Abstract
S100 proteins are EF hand type Ca2+ binding proteins thought to function in stimulus-response coupling by binding to and thereby regulating cellular targets in a Ca2+-dependent manner. To isolate such target(s) of the S100P protein we devised an affinity chromatography approach that selects for S100 protein ligands requiring the biologically active S100 dimer for interaction. Hereby we identify ezrin, a membrane/F-actin cross-linking protein, as a dimer-specific S100P ligand. S100P-ezrin complex formation is Ca2+ dependent and most likely occurs within cells because both proteins colocalize at the plasma membrane after growth factor or Ca2+ ionophore stimulation. The S100P binding site is located in the N-terminal domain of ezrin and is accessible for interaction in dormant ezrin, in which binding sites for F-actin and transmembrane proteins are masked through an association between the N- and C-terminal domains. Interestingly, S100P binding unmasks the F-actin binding site, thereby at least partially activating the ezrin molecule. This identifies S100P as a novel activator of ezrin and indicates that activation of ezrin's cross-linking function can occur directly in response to Ca2+ transients.
INTRODUCTION
The plasma membrane-cytoskeleton interface is a dynamic structure participating in a variety of cellular events ranging from the organization of cell-matrix and cell-cell adhesion sites to the stabilization and regulation of cell surface structures like microvilli and filipodia and the control of cell migration and intracellular vesicle transport. Actin filaments and associated proteins are the major constituents of the membrane-underlying cytoskeleton and their contribution to the abovementioned processes has been established in a number of cell systems. Although several proteins modulating the state of actin filaments have been described only a few components providing a direct linkage between actin filaments and the membrane have been identified in regions outside the well-organized adhesion sites. Among these linking proteins members of the ezrin/radixin/moesin (ERM) family are probably the best characterized. ERM proteins typically contain a N-terminal membrane binding domain (N-ERMAD), which can interact with phosphatidylinositol 4,5-bisphosphate (PIP2) and a number of membrane proteins, and a C-terminal domain (C-ERMAD), which contains a F-actin binding site. Because of interactions between these two domains, F-actin and membrane protein binding sites are usually masked in the so-called dormant ERM proteins. They can be activated after threonine phosphorylation in the C-terminal domain and a resulting conformational change in the ERM molecule (for reviews see Bretscher et al., 2000, 2002; Tsukita et al., 1997; Mangeat et al., 1999; Gautreau et al., 2002). Thus, activation of the membrane-cytoskeleton linking function in ERM proteins requires cell signaling culminating in, e.g., threonine phosphorylation of ERM proteins (Simons et al., 1998).
Although not shown in the case of ERM proteins, Ca2+ ions are known to serve important messenger functions in the regulation of membrane-cytoskeleton contacts and the membrane-underlying cytoskeleton. Among other things Ca2+ transients regulate the spectrin binding to NMDA membrane receptors (Wechsler and Teichberg, 1998), trigger through activation of calmodulin a disassembly of cortical F-actin in mast cells (Sullivan et al., 2000), and induce cell shape changes in fertilized Xenopus eggs (Muto and Mikoshiba, 1998). Information on proteins transmitting these Ca2+ signals is scarce, although members of two multigene families of Ca2+-binding proteins, which are found in the membrane-underlying cytoskeleton of numerous cells, have been implicated in Ca2+ stimulus-response coupling in the cell cortex. Examples are several Ca2+/phospholipid-binding proteins of the annexin family, in particular annexins 2 and 6, which are enriched at sites of cholesterol-rich membrane microdomains (Oliferenko et al., 1999; Babiychuk and Draeger, 2000; Zobiack et al., 2002), and EF hand type Ca2+-binding proteins like calmodulin (Rogers and Strehler, 1996), α-actinin (Noegel, 1996), the neuronal EF hand protein VILIP (Lenz et al., 1996), and the penta EF hand protein peflin (Kitaura et al., 2001).
Some evidence also links S100 proteins, which constitute a distinct subfamily of EF hand type Ca2+-binding proteins to the regulation of cytoskeleton dynamics and membrane-cytoskeleton interactions. S100 A1 and B, for example, bind to and possibly regulate actin-associated proteins like caldesmon and CapZ, S100A10 is required for targeting the membrane and F-actin binding protein annexin 2 to the cortical cytoskeleton and S100A8, 9, and 12 translocate to the plasma membrane upon Ca2+ elevation in leukocytes (for reviews see Donato, 2001; Heizmann et al., 2002). However, functional information on the role of S100 proteins in membrane-cytoskeleton dynamics is very limited. Moreover, most binding partners of S100 proteins have so far only been described in in vitro studies with intrinsic limitations as to the specificity of the interactions. Despite this lack of functional information S100 proteins are structurally well characterized. They consist of two EF hand Ca2+-binding motifs that are connected via a flexible linker and flanked by N- and C-terminal extensions. Typically, S100 proteins form homodimers and structural evidence strongly indicates that it is only the S100 dimer that interacts with and thereby regulates target proteins in a Ca2+-dependent manner (Rety et al., 1999; Rety et al., 2000; Rustandi et al., 2000).
These structural findings led us develop an approach for identifying specific binding partners of S100 proteins, i.e., those that will only interact with the biologically active S100 dimer. We constructed affinity matrices, based on our identification of a mutant S100P protein that fails to engage in dimer formation (Koltzscher and Gerke, 2000), containing either the wild-type dimeric or mutant monomeric S100 protein and identified in a placental extract ezrin as a specific binding partner only interacting with dimeric S100P. The interaction is Ca2+ dependent, observed for dormant ezrin, and mediated through the N-terminal ezrin domain. Importantly, binding to S100P unmasks the F-actin binding site in dormant ezrin. Using GFP-tagged S100P derivatives we also show that upon Ca2+ elevation in human A431 cells the dimeric but not the monomeric S100 protein translocates to the cell cortex showing a colocalization with ezrin. Thus, by means of triggering complex formation between S100P and ezrin elevations in cellular Ca2+ levels could directly affect ezrin function in the cell cortex.
MATERIALS AND METHODS
Construction of Plasmids
The cDNAs encoding WT human S100P, WT human ezrin, and human ezrin deletion mutants (N-ERMAD, aa 1–323, and C-ERMAD, aa 461–586) were generated by polymerase chain reactions using a human placental MATCHMAKER cDNA library in pACT2 (Clontech, Palo Alto, CA) as template. PCR was carried out using high fidelity Pfu DNA polymerase (Stratagene, La Jolla, CA) and oligonucleotide primers containing appropriate restriction enzyme sites. The cDNA encoding the F15A S100P mutant protein was generated by using mutated primers in the PCR reaction. cDNA products of the PCRs were cloned into the bacterial expression vector pET28a+ (Novagen, Madison, WI) or into pEGFP-C2 (Clontech) in frame with either the 6× His-Tag or the green fluorescent protein, respectively. The correctness of all constructs was verified by sequence analysis.
Recombinant Expression and Purification of S100P and Ezrin
pET 28a+ constructs encoding WT S100P, F15A S100P, WT ezrin, and ezrin deletion mutants with N-terminal histidine tags were used to transform Escherichia coli cells [strain BL21(DE3)pLysS]. Transformed bacteria were grown to an OD600 of 0.6, and recombinant protein expression was then induced by adding IPTG to a concentration of 1 mM. After incubation for an additional 3 h, cells were harvested by centrifugation (5000 × g; 10 min) and resuspended in lysis buffer (50 mM Tris-Cl, pH 7.5; 300 mM NaCl; 20 mM imidazole-HCl, pH 7.5; 1 mM EDTA; 10 mM β-mercaptoethanol; 1 mM PMSF; 10 μM leupeptin). For preparation of WT S100P and F15A S100P cells were lysed by repeated freeze/thaw cycles (3 times) and sonication. The lysate was centrifuged for 1 h at 100,000 × g, and the remaining supernatant was made 5 mM in Ca2+ and applied to a phenyl-sepharose (Pharmacia, Freiburg, Germany) column equilibrated in lysis buffer containing 0.5 mM Ca2+ and no EDTA. After extensive washing with the same buffer, bound WT S100P protein was eluted with lysis buffer containing 1 mM EGTA, whereas F15A S100P protein was eluted with 30% isopropanole in lysis buffer containing 1 mM EGTA. S100P-containing fractions were pooled, dialyzed against 20 mM imidazole, pH 7.5, 300 mM NaCl, 10 mM β-mercaptoethanol, and 1 mM PMSF, and applied to a Ni-NTA-agarose (Qiagen, Hilden, Germany) column equilibrated in the same buffer. S100P proteins were eluted with 250 mM imidazole, pH 7.5, 300 mM NaCl, 10 mM β-mercaptoethanol, and 1 mM PMSF.
For preparation of WT ezrin and ezrin deletion mutants cells were lysed only by sonication. The lysates were centrifuged for 1 h at 100,000 × g, and the supernatants were applied to a Ni-NTA agarose column equilibrated in lysis buffer. After washing with lysis buffer containing 35 mM imidazole, pH 7.5, bound ezrin proteins were eluted with 300 mM imidazole, pH 7.5, 300 mM NaCl, 10 mM β-mercaptoethanol, and 1 mM PMSF.
Preparation of Placental Protein Extracts
Twenty-five grams of frozen placenta were homogenized using a Waring blender in 180 ml of homogenization buffer (30 mM HEPES, pH 7.2; 140 mM NaCl; 2 mM MgCl2; 1% Triton X-100; 1 mM DTT; 1 mM EDTA; 10 μg/ml pepstatin; 35 μg/ml aprotinin; 3 μM leupeptin; 0.5 μg/ml TPCK; 1.5 mM PMSF). The homogenate was clarified by centrifugation at 22,000 × g for 30 min at 4°C, and the resulting supernatant was further centrifuged at 100,000 × g for 1 h at 4°C. The lysate was dialyzed three times against 50 volumes of buffer D (30 mM HEPES, pH 7.2; 20 mM imidazole, pH 7.2; 300 mM NaCl; 2 mM MgCl2; 10 mM β-mercaptoethanol; 10 μg/ml pepstatin; 35 μg/ml aprotinin; 3 μM leupeptin; 0.5 μg/ml TPCK; 1.5 mM PMSF; 2 mM NaN3). The extract obtained typically had a protein concentration of ∼5 mg/ml.
Affinity Chromatography of Placental Proteins on Immobilized S100P
To generate an affinity column, His-tagged WT S100P protein was immobilized on Ni-NTA agarose. In parallel a second affinity column was prepared using the His-tagged mutant protein F15A S100P. The latter served as a control, because the mutant albeit folding correctly was incapable of forming dimers neither with itself nor with the WT S100P protein (Koltzscher and Gerke, 2000).
Five milligrams of purified, His-tagged WT S100P or F15A S100P, respectively, was dialyzed against buffer C (30 mM HEPES, pH 7.2; 20 mM imidazole, pH 7.2; 300 mM NaCl; 2 mM MgCl2; 0.5 mM CaCl2; 10 mM β-mercaptoethanol; 10 μg/ml pepstatin; 35 μg/ml aprotinin; 3 μM leupeptin; 0.5 μg/ml TPCK; 1.5 mM PMSF; 2 mM NaN3), added to 2 ml Ni-NTA-agarose equilibrated in the same buffer and incubated overnight at 4°C with gentle inversion. The Ni-NTA-agarose/S100P protein slurries were then transferred to 10-ml polypropylene columns (Pierce, Rockford, IL) and washed with 10 column volumes of buffer C to remove unbound protein. Before loading and while gentle stirring, CaCl2 was added carefully to the placental protein extract to a final concentration of 0.7 mM. Thirty milligrams of placental protein extract was then loaded onto each chromatography column. The columns were washed with 10 column volumes of buffer C, 10 column volumes buffer C1 (same as buffer C except that it contained 0.7 mM EGTA instead of 0.5 mM CaCl2) to elute proteins bound in a Ca2+-dependent manner and finally with 10 column volumes of buffer C2 (same as buffer C1 but with 250 mM imidazole, pH 7.2, instead of only 20 mM) to remove from the columns the His-tag bound proteins and Ca2+-independent interaction partners. Fractions were collected throughout, and aliquots were analyzed in gradient (7–15%) SDS-polyacrylamide gels stained with Coomassie brilliant blue (Laemmli, 1970).
MALDI-TOF Analysis and Database Searches
After SDS-PAGE the protein band of interest was prepared for mass spectrometry in a procedure modified from Shevchenko et al. (1996) and Zhang et al. (1998). A gel slice containing the protein band was excised and destained in 25 mM NH4HCO3 containing 50% methanol. It was washed in acetic acid/methanol/water (10/45/45 vol/vol/vol) for 30 min and in water for 30 min. Subsequently it was shrunk in acetonitrile and dried. The gel pieces were then reswollen in 20 μl of 40 mM NH4HCO3 containing 400 ng trypsin (BoehringerMannheim, Mannheim, Germany). After 30 min of incubation, extra trypsin solution was removed, and 50 mM NH4HCO3 was added to cover the gel. Digestion was carried out at 37°C overnight. The supernatant was then transferred to a clean Eppendorf tube and peptides were extracted three times with 70 μl acetonitrile/water/formic acid (50/45/5 vol/vol/vol). Supernatants were pooled and lyophilized. The dried extract was redissolved in 7 μl water/acetonitrile (95/5 vol/vol) containing 0.1% trifluoric acid (TFA; Merck, Darmstadt, Germany) and purified using ZipTips (Millipore, Bedford, MA). Peptides were eluted with 8 μl acetonitrile/0.1% aqueous TFA (70/30 vol/vol). For matrix preparation, 10 mg α-cyano-4-hydroxycinnamic acid (α-cyano; Sigma, Deisenhofen, Germany) were washed with acetone and dissolved in 1 ml of 49.5/49.5/1 (vol/vol/vol) acetonitrile/ethanol/0.1% aqueous TFA. 0.5 μl of this matrix preparation was spotted onto the target, followed by the same amount of peptide sample, and both solutions were mixed directly on the target for MALDI peptide mapping on TofSpec 2E (Micromass Ltd., Manchester, UK). Digests were run in positive ion reflectron mode using a matrix suppression of 500. Masses were externally calibrated and internally corrected using the lock mass option of the instrument providing m/z values better than 50 ppm up to m/z 2500. Nanospray-MS/MS of tryptic peptides was performed using iontrap (Bruker Daltonics, Bremen, Germany) and Q-TOF (Micromass, Manchester, UK), kindly made available to us by Prof. J. Peter-Katalinic (Institute of Medical Physics and Biophysics, University of Münster).
Database searches were performed using either ProteinProbe (Micromass) locally or Mascot (Matrix Science, London, UK) and Protein Prospector (University of California, San Francisco, CA), available to the public on the internet. Databases searched were SwissProt (University of Geneva and European Bioinformatics Institut), NCBI (National Center of Bioinformatics, Bethesda, MD), or OWL (University of Leeds, UK). BLAST2.0 searches were carried out at the Swiss Institute of Bioinformatics (SIB).
In Vitro Binding Assays
The interaction between S100P and ezrin was analyzed by using affinity columns containing one of the binding partners in an immobilized form. To prepare affinity columns, purified His-tagged proteins (S100P, ezrin, and their mutant derivatives) were dialyzed against buffer A (30 mM HEPES, pH 7.2; 25 mM imidazole, pH 7.2; 150 mM NaCl; 1 mM MgCl2; 0.5 mM CaCl2; 10 mM β-mercaptoethanol; 1.5 mM PMSF) and rebound to Ni-NTA agarose. In each case 300 μg of purified His-tagged protein, WT S100P, F15A S100P, WT ezrin, N-ERMAD, and C-ERMAD, respectively, were mixed with 250 μl of a 50% Ni-NTA agarose slurry equilibrated in buffer A and incubated for at least 2 h at 4°C with gentle agitation. The mixtures were then transferred to 2-ml polypropylene columns (Pierce) and washed with buffer A to remove unbound protein. In the case of immobilized WT S100P or F15A S100P, purified and thrombin-cleaved WT ezrin, N-ERMAD, or C-ERMAD protein (∼100 μg each, in buffer A) were added to the column in the fluid phase. After washing with buffer A (10 column volumes), Ca2+-dependently bound proteins were eluted with 10 column volumes of buffer B (same as buffer A but containing 0.7 mM EGTA instead of 0.5 mM CaCl2), and the columns were then stripped of His-tagged proteins by elution with 10 volumes of buffer E (same as buffer B but with 250 mM imidazole instead of only 25 mM). The vice versa experiments, using immobilized WT ezrin, were performed in a corresponding manner using in the fluid phase thrombin-cleaved WT S100P or F15A S100P and as further controls recombinantly expressed untagged WT S100P (Becker et al., 1992), untagged WT S100A1 (Osterloh et al., 1998), and untagged WT S100A11 (Seemann et al., 1996). Aliquots of each wash and elution fraction were analyzed by 12.5% Tris/tricine SDS-PAGE (Schägger and von Jagow, 1987), and the gels were stained with Coomassie.
Thrombin Cleavage of Purified Proteins
About 0.2–1 mg of purified His-tagged proteins were mixed in buffer A (without PMSF) with 0.1–0.5 ml Ni-NTA agarose, and proteins were allowed to bind at 4°C for at least 2 h. The mixtures were then centrifuged at 2500 rpm in a microcentrifuge for 5 min at 4°C, the supernatants removed, and the pellets washed two times with buffer A (without PMSF). The final pellets were resuspended in buffer A (without PMSF) at a protein concentration of 3 μg/μl and 0.2–0.8 U thrombin (Sigma) were added to the slurry. After incubation at 37°C for 2 h, the Ni-NTA agarose was pelleted in a microcentrifuge (2500 rpm, 5 min, 4°C), and the thrombin-cleaved proteins lacking the His-tag were recovered in the supernatants. To inhibit further thrombin activity the supernatants were made 1.5 mM in PMSF. Recovery of cleaved protein was ∼90% as judged by SDS-PAGE.
F-actin Binding Assay
G-actin, 100 μg, in buffer G (5 mM Tris-HCl, pH 8.0; 0.2 mM ATP; 0.5 mM DTT; 0.2 mM CaCl2) were mixed with 10 μg WT ezrin either alone or in the presence of 10 μg WT S100P or F15A S100P, respectively, in a total volume of 50 μl. Experiments were carried out in the presence of Ca2+ by adding 4 μl of 45 mM CaCl2 (final concentration ∼3 mM) or in the absence of Ca2+ by adding 4 μl 100 mM EGTA (final concentration ∼6.6 mM). F-actin polymerization was then induced by adding 6 μl of 10× polymerization buffer (20 mM Tris-HCl, pH 8.8; 500 mM KCl; 10 mM ATP; 20 mM MgCl2) and the reaction was incubated for 3 h at 20°C. After high-speed centrifugation (150,000 × g, 1 h, 4°C), the supernatant was collected and the F-actin pellet was resuspended in 60 μl 1 × polymerization buffer (with or without Ca2+/EGTA) and centrifuged again (150,000 × g, 1 h, 4°C). The supernatant was removed and the pellet was resuspended in 60 μl of SDS-PAGE sample buffer. Equivalent aliquots of the supernatant and pellet fractions were subjected to SDS-PAGE in 12.5% Tris/tricine gels followed by Coomassie brilliant blue staining. Dried gels were then analyzed on a Lumi-Imager F1 using the LumiAnalyst software (Boehringer Mannheim) for quantification of visible bands.
Cell Transfection and Immunofluorescence Microscopy
A431 cells were maintained in DME medium supplemented with 10% FCS, 1% l-glutamine and antibiotics. Transient transfection of pEGFP plasmids encoding the fusion proteins GFP-WT S100P and GFP-F15A S100P, respectively, was carried out by electroporation (250 V, 900 μF) of trypsinized cells in 4-mm cuvettes. After transfection cells were seeded on coverslips and allowed to express the recombinant protein for 3 d. Cells were then fixed with 3% formaldehyde in PBS for 20 min at room temperature. After quenching in 50 mM NH4Cl for 10 min cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min and incubated in PBS supplemented with 5% BSA for 30 min. Cells were then incubated with affinity-purified antiezrin antibodies (B23, gift from A. Bretscher) for 30 min and washed three times with PBS/5% BSA, and primary antibodies were stained with Cy3-conjugated goat anti-rabbit secondary antibodies (Jackson ImmunoResearch, Dianova, Hamburg, Germany). Cells were washed three times with PBS, once with distilled H2O, mounted in Mowiol, and analyzed using a confocal laser scanning microscope (Zeiss, Jena, Germany; LSM 510).
EGF and A23187 Treatment of A431 Cells
Transiently transfected cells grown on coverslips were transferred to DMEM without FCS and cultured for 6–12 h. EGF or A23187 were then added at a final concentration of 10 μg/ml and 1 μM, respectively. Cells were incubated for 2–15 min (EGF) or 1 min (A23187), fixed, and processed for immunofluorescence microscopy.
RESULTS
An Affinity Chromatography Approach for Identifying Target Proteins of the S100P Dimer
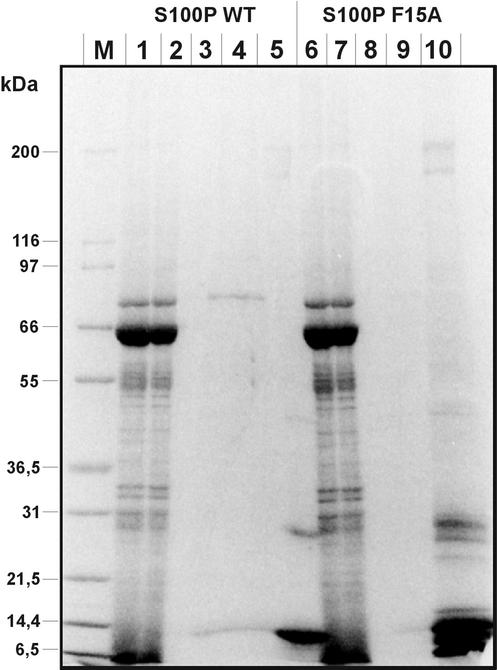
S100P is a member of the S100 protein family initially identified in human placenta (Becker et al., 1992; Emoto et al., 1992). Though its biochemical properties including the affinities of the two EF hand Ca2+-binding sites were elucidated to some extent (Becker et al., 1992; Gribenko and Makhatadze, 1998; Gribenko et al., 2002), cellular target proteins of S100P and thus its physiological role had not been characterized up to now. In an attempt to identify such targets we subjected a placental protein extract to Ca2+-dependent affinity chromatography on His-tagged S100P immobilized on Ni-Agarose. S100 proteins display hydrophobic surfaces in their Ca2+-bound conformation (for reviews see Kligman and Hilt, 1988; Krebs et al., 1995) and thus have the tendency to engage in nonspecific though still Ca2+-dependent interactions. To verify that S100P binding proteins selected in our affinity approach would be specific targets and not retained due to nonspecific hydrophobic interactions, we used as a negative control a novel S100 mutant protein incapable of forming dimers. This mutant, F15A S100P, was identified in a previous two-hybrid screen as a monomeric molecule that was unable to form a dimer even with a wild-type (WT) S100P chain. However, F15A S100P still exhibited a Ca2+-triggered increase in hydrophobicity (Koltzscher and Gerke, 2000). Because high-resolution structural analyses of other S100 proteins had revealed that dimers form the entities capable of target protein binding (Rety et al., 1999, 2000; Rustandi et al., 2000), the monomeric S100P variant should not be able to interact with specific targets. Therefore immobilized His-tagged F15A S100P was used as a negative control in a parallel affinity chromatography of placental proteins. Figure 1 reveals that a number of minor bands are retained on both, the WT and the F15A S100P matrices, most likely due to nonspecific hydrophobic interactions. Moreover, when the His-tagged S100P proteins are released from the matrices by treatment with imidazole an identical set of bands is coeluted in both cases, i.e., together with WT and F15A S100P (Figure 1, lanes 5 and 10). However, when before imidazole stripping proteins bound Ca2+-dependently to immobilized S100P are eluted with EGTA, a polypeptide of ∼80 kDa is recovered only in the eluate of the WT S100P column (Figure 1, compare lanes 4 and 9). This 80-kDa protein therefore represents the candidate of a specific target only interacting with the dimeric S100P in a Ca2+-dependent manner.
Figure 1.
Affinity chromatography of placental proteins on immobilized WT S100P (lanes 1–5) and F15A S100P (lanes 6–10). Placental protein extract (lanes 1 and 6) was loaded onto each column in a Ca2+-containing buffer. The flow through was collected (lanes 2 and 7), and columns were then washed in the presence of Ca2+ (lanes 3 and 8). Subsequently Ca2+-dependently bound proteins were eluted with a buffer containing EGTA (lanes 4 and 9). Finally the columns were treated with an imidazole-containing buffer to strip off all bound proteins (lanes 5 and 10). M, molecular weight markers. Proteins present in the different fractions were subjected to SDS-PAGE and then stained with Coomassie. Note the presence of an 80-kDa polypeptide in the EGTA eluate of the WT S100P column (lane 4), which is not found in the F15A S100P eluate (lane 9).
To identify the nature of the 80-kDa band, it was excised from the gel and subjected to in-gel trypsin digestion and subsequent MALDI-MS for peptide mass fingerprinting (our unpublished results). Database searches of the mass profiles obtained revealed that they matched those of human ezrin. This identification was corroborated by sequence information of a number of peptides analyzed by nanospray MS/MS. None of the peptide sequences obtained matched those of other members of the ERM family which are also present in placental extracts (Bretscher, 1989; Lankes and Furthmayr, 1991).
Ca2+-bound S100P Directly Interacts with Ezrin
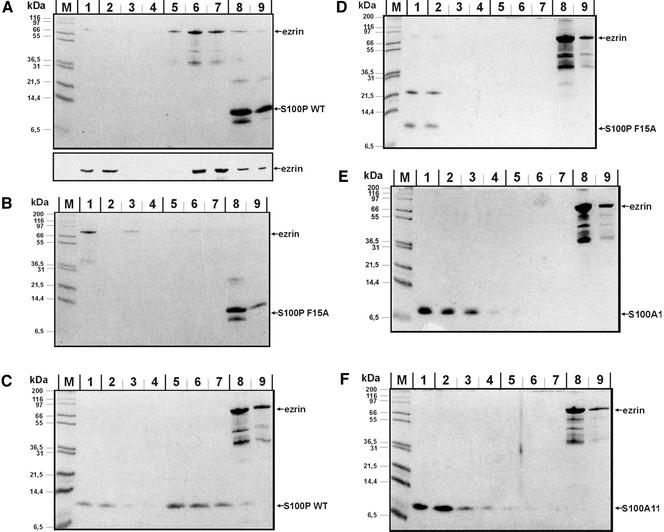
To elucidate whether S100P and ezrin directly interact with one another or whether other placental proteins participate in forming a higher-order complex retained on the affinity column, we performed binding assays with purified ezrin and S100P. In a first set of experiments immobilized S100P (WT or F15A mutant) was incubated in the presence of Ca2+ with purified ezrin generated by recombinant expression of a His-tagged version and subsequent removal of the His tag through thrombin cleavage. Figure 2, A and B, reveals that ezrin is retained in a Ca2+-dependent manner, i.e., can be eluted with an EGTA-containing buffer, on the WT but not the F15A S100P column. This shows that ezrin binds directly to S100P with the interaction being specific for the dimeric S100P protein. We also used lysates from bacteria expressing a nontagged version of WT ezrin instead of the purified and thrombin-cleaved protein in the same set of experiments. Identical results regarding the dimer-specific and Ca2+-dependent binding to S100P were obtained, revealing that the authentic ezrin is capable of binding S100P directly (our unpublished results).
Figure 2.
Direct interaction between ezrin and S100P derivatives analyzed by affinity chromatography approaches. In each experiment, His-tagged protein (WT S100P, F15A S100P, or ezrin) was bound to the Ni-matrix, and the potential ligand (ezrin, WT S100P, F15A S100P, S100A1, or S100A11) was added as purified untagged protein in the fluid phase. Binding reactions were carried out in the presence of Ca2+ and columns were then washed with a Ca2+-containing buffer. Subsequently the columns were treated with an EGTA-containing buffer to elute Ca2+-dependently bound proteins, and finally they were stripped with imidazole-containing buffer. Equivalent amounts of all fractions were subjected to SDS-PAGE, and proteins were visualized by Coomassie staining. To unambiguously identify the recombinantly expressed ezrin, samples of an independent experiment shown in the bottom part of panel A were analyzed by immunoblotting with antiezrin antibodies. Lanes 1 always show the flow-through, lanes 2–4 washing steps in the presence of Ca2+, lanes 5–7 EGTA elution steps, and lanes 8 and 9 fractions of the imidazole stripping, respectively. M, molecular weight markers. (A) Immobilized His-WT S100P with thrombin-cleaved ezrin in the fluid phase; (B) immobilized His-F15A S100P with thrombin-cleaved ezrin in the fluid phase; (C) immobilized His-ezrin with thrombin-cleaved WT S100P in the fluid phase; (D) immobilized His-ezrin with thrombin-cleaved F15A S100P in the fluid phase; (E) immobilized His-ezrin with untagged S100A1 in the fluid phase; (F) immobilized His-ezrin with untagged S100A11 in the fluid phase.
Next, we performed the binding experiments in the opposite order, i.e., His-tagged ezrin was immobilized on Ni-Agarose beads and incubated in the presence of Ca2+ with purified WT or mutant S100P. Again, a specific and direct interaction can be observed. Although WT S100P binds to the ezrin matrix in the presence of Ca2+ and is released upon treatment with EGTA, F15A S100P fails to be retained and is recovered exclusively in the flow through and wash fractions (Figure 2, C and D). To include additional specificity controls, we tested whether other dimeric S100 proteins are capable of binding ezrin. Therefore purified S100A1 and S100A11 were incubated with the ezrin matrix and any Ca2+-dependently bound protein was released with EGTA. Figure 2, E and F, reveals that in both cases no protein is retained on the matrix, indicating that the interaction of ezrin with S100P is specific for this member of the S100 protein family.
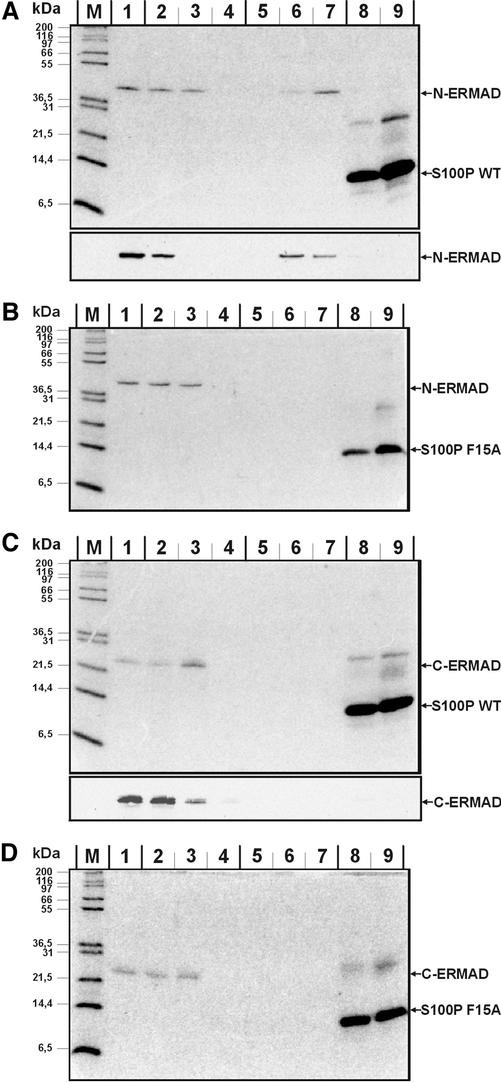
S100P Binds to the N-terminal Domain of Ezrin
Ezrin consists of two principal domains, an N-terminal domain (N-ERMAD) capable of binding EBP50 (ERM-binding phosphoprotein 50) and a number of membrane proteins and a C-terminal domain (C-ERMAD) harboring the F-actin binding site (for reviews see Bretscher et al., 2000, 2002; Tsukita et al., 1997; Mangeat et al., 1999). To elucidate whether the S100P binding site maps to one of these domains, N-ERMAD and C-ERMAD of ezrin were expressed recombinantly, purified, and applied to columns containing immobilized WT or F15A S100P, respectively. Incubations were carried out in the presence of Ca2+ and after extensive washing the columns were eluted with EGTA- and then imidazole-containing buffers, i.e., experimental conditions were identical to those applied for full length ezrin. Figure 3 shows that only N-ERMAD is retained on the column in a Ca2+-dependent manner, thereby identifying N-ERMAD as the site of S100P interaction. Again the interaction is specific for the dimeric S100P molecule.
Figure 3.
Interaction of S100P derivatives with the N- and C-terminal domains of ezrin (N- and C-ERMAD). Experiments were performed as those described in Figure 2 using immobilized S100P derivatives and the ezrin domains in the fluid phase. Equivalent amounts of flow through, wash, and eluate fractions were analyzed by SDS-PAGE and subsequent Coomassie staining. In the bottom parts of A and C, samples of an independent experiment were subjected to immunoblotting with antibodies recognizing the T7 tag of the recombinantly expressed N-ERMAD and C-ERMAD. Lanes 1 always show the flow-through, lanes 2–4 washing steps in the presence of Ca2+, lanes 5–7 EGTA elution steps and lanes 8 and 9 fractions of the imidazole stripping, respectively. M, molecular weight markers. (A) Immobilized His-WT S100P with thrombin-cleaved N-ERMAD in the fluid phase; (B) immobilized His-F15A S100P with thrombin-cleaved N-ERMAD in the fluid phase; (C) immobilized His-WT S100P with thrombin-cleaved C-ERMAD in the fluid phase; (D) immobilized His-F15A S100P with thrombin-cleaved C-ERMAD in the fluid phase. Note the Ca2+-dependent binding of N-ERMAD to immobilized WT but not F15A S100P. No interaction is seen in the case of C-ERMAD with the bands visible in the imidazole eluate of C and D most likely representing contaminating bacterial proteins because they are not recognized by the anti-T7 tag antibody (bottom part of C).
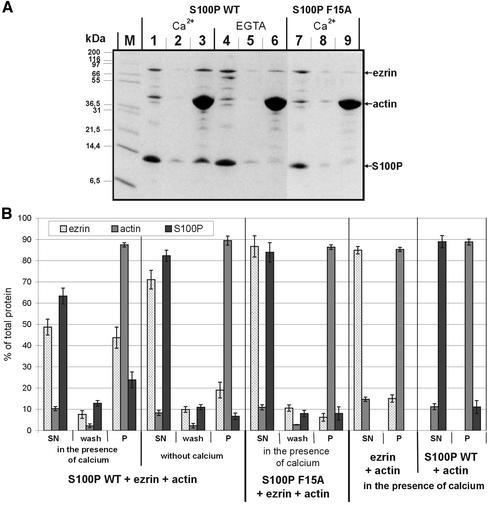
The Ezrin–F-actin Interaction Is Regulated by S100P
The binding assays described above were carried out with nonphosphorylated and thus nonactive or dormant ezrin and revealed that S100P in contrast to most other protein-binding partners of ezrin is capable of interacting with the dormant molecule. Because S100 proteins are thought to participate in stimulus-response coupling by regulating activities of their respective targets upon Ca2+-triggered binding, we next sought to determine whether the Ca2+-dependent binding of S100P to ezrin results in an altered activity of the dormant ezrin. A property typically affected upon phosphorylation-dependent activation of ezrin is its F-actin binding. Although dormant ezrin does not interact with F-actin, threonine phosphorylation in the C-terminal domain unmasks the F-actin binding site (Simons et al., 1998; Nakamura et al., 1999). Therefore we analyzed whether the ability of ezrin to interact with F-actin is affected when it resides in a complex with S100P. F-actin cosedimentation assays were carried out with purified ezrin and WT or F15A S100P in the presence or absence of Ca2+. Figure 4 reveals that a significant fraction of ezrin cosediments with F-actin in the presence of WT but not F15A S100P. This cosedimentation is not observed in the absence of S100P, thus revealing that the ezrin used in the assays is in its dormant conformation and not activated by mild proteolysis (Figure 4B). As estimated by densitometric scanning, the stoichiometry of ezrin to actin in the pellet fraction (lane 3) is approximately 1:10 and thus in the range of what has been observed previously in F-actin binding experiments (Yao et al., 1996; Roy et al., 1997). Increasing in the cosedimentation assay the amount of actin also leads to an increased amount of copelleted ezrin, with the estimated stoichiometry remaining at 1:10 (ezrin-actin; our unpublished results). The ezrin-F-actin cosedimentation is Ca2+ dependent and most likely due to S100P-ezrin complex formation because a fraction of WT S100P but not the monomeric mutant molecule is recovered together with ezrin in the F-actin sediment. Such F-actin cosedimentation of S100P is not seen in the absence of ezrin or Ca2+ (Figure 4B). Thus, the Ca2+-regulated binding of S100P to ezrin can expose the F-actin binding site in a manner not depending on PIP2 binding and threonine phosphorylation.
Figure 4.
F-actin cosedimentation assays. (A) WT S100P (lanes 1–6) or F15A S100P (lanes 7–9) were mixed with ezrin and F-actin either in the presence of Ca2+ (lanes 1–3 for WT S100P, and lanes 7–9 for F15A S100P) or in the presence of EGTA (lanes 4–6). After incubation and high-speed centrifugation the resulting supernatants were collected (lanes 1, 4, and 7). The remaining pellets were washed once with incubation buffer (lanes 2, 5, and 8 represent the supernatants of these washes), and the final pellets (lanes 3, 6, and 9) were dissolved by boiling in SDS-sample buffer. Equivalent aliquots of all fractions were subjected to SDS-PAGE. M, molecular weight markers. (B) Quantification of the cosedimentation data. Coomassie-stained gels containing the different fractions of F-actin cosedimentation experiments (one example is shown in A) were analyzed on a Lumi-Imager (Boehringer Mannheim) for quantification of the stained protein bands. For each protein, the signal obtained was normalized to the total amount of the respective protein added to reaction, which was set arbitrarily to 100%. Error bars indicate SD of the eight independent experiments analyzed. SN is the supernatant after the first centrifugation, i.e., the protein not bound to or incorporated into F-actin filaments, wash is the supernatant after washing of the F-actin pellet in incubation buffer, and P represents the protein fraction remaining in the final pellet, i.e., the proteins incorporated into (actin) or bound to (ezrin, S100P) F-actin filaments. Note the F-actin-ezrin cosedimentation in the presence of WT but not F15A S100P, which is only observed in the presence of Ca2+. The amount of ezrin copelleted in the absence of Ca2+ (EGTA, lane 6) is not increased with increasing actin concentrations (in contrast to reactions performed in the presence of Ca2+, unpublished results) and thus most likely represents a nonspecific background. Cosedimentation experiments performed only with ezrin and F-actin reveal no appreciable ezrin pelleting, indicating that the ezrin preparation used is in the dormant conformation and not activated by mild proteolysis (B).
S100P and Ezrin Colocalize in Stimulated A431 Cells
In a number of cells ezrin has been localized to the plasma membrane, in particular to areas where actin filaments are concentrated, e.g., microvilli, membrane ruffles, and cell adhesion sites (see, for example, Bonilha et al., 1999; Bretscher, 1999; Yonemura et al., 1999; Gautreau et al., 2000). This localization is dependent on an activation of the dormant ezrin that resides in the cytosol. In A431 human epidermoid cells a cytosol-membrane translocation of ezrin is observed in response to EGF, and EGF also triggers an elongation of microvilli in A431 cells overexpressing ERM binding membrane proteins (Bretscher, 1989; Yonemura et al., 1999). Therefore we chose A431 cells cultivated in the absence or presence of EGF to study the intracellular localization of S100P in comparison to that of ezrin. Although we used different antigen preparations and immunization strategies, we could not obtain high-affinity anti-S100P antibodies not cross-reacting with other members of the S100 family. Therefore we recorded the distribution of S100P within A431 cells by ectopically expressing a GFP-S100P fusion protein. In this chimera the GFP was fused to the N-terminus of S100P because we had shown previously for another S100 protein (S100A10) that an N-terminal GFP tag did not affect biochemical properties or intracellular localization of the S100 protein (Zobiack et al., 2001).
Cells expressing GFP-WT S100P show a diffuse general GFP signal when kept in serum-free medium. Likewise, ezrin is found throughout the cytoplasm and is not concentrated at the plasma membrane under these conditions (Figure 5). When cells are exposed to EGF for 2 or 15 min, a considerable fraction of the ezrin translocates to areas beneath the plasma membrane, showing a particular enrichment at sites of membrane ruffles and microvilli. GFP-WT S100P also shows a partial membrane localization in the EGF-treated cells, and this membrane-associated GFP-WT S100P colocalizes with ezrin (Figure 6, top panels). In contrast, a GFP chimera of the F15A S100P mutant protein (GFP-F15A S100P) remains cytosolic after EGF stimulation (Figure 6, bottom panels). The intracellular distribution of the different GFP-S100P derivatives (WT and F15A) was also recorded in cells cultivated in serum. Similar to the EGF stimulus, these conditions induce a partial membrane localization of WT S100P that is not observed for the F15A S100P mutant. Again staining of the membrane associated S100P overlaps with that of ezrin (our unpublished results).
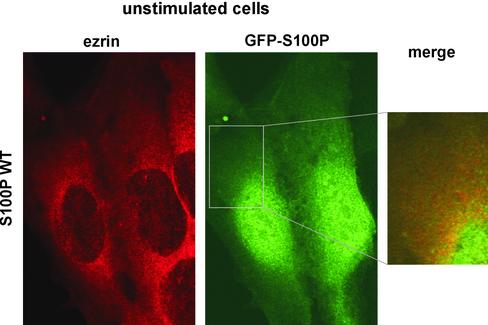
Figure 5.
Localization of ezrin and GFP-tagged WT S100P in unstimulated A431 cells. A431 cells were transiently transfected with a plasmid encoding GFP-WT S100P and were allowed to express the GFP-fusion protein for 3 d. Subsequently, the cells were fixed with PFA, permeabilized, and stained with antibodies against human ezrin (left panel). A merged image of the ezrin and GFP-S100P signals is shown in the inset. Note that both proteins show a diffuse distribution with no obvious colocalization. A substantial fraction of the GFP-S100P is present in the nucleus, most likely a consequence of the overexpression and a resulting lack of cytosolic binding partners.
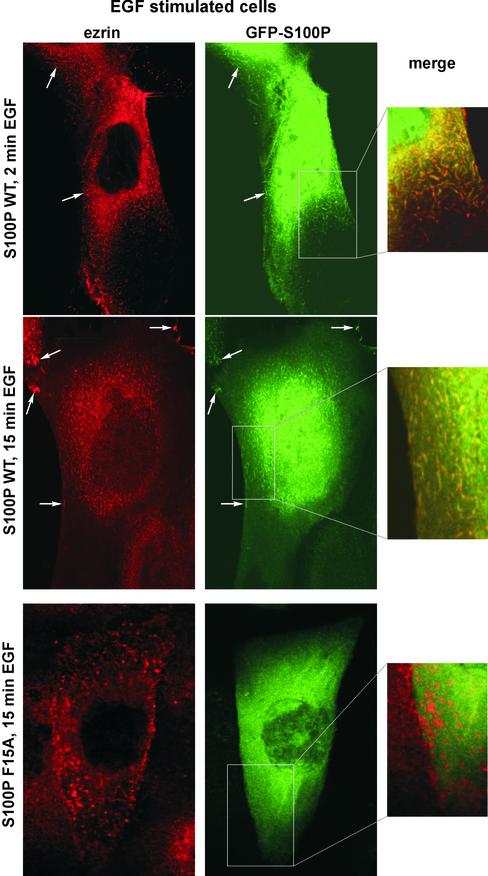
Figure 6.
EGF treatment of A431 cells induces a colocalization of ezrin and GFP-WT S100P to microvillar protrusions. A431 cells ectopically expressing GFP-WT S100P or GFP-F15A S100P were transferred to serum-free medium and cultivated for 12 h. Subsequently, the cells were stimulated with EGF for 2 or 15 min, respectively, and then fixed with PFA, permeabilized, and stained with antibodies against human ezrin. As compared with unstimulated cells (Figure 5), EGF stimulation induces a colocalization of ezrin and GFP-WT S100P to microvillar membrane protrusions and the plasma membrane (top panels, arrows). This is clearly seen in the merged image depicting a higher magnification of one area of the cell. In contrast, the GFP-F15A S100P mutant is not found in the microvilli of EGF-treated A431 cells that are positive for ezrin (bottom panels).
The experiments described above establish that ezrin and S100P show partially overlapping distributions in serum- and EGF-stimulated cells and that this requires an intact ezrin binding site in S100P, i.e., formation of the biologically active dimer. The EGF treatment of A431 cells carried out here is known to elicit a rapid and sustained rise in intracellular Ca2+ but also triggers other signaling events (Hepler et al., 1987; Hughes et al., 1991). Thus, the conditions chosen do not allow to analyze directly the contribution of Ca2+ signaling to the translocation of S100P and ezrin. To address this point we treated A431 cells with the Ca2+ ionophore A23187, which enabled us to increase intracellular Ca2+ concentrations without initiating signaling events through the activation of growth factor receptors. Figure 7 reveals that treatment with the Ca2+ ionophore also induces a significant recruitment of ezrin to the cell periphery. Moreover, a marked increase in the number of microvilli and membrane ruffles is observed in phase images of the ionophore-treated cells (our unpublished results) with the membrane extensions being positive for ezrin. Concomitantly, some WT but not F15A S100P is translocated to the cell periphery and the microvillar protrusions (Figure 7). This parallel and Ca2+-induced translocation indicates that the Ca2+-regulated binding of S100P to ezrin could directly activate dormant ezrin molecules within cells.
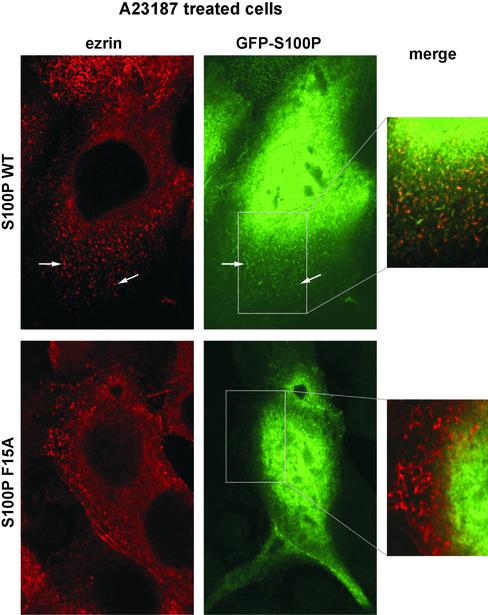
Figure 7.
GFP-WT S100P colocalizes with ezrin in microvilli induced by A23187 treatment. After transfection with the GFP-WT S100P or GFP-F15A S100P encoding plasmids and serum starvation (see Figure 6), A431 cells were treated for 1 min with the Ca2+ ionophore A23187. Cells were then fixed, permeabilized and stained for ezrin using a specific antibody. Note that the A23187 treatment induces an efficient formation of microvillar protrusions that are positive for ezrin and GFP-WT S100P (top panels) but not GFP-F15A S100P (bottom panels).
DISCUSSION
Similar to other EF hand proteins, members of the S100 family are thought to function in Ca2+ stimulus-response coupling by regulating cellular target proteins through direct and Ca2+-dependent interaction. Evidence supporting this view has remained scarce, mainly because of the problem of identifying specific targets. Although a number of such targets have been described, they often lack specificity for a given member of the S100 family, with the interaction possibly being mediated through hydrophobic residues that become exposed on the surface of a Ca2+-bound S100 protein (for reviews, see Donato, 2001; Heizmann et al., 2002). To circumvent this problem of rather nonspecific hydrophobic interactions possibly masking a more specific protein binding, we introduced an affinity approach that distinguishes between S100 protein ligands binding solely because of a Ca2+-induced increase in hydrophobicity and those requiring formation of the biologically active dimer for binding. Such comparative analysis was made possible by our identification of a mutant S100P protein (F15A S100P) in which a single amino acid substitution had abolished the capability of forming a dimer even with the WT S100P chain. Despite its lack of dimer formation the F15A S100P mutant was still able to bind to hydrophobic matrices in the presence of Ca2+, thus exposing a hydrophobic surface, typically mediating the less specific protein interactions (Koltzscher and Gerke, 2000). Using WT and F15A S100P as affinity probes, we now identify ezrin as a specific S100P target that only interacts with the S100P dimer and not with the monomeric mutant derivative.
The specific interaction between dimeric S100P and ezrin not only underscores the validity of our approach but is also the first identification of a target protein for S100P. S100P is a member of the family initially isolated from placental tissue (Becker et al., 1992; Emoto et al., 1992). Through RT-PCR and EST analyses, it has recently also been identified in a number of other tissues and cells, including A431 (Amler et al., 2000; Guerreiro Da Silva et al., 2000; M. Koltzscher and V. Gerke, unpublished observation). All S100P-expressing tissues reported so far contain ezrin at least in their epithelial cell layers (Berryman et al., 1993), thus indicating that the interaction reported here could be of relevance in a number of cells.
Our analysis identifies S100P as one of the few ligands capable of interacting with ezrin in its dormant conformation. This interaction is seen with the two proteins expressed recombinantly in bacteria and also in placental extracts when endogenous ezrin is affinity selected by immobilized S100P (see Figure 1). Binding of S100P occurs via the N-ERMAD of ezrin, which also contains the binding sites for membrane proteins and EBP-50. However, it appears unlikely that the binding sites for these ligands overlap with that of S100P to a substantial degree because the former but not the latter are masked in dormant ezrin. Another binding partner of dormant ezrin is the regulatory subunit of type II cAMP-dependent protein kinase. Here the binding region has been mapped to residues 373–439, i.e., to a sequence outside of N-ERMAD, thus indicating that dormant ezrin can engage in different types of protein interactions (Dransfield et al., 1997). Unmasking of the binding sites in dormant ezrin and the resulting activation of the membrane-cytoskeleton linking function of ezrin requires conformational changes that can be triggered by binding of phosphatidylinositol 4,5-bisphosphate to the N-terminal domain and/or phosphorylation of a conserved threonine in the C-ERMAD (Niggli et al., 1995; Nakamura et al., 1999; Hamada et al., 2000). Because the interaction with S100P also activates ezrin's F-actin binding, similar conformational changes are likely to occur as a consequence of but not before ezrin-S100P complex formation.
In several cells including A431 signaling via activated growth factor receptors has been shown to lead to ezrin phosphorylation, in this case on tyrosine residues, and to a redistribution of ezrin to microvilli, which are induced by this treatment (Bretscher, 1989; Krieg and Hunter, 1992). However, replacement of the tyrosine phosphorylation sites by phenylalanine does not alter the microvillar localization of ezrin in EGF-stimulated cells indicating that tyrosine phosphorylation does not directly activate dormant ezrin and thus its membrane/cytoskeleton association (Crepaldi et al., 1997). Because EGF stimulation also triggers an increase in intracellular Ca2+, the Ca2+-dependent activation of ezrin by S100P could participate in mediating morphological changes induced by ezrin translocation in EGF-treated cells. Such Ca2+-triggered activation of ezrin's cross-linking function is supported by experiments using the Ca2+ ionophore A23187 to elevate intracellular Ca2+-independent of an activation of growth factor receptors. Ionophore treatment and the resulting Ca2+ rise induce not only a recruitment of ezrin to the cell periphery but also an increase in the number of microvillar protrusions (Figure 7). To some extent this resembles the scenario observed in retinal pigment epithelial cells overexpressing transfected ezrin (Bonilha et al., 1999) and in LLCPK cells ectopically expressing a (presumably permanently activated) T567D ezrin mutant (Gautreau et al., 2000). Thus the induction of microvilli and membrane ruffles observed in Ca2+ ionophore–treated cells is likely to be a consequence of ezrin activation, which in turn could be mediated by Ca2+-bound S100P.
Complex formation between Ca2+-bound S100P and ezrin could be stable or transient in nature. A more transient interaction would be in line with the observation that colocalization of ezrin and S100P in the cell periphery is limited in stimulated A431 cells (Figures 6 and 7). What parameters could participate in rendering the S100P-ezrin interaction transient? Our biochemical data only reveal stable complexes capable of binding F-actin. Thus the activation of ezrin's cytoskeleton binding does not alter the affinity for S100P. However, S100P-dependent activation of the F-actin binding site in C-ERMAD could result from a conformational change resembling the separation of N- and C-ERMAD induced by Thr-567 phosphorylation or PIP2 binding (Hamada et al., 2000; Pearson et al., 2000). As a consequence the N-ERMAD and the central α-helical domain of S100P-bound ezrin will be accessible to binding partners like type-1 transmembrane proteins, EBP-50, and the recently identified palladin (Mykkänen et al., 2001), and such binding could in turn render the binding of S100P less stable. However, at present we cannot exclude the possibility that S100P binding to ezrin only unmasks the F-actin binding site without disrupting the N-/C-ERMAD interaction. Future experiments analyzing ezrin's conformational changes induced by S100P binding and reconstituting multiprotein complexes of ezrin with various ligands under defined conditions have to address this question.
Acknowledgments
We thank Anthony Bretscher (Cornell University, Ithaca, NY) for kindly providing ezrin antibodies and an ezrin cDNA construct. S.K. acknowledges a research grant from the German Department of Research and Technology (No. 01KX9820/B) and the Institute of Medical Physics and Biophysics (University of Münster) for hosting the IZKF-proteomics laboratory.
References
- Amler, L.C. et al. (2000). Dysregulated expression of androgen-responsive and nonresponsive genes in the androgen-independent prostate cancer xenograft model CWR22–R1. Cancer Res. 60, 134-141. [PubMed] [Google Scholar]
- Babiychuk, E.B., and Draeger, A. (2000). Annexins in cell membrane dynamics: Ca(2+)-regulated association of lipid microdomains. J. Cell Biol. 150, 1113-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, T., Gerke, V., Kube, E., and Weber, K. (1992). S100P, a novel Ca(2+)-binding protein from human placenta. cDNA cloning, recombinant protein expression and Ca2+ binding properties. Eur. J. Biochem. 207, 541-547. [DOI] [PubMed] [Google Scholar]
- Berryman, M., Franck, Z., and Bretscher, A. (1993). Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J. Cell Sci. 105, 1025-1043. [DOI] [PubMed] [Google Scholar]
- Bonilha, V.L., Finnemann, S.C., and Rodriguez-Boulan, E. (1999). Ezrin promotes morphogenesis of apical microvilli and basal infoldings in retinal pigment epithelium. J. Cell Biol. 147, 1533-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher, A. (1989). Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A431 cells by epidermal growth factor. J. Cell Biol. 108, 921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher, A. (1999). Regulation of cortical structure by the ezrinradixin-moesin protein family. Curr. Opin. Cell Biol. 11, 109-116. [DOI] [PubMed] [Google Scholar]
- Bretscher, A., Edwards, K., and Fehon, R.G. (2002). ERM proteins and merlin: integrators at the cell cortex. Nat. Rev. Mol. Cell. Biol. 3, 586-599. [DOI] [PubMed] [Google Scholar]
- Bretscher, A., Chambers, D., Nguyen, R., and Reczek, D. (2000). ERM-merlin and EBP50 protein families in plasma membrane organization and function. Annu. Rev. Cell Dev. Biol. 16, 113-143. [DOI] [PubMed] [Google Scholar]
- Crepaldi, T., Gautreau, A., Comoglio, P.M., Louvard, C., and Arpin, M. (1997). Ezrin is an effector of hepatocyte growth factor-mediated migration and morphogenesis in epithelial cells. J. Cell Biol. 138, 423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato, R. (2001). S100: a multigene family of calcium-modulated proteins of the EF hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 33, 637-668. [DOI] [PubMed] [Google Scholar]
- Dransfield, D.T., Bradford, A.J., Smith, J., Martin, M., Roy, C., Mangeat, P.H., and Goldenring, J.R. (1997). Ezrin is a cyclic AMP-dependent protein kinase anchoring protein. EMBO J. 16, 35-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto, Y., Kobayashi, R., Akatsika, H., and Hidaka, H. (1992). Purification and characterization of a new member of the S-100 protein family from human placenta. Biochem. Biophys. Res. Commun. 182, 1246-1253. [DOI] [PubMed] [Google Scholar]
- Gautreau, A., Louvard, D., and Arpin, M. (2002). ERM proteins and the NF2 tumor suppressor: the Yin and Yang of cortical actin organization and cell growth signaling. Curr. Opin. Cell Biol. 14, 104-109. [DOI] [PubMed] [Google Scholar]
- Gautreau, A., Louvard, D., and Arpin, M. (2000). Morphogenic effects of ezrin require a phosphorylation-induced transition from oligomers to monomers at the plasma membrane. J. Cell Biol. 150, 193-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribenko, A.V., Guzman-Casado, M., Lopez, M.M., and Makhatadze, G.I. (2002). Conformational and thermodynamic properties of peptide binding to the human S100P protein. Protein Sci. 11, 1367-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribenko, A.V., and Makhatadze, G.I. (1998). Oligomerization and divalent ion binding properties of the S100P protein: a Ca2+/Mg2+-switch model. J. Mol. Biol. 283, 679-694. [DOI] [PubMed] [Google Scholar]
- Guerreiro Da Silva, I.D., Hu, Y.F., Russo, I.H., Ao, X., Salicioni, A.M., Yang, X., and Russo, J. (2000). S100P calcium binding protein overexpression is associated with immortalization of human breast epithelial cells in vitro and early stages of breast cancer development in vivo. Int. J. Oncol. 16, 231-240. [PubMed] [Google Scholar]
- Hamada, K., Shimizu, T., Matsui, T., Tsukita, S., and Hakoshima, T. (2000). Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J. 19, 4449-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heizmann, C.W., Fritz, G., and Schäfer, B.W. (2002). S100 proteins: structure, functions and pathology. Frontiers Biosci. 7, 1356-1368. [DOI] [PubMed] [Google Scholar]
- Hepler, J.R., Nakahata, N., Lovenberg, T.W., DiGuiseppi, J., Herman, B., Earp, H.S., and Harden, T.K. (1987). Epidermal growth factor stimulates the rapid accumulation of inositol (1,4,5)-trisphosphate and a rise in cytosolic calcium mobilized from intracellular stores in A431 cells. J. Biol. Chem. 262, 2951-2956. [PubMed] [Google Scholar]
- Hughes, A.R., Bird, G.S., Obie, J.F., Thastrup, O., and Putney, J.W. (1991). Role of inositol (1,4,5)trisphosphate in epidermal growth factor-induced Ca2+ signaling in A431 cells. Mol. Pharmacol. 40, 254-262. [PubMed] [Google Scholar]
- Kitaura, Y., Matsumoto, S., Satoh, H., Hitomi, K., and Maki, M. (2001). Peflin and ALG-2, members of the penta-EF-hand protein family, form a heterodimer that dissociates in a Ca2+-dependent manner. J. Biol. Chem. 276, 14053-14058. [DOI] [PubMed] [Google Scholar]
- Kligman, D., and Hilt, D.C. (1988). The S-100 protein family. Trends Biochem. Sci. 13, 437-443. [DOI] [PubMed] [Google Scholar]
- Koltzscher, M., and Gerke, V. (2000). Identification of hydrophobic amino acid residues involved in the formation of S100P homodimers in vivo. Biochemistry 39, 9533-9539. [DOI] [PubMed] [Google Scholar]
- Krebs, J., Quadroni, M., and van Eldik, L.J. (1995). Dance of the dimers. Nat. Struct. Biol. 2, 711-714. [DOI] [PubMed] [Google Scholar]
- Krieg, J., and Hunter, T. (1992). Identification of two major epidermal growth factor-induced tyrosine phosphorylation sites in the microvillar core protein ezrin. J. Biol. Chem. 267, 19258-19265. [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- Lankes, W.T., and Furthmayr, H. (1991). Moesin: a member of the protein 4.1-talin-ezrin family of proteins. Proc. Natl. Acad. Sci. USA 88, 8297-8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz, S.E., Braunawell, K.H., Weise, C., Nedlina-Chittka, A., and Gundelfinger, E. (1996). The neuronal EF-hand Ca(2+)-binding protein VILIP: interaction with cell membrane and actin-based cytoskeleton. Biochem. Biophys. Res. Commun. 225, 1078-1083. [DOI] [PubMed] [Google Scholar]
- Mangeat, P., Roy, C., and Martin, M. (1999). ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol. 9, 187-192. [DOI] [PubMed] [Google Scholar]
- Muto, A., and Mikoshiba, K. (1998). Activation of inositol 1, 4, 5-triphosphate receptors induces transient changes in cell shape of fertilized Xenopus eggs. Cell Motil. Cytoskel. 39, 201-208. [DOI] [PubMed] [Google Scholar]
- Mykkänen, O.-M., Grönholm, M., Rönty, M., Lalowski, M., Salmikangas, P., Suila, H., and Carpen, O. (2001). Characterization of human palladin, a microfilament-associated protein. Mol. Biol. Cell 12, 3060-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, F., Huang, L., Pestonjamasp, K., Luna, E.J., and Furthmayr, H. (1999). Regulation of F-actin binding to platelet moesin in vitro by both phosphorylation of threonine 558 and polyphosphatidylinositides. Mol. Biol. Cell 10, 2669-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggli, V., Andreoli, C., Roy, C., and Mangeat, P. (1995). Identification of a phosphatidylinositol-4, 5-biphosphate-binding domain in the N-terminal region of ezrin. FEBS Lett. 376, 172-176. [DOI] [PubMed] [Google Scholar]
- Noegel, A.A. (1996). Alpha-actinin. In: Guidebook to the Calcium Binding Proteins, ed. M.R. Celio, Oxford: Oxford University Press, 21-23.
- Oliferenko, S., Paiha, K., Harder, T., Gerke, V., Schwarzler, C., Schwarz, H., Beug, H., Günthert, U., and Huber, L. (1999). Analysis of CD44-containing lipid rafts: recruitment of annexin II and stabilization by the actin cytoskeleton. J. Cell Biol. 146, 843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterloh, D., Ivanenkov, V., and Gerke, V. (1998). Hydrophobic residues in the C-terminal extension of S100A1 are essential for target protein binding but not for dimerization. Cell Calcium 24, 137-151. [DOI] [PubMed] [Google Scholar]
- Pearson, M., Reczek, D., Bretscher, A., and Karplus, P. (2000). Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell 101, 259-270. [DOI] [PubMed] [Google Scholar]
- Rety, S., Osterloh, D., Arie, J.-P., Tabaries, S., Seemann, J., Russo-Marie, F., Gerke, V., and Lewit-Bentley, A. (2000). Structural basis of the Ca(2+)-dependent association between S100C (S100A11) and its target, the N-terminal part of annexin I. Structure 8, 175-184. [DOI] [PubMed] [Google Scholar]
- Rety, S., Sopkova, J., Renouard, M., Osterloh, D., Tabaries, S., Russo-Marie, F., and Lewit-Bentley, A. (1999). The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat. Struct. Biol. 6, 89-95. [DOI] [PubMed] [Google Scholar]
- Rogers, M.S., and Strehler, E.E. (1996). Calmodulin. ed. M.R. Celio, Oxford: Oxford University Press.
- Roy, C., Martin, M., and Mangeat, P. (1997). A dual involvement of the amino-terminal domain of ezrin in F- and G-actin binding. J. Biol. Chem. 272, 20088-20095. [DOI] [PubMed] [Google Scholar]
- Rustandi, R.R., Baldisseri, D.M., and Weber, D.J. (2000). Structure of the negative regulatory domain of p53 bound to S100B. Nat. Struct. Biol. 7, 570-574. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368-379. [DOI] [PubMed] [Google Scholar]
- Seemann, J., Weber, K., and Gerke, V. (1996). Structural requirements for annexin I-S100C complex formation. Biochem. J. 319, 123-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Anal. Chem. 68: 850-858. [DOI] [PubMed] [Google Scholar]
- Simons, P.C., Pietromonaco, S.F., Reczek, D., Bretscher, A., and Elias, L. (1998). C-terminal threonine phosphorylation activates ERM proteins to link the cell's cortical lipid bilayer to the cytoskeleton. Biochem. Biophys. Res. Commun. 253, 561-565. [DOI] [PubMed] [Google Scholar]
- Sullivan, R., Burnham, M., Torok, K., and Koffer, A. (2000). Calmodulin regulates the disassembly of cortical F-actin in mast cells but is not required for secretion. Cell Calcium 28, 33-46. [DOI] [PubMed] [Google Scholar]
- Tsukita, S., Yonemura, S., and Tsukita, S. (1997). ERM proteins: head-to-tail regulation of actin-plasma membrane interactions. Trends Biochem. Sci. 22, 53-58. [DOI] [PubMed] [Google Scholar]
- Wechsler, A., and Teichberg, V.I. (1998). Brain spectrin binding to the NMDA receptor is regulated by phosphorylation, calcium and calmodulin. EMBO J. 17, 3931-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X., Cheng, L., and Forte, J.G. (1996). Biochemical characterization of ezrin-actin interaction. J. Biol. Chem. 271, 7224-7229. [DOI] [PubMed] [Google Scholar]
- Yonemura, S., Tsukita, S., and Tsukita, S. (1999). Direct involvement of ezrin/radixin/moesin (ERM)-binding membrane proteins in the organization of microvilli in collaboration with activated ERM proteins. J. Cell Biol. 145, 1497-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Herring, C.J., Romano, P.R., Szczepanowska, J., Brzeska, H., Hinnebusch, A.G., and Qin, J. (1998). Anal. Chem. 70: 2050-2059. [DOI] [PubMed] [Google Scholar]
- Zobiack, N., Gerke, V., and Rescher, U. (2001). Complex formation and submembranous localization of annexin 2 and S100A10 in live HepG2 cells. FEBS Lett. 500, 137-140. [DOI] [PubMed] [Google Scholar]
- Zobiack, N., Rescher, U., Laarmann, S., Michgehl, S., Schmidt, M.A., and Gerke, V. (2002). Cell surface attachment of pedestal forming enteropathogenic E. coli induces a clustering of raft components and a recruitment of annexin 2. J. Cell Sci. 115, 91-98. [DOI] [PubMed] [Google Scholar]