nelson et al. 2006). PCR reactions were performed using 5 μL of the eluted material in 25-μL reactions, and products were visualized on 1% agarose gels run in 1× TBE buffer. Nuclear ChIP was utilized to examine pseudo-NORs and was performed as described previously (Mais et al. 2005). Precise PCR conditions and sequence of primer pairs are available upon request.
Acknowledgments
We thank Nick Watkins for hU3–55K antibodies, the U3-snoRNA probe, useful discussions, and advice. We thank Mike Dixon for Treacle antibodies, Tom Meier for Nopp140 and Nap57 antibodies, and Ger Pruijn for Mpp10/Imp3/Imp4 GFP expression constructs. This work was supported by the MRC UK and Tenovus Scotland.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.436707
References
- Andersen J.S., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I., Lyon C.E., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I., Fox A.H., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I., Leung A.K., Lam Y.W., Steen H., Mann M., Lamond A.I., Lam Y.W., Steen H., Mann M., Lamond A.I., Steen H., Mann M., Lamond A.I., Mann M., Lamond A.I., Lamond A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Azuma M., Toyama R., Laver E., Dawid I.B., Toyama R., Laver E., Dawid I.B., Laver E., Dawid I.B., Dawid I.B. Perturbation of rRNA synthesis in the bap28 mutation leads to apoptosis mediated by p53 in the zebrafish central nervous system. J. Biol. Chem. 2006;281:13309–13316. doi: 10.1074/jbc.M601892200. [DOI] [PubMed] [Google Scholar]
- Chagnon P., Michaud J., Mitchell G., Mercier J., Marion J.F., Drouin E., Rasquin-Weber A., Hudson T.J., Richter A., Michaud J., Mitchell G., Mercier J., Marion J.F., Drouin E., Rasquin-Weber A., Hudson T.J., Richter A., Mitchell G., Mercier J., Marion J.F., Drouin E., Rasquin-Weber A., Hudson T.J., Richter A., Mercier J., Marion J.F., Drouin E., Rasquin-Weber A., Hudson T.J., Richter A., Marion J.F., Drouin E., Rasquin-Weber A., Hudson T.J., Richter A., Drouin E., Rasquin-Weber A., Hudson T.J., Richter A., Rasquin-Weber A., Hudson T.J., Richter A., Hudson T.J., Richter A., Richter A. A missense mutation (R565W) in cirhin (FLJ14728) in North American Indian childhood cirrhosis. Am. J. Hum. Genet. 2002;71:1443–1449. doi: 10.1086/344580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.K., Pai C.Y., Huang J.Y., Yeh N.H., Pai C.Y., Huang J.Y., Yeh N.H., Huang J.Y., Yeh N.H., Yeh N.H. Human Nopp140, which interacts with RNA polymerase I: Implications for rRNA gene transcription and nucleolar structural organization. Mol. Cell. Biol. 1999;19:8536–8546. doi: 10.1128/mcb.19.12.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur W.A., Fournier M.J., Fournier M.J. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., Settlage R.E., Shabanowitz J., Osheim Y., Shabanowitz J., Osheim Y., Osheim Y., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Hoffmann-Rohrer U., Hu Q., Grummt I., Rothblum L.I., Phair R.D., Misteli T., Hoffmann-Rohrer U., Hu Q., Grummt I., Rothblum L.I., Phair R.D., Misteli T., Hu Q., Grummt I., Rothblum L.I., Phair R.D., Misteli T., Grummt I., Rothblum L.I., Phair R.D., Misteli T., Rothblum L.I., Phair R.D., Misteli T., Phair R.D., Misteli T., Misteli T. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- Gallagher J.E., Dunbar D.A., Granneman S., Mitchell B.M., Osheim Y., Beyer A.L., Baserga S.J., Dunbar D.A., Granneman S., Mitchell B.M., Osheim Y., Beyer A.L., Baserga S.J., Granneman S., Mitchell B.M., Osheim Y., Beyer A.L., Baserga S.J., Mitchell B.M., Osheim Y., Beyer A.L., Baserga S.J., Osheim Y., Beyer A.L., Baserga S.J., Beyer A.L., Baserga S.J., Baserga S.J. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes & Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales B., Henning D., So R.B., Dixon J., Dixon M.J., Valdez B.C., Henning D., So R.B., Dixon J., Dixon M.J., Valdez B.C., So R.B., Dixon J., Dixon M.J., Valdez B.C., Dixon J., Dixon M.J., Valdez B.C., Dixon M.J., Valdez B.C., Valdez B.C. The Treacher Collins syndrome (TCOF1) gene product is involved in pre-rRNA methylation. Hum. Mol. Genet. 2005;14:2035–2043. doi: 10.1093/hmg/ddi208. [DOI] [PubMed] [Google Scholar]
- Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D., Bassler J., Petfalski E., Strauss D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D., Petfalski E., Strauss D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D., Strauss D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D., Schafer T., Kuster B., Tschochner H., Tollervey D., Kuster B., Tschochner H., Tollervey D., Tschochner H., Tollervey D., Tollervey D., et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- Granneman S., Gallagher J.E., Vogelzangs J., Horstman W., van Venrooij W.J., Baserga S.J., Pruijn G.J., Gallagher J.E., Vogelzangs J., Horstman W., van Venrooij W.J., Baserga S.J., Pruijn G.J., Vogelzangs J., Horstman W., van Venrooij W.J., Baserga S.J., Pruijn G.J., Horstman W., van Venrooij W.J., Baserga S.J., Pruijn G.J., van Venrooij W.J., Baserga S.J., Pruijn G.J., Baserga S.J., Pruijn G.J., Pruijn G.J. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60-80S ribonucleoprotein complexes. Nucleic Acids Res. 2003;31:1877–1887. doi: 10.1093/nar/gkg300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolova K.V., Nicoloso M., Mazan S., Hadjiolov A.A., Bachellerie J.P., Nicoloso M., Mazan S., Hadjiolov A.A., Bachellerie J.P., Mazan S., Hadjiolov A.A., Bachellerie J.P., Hadjiolov A.A., Bachellerie J.P., Bachellerie J.P. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. J. Biochem. 1993;212:211–215. doi: 10.1111/j.1432-1033.1993.tb17652.x. [DOI] [PubMed] [Google Scholar]
- Hayano T., Yanagida M., Yamauchi Y., Shinkawa T., Isobe T., Takahashi N., Yanagida M., Yamauchi Y., Shinkawa T., Isobe T., Takahashi N., Yamauchi Y., Shinkawa T., Isobe T., Takahashi N., Shinkawa T., Isobe T., Takahashi N., Isobe T., Takahashi N., Takahashi N. Proteomic analysis of human Nop56p-associated pre-ribosomal ribonucleoprotein complexes. Possible link between Nop56p and the nucleolar protein treacle responsible for Treacher Collins syndrome. J. Biol. Chem. 2003;278:34309–34319. doi: 10.1074/jbc.M304304200. [DOI] [PubMed] [Google Scholar]
- Jordan P., Mannervik M., Tora L., Carmo-Fonseca M., Mannervik M., Tora L., Carmo-Fonseca M., Tora L., Carmo-Fonseca M., Carmo-Fonseca M. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Craig N., Sollner W.B., Craig N., Sollner W.B., Sollner W.B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol. Cell. Biol. 1987;7:2891–2898. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Tyc K., Steitz J.A., Sollner W.B., Tyc K., Steitz J.A., Sollner W.B., Steitz J.A., Sollner W.B., Sollner W.B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Kopp K., Gasiorowski J.Z., Chen D., Gilmore R., Norton J.T., Wang C., Leary D.J., Chan E.K., Dean D.A., Huang S., Gasiorowski J.Z., Chen D., Gilmore R., Norton J.T., Wang C., Leary D.J., Chan E.K., Dean D.A., Huang S., Chen D., Gilmore R., Norton J.T., Wang C., Leary D.J., Chan E.K., Dean D.A., Huang S., Gilmore R., Norton J.T., Wang C., Leary D.J., Chan E.K., Dean D.A., Huang S., Norton J.T., Wang C., Leary D.J., Chan E.K., Dean D.A., Huang S., Wang C., Leary D.J., Chan E.K., Dean D.A., Huang S., Leary D.J., Chan E.K., Dean D.A., Huang S., Chan E.K., Dean D.A., Huang S., Dean D.A., Huang S., Huang S. Pol I transcription and pre-rRNA processing are coordinated in a transcription-dependent manner in mammalian cells. Mol. Biol. Cell. 2007;18:394–403. doi: 10.1091/mbc.E06-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Peng W.T., Cagney G., Robinson M.D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P., Peng W.T., Cagney G., Robinson M.D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P., Cagney G., Robinson M.D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P., Robinson M.D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P., Guo X., Zhang X., Canadien V., Richards D.P., Zhang X., Canadien V., Richards D.P., Canadien V., Richards D.P., Richards D.P., et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Leung A.K., Gerlich D., Miller G., Lyon C., Lam Y.W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A.I., Gerlich D., Miller G., Lyon C., Lam Y.W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A.I., Miller G., Lyon C., Lam Y.W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A.I., Lyon C., Lam Y.W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A.I., Lam Y.W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A.I., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A.I., Daigle N., Zomerdijk J., Ellenberg J., Lamond A.I., Zomerdijk J., Ellenberg J., Lamond A.I., Ellenberg J., Lamond A.I., Lamond A.I. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubben B., Marshallsay C., Rottmann N., Luhrmann R., Marshallsay C., Rottmann N., Luhrmann R., Rottmann N., Luhrmann R., Luhrmann R. Isolation of U3 snoRNP from CHO cells: A novel 55 kDa protein binds to the central part of U3 snoRNA. Nucleic Acids Res. 1993;21:5377–5385. doi: 10.1093/nar/21.23.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mais C., Wright J.E., Prieto J.L., Raggett S.L., McStay B., Wright J.E., Prieto J.L., Raggett S.L., McStay B., Prieto J.L., Raggett S.L., McStay B., Raggett S.L., McStay B., McStay B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes & Dev. 2005;19:50–64. doi: 10.1101/gad.310705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier U.T. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier U.T., Blobel G., Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J. Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller O.L., Beatty B.R., Beatty B.R. Visualization of nucleolar genes. Science. 1969;164:955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
- Mougey E.B., O’Reilly M., Osheim Y., Miller O.J., Beyer A., Sollner W.B., O’Reilly M., Osheim Y., Miller O.J., Beyer A., Sollner W.B., Osheim Y., Miller O.J., Beyer A., Sollner W.B., Miller O.J., Beyer A., Sollner W.B., Beyer A., Sollner W.B., Sollner W.B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes & Dev. 1993;7:1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Smetana K., Busch H., Smetana K., Busch H., Busch H. Quantitative aspects of isolation of nucleoli of the Walker carcinosarcoma and liver of the rat. Cancer Res. 1963;23:510–522. [PubMed] [Google Scholar]
- Nelson J.D., Denisenko O., Sova P., Bomsztyk K., Denisenko O., Sova P., Bomsztyk K., Sova P., Bomsztyk K., Bomsztyk K. Fast chromatin immunoprecipitation assay. Nucleic Acids Res. 2006;34:e2. doi: 10.1093/nar/gnj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.O. Sensing cellular stress: Another new function for the nucleolus? Sci. STKE. 2004;2004:pe10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- O’Sullivan A.C., Sullivan G.J., McStay B., Sullivan G.J., McStay B., McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto J.L., McStay B., McStay B. Nucleolar biogenesis: The first small steps. Biochem. Soc. Trans. 2005;33:1441–1443. doi: 10.1042/BST0331441. [DOI] [PubMed] [Google Scholar]
- Raska I., Shaw P.J., Cmarko D., Shaw P.J., Cmarko D., Cmarko D. Structure and function of the nucleolus in the spotlight. Curr. Opin. Cell Biol. 2006;18:325–334. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Roussel P., Andre C., Comai L., Hernandez-Verdun D., Andre C., Comai L., Hernandez-Verdun D., Comai L., Hernandez-Verdun D., Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J. Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J., Zomerdijk J.C., Zomerdijk J.C. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D.A., Michel A., Sikes M.L., Vu L., Dodd J.A., Salgia S., Osheim Y.N., Beyer A.L., Nomura M., Michel A., Sikes M.L., Vu L., Dodd J.A., Salgia S., Osheim Y.N., Beyer A.L., Nomura M., Sikes M.L., Vu L., Dodd J.A., Salgia S., Osheim Y.N., Beyer A.L., Nomura M., Vu L., Dodd J.A., Salgia S., Osheim Y.N., Beyer A.L., Nomura M., Dodd J.A., Salgia S., Osheim Y.N., Beyer A.L., Nomura M., Salgia S., Osheim Y.N., Beyer A.L., Nomura M., Osheim Y.N., Beyer A.L., Nomura M., Beyer A.L., Nomura M., Nomura M. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol. Cell. 2007;26:217–229. doi: 10.1016/j.molcel.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky V., Langlois F., Gagnon-Kugler T., Rothblum L.I., Moss T., Langlois F., Gagnon-Kugler T., Rothblum L.I., Moss T., Gagnon-Kugler T., Rothblum L.I., Moss T., Rothblum L.I., Moss T., Moss T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell. 2006;21:629–639. doi: 10.1016/j.molcel.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Tavner F.J., Simpson R., Tashiro S., Favier D., Jenkins N.A., Gilbert D.J., Copeland N.G., Macmillan E.M., Lutwyche J., Keough R.A., Simpson R., Tashiro S., Favier D., Jenkins N.A., Gilbert D.J., Copeland N.G., Macmillan E.M., Lutwyche J., Keough R.A., Tashiro S., Favier D., Jenkins N.A., Gilbert D.J., Copeland N.G., Macmillan E.M., Lutwyche J., Keough R.A., Favier D., Jenkins N.A., Gilbert D.J., Copeland N.G., Macmillan E.M., Lutwyche J., Keough R.A., Jenkins N.A., Gilbert D.J., Copeland N.G., Macmillan E.M., Lutwyche J., Keough R.A., Gilbert D.J., Copeland N.G., Macmillan E.M., Lutwyche J., Keough R.A., Copeland N.G., Macmillan E.M., Lutwyche J., Keough R.A., Macmillan E.M., Lutwyche J., Keough R.A., Lutwyche J., Keough R.A., Keough R.A., et al. Molecular cloning reveals that the p160 Myb-binding protein is a novel, predominantly nucleolar protein which may play a role in transactivation by Myb. Mol. Cell. Biol. 1998;18:989–1002. doi: 10.1128/mcb.18.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez B.C., Henning D., So R.B., Dixon J., Dixon M.J., Henning D., So R.B., Dixon J., Dixon M.J., So R.B., Dixon J., Dixon M.J., Dixon J., Dixon M.J., Dixon M.J. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc. Natl. Acad. Sci. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Query C.C., Meier U.T., Query C.C., Meier U.T., Meier U.T. Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol. Cell. Biol. 2002;22:8457–8466. doi: 10.1128/MCB.22.24.8457-8466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Warner J.R. Nascent ribosomes. Cell. 2001;107:133–136. doi: 10.1016/s0092-8674(01)00531-1. [DOI] [PubMed] [Google Scholar]
- Yang Y., Isaac C., Wang C., Dragon F., Pogacic V., Meier U.T., Isaac C., Wang C., Dragon F., Pogacic V., Meier U.T., Wang C., Dragon F., Pogacic V., Meier U.T., Dragon F., Pogacic V., Meier U.T., Pogacic V., Meier U.T., Meier U.T. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol. Biol. Cell. 2000;11:567–577. doi: 10.1091/mbc.11.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Mitchell G.A., Richter A., Mitchell G.A., Richter A., Richter A. Nucleolar localization of cirhin, the protein mutated in North American Indian childhood cirrhosis. Exp. Cell Res. 2005;311:218–228. doi: 10.1016/j.yexcr.2005.08.012. [DOI] [PubMed] [Google Scholar]