Abstract
Several positive regulators of phytochrome A signaling—e.g., LAF1, HFR1, and HY5—operate downstream from the photoreceptor, but their relative sites of action in the transduction pathway are unknown. Here, we show that HFR1RNAi/laf1 or hfr1-201/LAF1RNAi generated by RNA interference (RNAi) has an additive phenotype under FR light compared with the single mutants, hfr1-201 or laf1. This result indicates that LAF1 and HFR1 function in largely independent pathways. LAF1, an R2R3-MYB factor, interacts with HFR1, a basic helix–loop–helix (bHLH) factor, and this interaction is abolished by the R97A mutation in the LAF1 R2R3 domain. Polyubiquitinations of LAF1 and HFR1 by the COP1 E3 ligase in vitro are inhibited by LAF1/HFR1 association. Consistent with this result, endogenous HFR1 is less stable in laf1 compared with wild type, and similarly, LAF1-3HA expressed from a transgene is also less stable in hfr1-201 than wild type. In transgenic plants, HFR1 levels are significantly elevated upon induced expression of LAF1 but not LAF1(R97A). Moreover, induced expression of LAF1 but not LAF1(R97A) delays post-translational HFR1 degradation in FR light. Constitutive coexpression of HFR1 and LAF1 but not HFR1 and LAF1 (R97A) confers FR hypersensitivity in double transgenic plants. Our results show that in addition to their independent functions in phyA signaling, LAF1 and HFR1 also cooperate post-translationally to stabilize each other through inhibition of ubiquitination by COP1, thereby enhancing phyA photoresponses.
Keywords: Light signaling, protein interaction, ubiquitination, proteasomal degradation, MG132
As sessile organisms, plants use light as an environmental cue to execute their growth and developmental program, from seed germination to stem elongation and even the transition to reproductive growth. Plants contain three known groups of photoreceptors: the phytochromes (phy), the cryptochromes, and the phototropins. The cryptochromes and phototropins are blue/ultraviolet-A (B/UV-A) light receptors, whereas the phytochromes sense primarily red (R) and far red (FR) light (Quail 2002). Of the five phytochromes that have been characterized in Arabidopsis, only phytochrome A (phyA) mediates the majority of the physiological responses involved in early developmental processes under FR light (Neff et al. 2000).
Genetic screens have proven to be a powerful approach to investigating the phyA signaling pathway. Under FR light, wild-type seedlings deetiolate and have short hypocotyls because of inhibition by FR. Mutants that are blocked in phyA signaling are impaired in this response, and their hypocotyls continue to elongate under FR. So far, >10 mutants (hy5, fhy1, fhy3, fin219, far1, pat1, rsf1/hfr1/rep1, laf1, laf3, and laf6) involved in phyA signaling have been reported (Oyama et al. 1997; Hudson et al. 1999; Bolle et al. 2000; Fairchild et al. 2000; Fankhauser and Chory 2000; Hsieh et al. 2000; Soh et al. 2000; Ballesteros et al. 2001; Desnos et al. 2001; Møller et al. 2001; Zeidler et al. 2001; Wang and Deng 2002; Hare et al. 2003a). All of these mutants display FR hyposensitivity with respect to hypocotyl elongation, indicating that their encoded factors likely function as positive regulators of phyA signaling. Nevertheless, none of these mutants has hypocotyls as long as a phyA-null mutant under FR light, indicating that phyA signal is distributed through more than one pathway downstream. This view is supported by the observation that a hy5-1hfr1-201 double mutant has an additive phenotype compared with the two single mutants (Kim et al. 2002). Other than the notion that HFR1 and HY5 control different pathways downstream from phyA, the relationship of the other positive regulators and their relative sites of action downstream from the photoreceptor are beginning to be characterized. Two signaling factors, FHY1 and its homolog FHL, likely operate in early steps of phyA signaling as hypocotyl lengths of fhy1 fhl double mutants are indistinguishable from those of phyA in FR (Zhou et al. 2005). Moreover, it has been reported that the light-induced translocation of phyA into the nucleus occurs through a direct interaction with FHY1/FHL (Hiltbrunner et al. 2005, 2006).
Post-translational degradation of positive regulators has recently emerged to be an important regulatory aspect of phyA signaling (Hare et al. 2003b). Previous genetic screens have yielded two genes, COP1 and SPA1, that encode negative regulators of phyA signaling since mutants deficient in these gene products are FR hypersensitive (McNellis et al. 1994; Hoecker et al. 1998). COP1 is a RING motif protein (von Arnim and Deng 1993) with E3 ligase activity, and at least three positive regulators of phyA responses, LAF1 (Seo et al. 2003), HY5 (Saijo et al. 2003), and HFR1 (Jang et al. 2005; Yang et al. 2005), have been shown to be ubiquitinated by this E3 ligase in vitro. The bZIP factor HY5 and the bHLH (basic helix–loop–helix) factor HFR1 interact with the COP1 WD40 domain, while the MYB factor LAF1 binds to its RING motif. Consistent with their being COP1 substrates, these three factors colocalize with the E3 ligase in nuclear bodies, which might represent storage sites or sites of factor ubiquitination.
In eukaryotes, the formation of multiprotein complexes involving combinatorial interactions between transcription factors of different families is an important aspect of gene regulation during signaling. The major families of transcription factors in Arabidopsis are AP2/EREBP (APETALA2/ethylene responsive element-binding protein), MYB-(R1)R2R3, and bHLH (Riechmann et al. 2000). Interaction between MYB and bHLH factors in the transcriptional control of genes was first reported for anthocyanin biosynthesis in maize (Goff et al. 1992) and subsequently found to operate for anthocyanin biosynthesis in dicots as well (Grotewold et al. 2000; Zimmermann et al. 2004; Quattrocchio et al. 2006). In all these cases, the MYB/bHLH heterodimers recognize novel cis-elements upstream of promoters. Other than binding to novel cis-elements, there is at least one report that transcription factor interaction (bHLH and Zinc finger protein) may affect their protein stability. Ubiquitination of the oncoprotein Myc by HectH9 E3 ligase is inhibited by the zinc finger protein Miz1 through competitive interaction with HectH9 (Adhikary et al. 2005).
Among the phyA signaling intermediates identified so far (Wang and Deng 2003), LAF1 is a MYB transcription activator (Ballesteros et al. 2001) while HFR1 is a bHLH protein. In this work, we investigated whether LAF1 and HFR1 control the same or different phyA signaling pathways. Because MYB/bHLH interaction has been previously reported for both plant (Grotewold et al. 2000; Zimmermann et al. 2004; Quattrocchio et al. 2006) and animal systems (Kaspar et al. 2005), we also examined possible LAF1/HFR1 interaction in phyA photoresponses. Here, we show that HFR1 and LAF1 regulate largely independent pathways downstream from phyA. Moreover, these two factors interact in vitro and in vivo, and one consequence of this interaction is an inhibition of their ubiquitination by the COP1 E3 ligase, leading to a post-translational stabilization of the factors.
Results
hfr1laf1 double mutant has additive phenotype compared with either single mutant
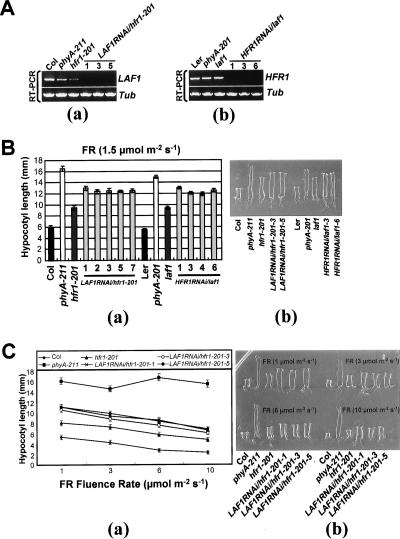
The seedling phenotype of hfr1-201(Col) and laf1(Ler) is not as severe as that of phyA-201(Ler) or phyA-211(Col), suggesting that LAF1 and HFR1 may control two largely distinct pathways. To examine this possibility, we created a hfr1laf1 double mutant for comparative analysis with single mutants hfr1-201 and laf1. Because the two single mutants are in different ecotype backgrounds, we decided to use hairpin (hp) RNA (RNAi [RNA interference]) to silence LAF1 expression in hfr1-201 and HFR1 expression in laf1. We designated these plants LAF1RNAi/hfr1-201 and HFR1RNAi/laf1 (hereafter referred to as double mutants) and selected three lines each for further analysis. Figure 1A shows that the RNAi-mediated silencing was effective in the selected transgenic lines with no detectable LAF1 or HFR1 transcript as monitored by RT–PCR. Phenotypic analysis shows that the double mutants displayed additive phenotype, with hypocotyl lengths longer than those of the single mutants but shorter than those of phyA mutants at low FR fluences (Fig. 1B). This was the case even at higher FR fluences (Fig. 1C). To further verify the specificity, we transformed wild-type plants with each RNAi vector. The hypocotyl lengths of single-gene RNAi lines were comparable to either hfr1-201 or laf1 mutants (data not shown).
Figure 1.
Phenotypes of hfr1laf1 double mutants under FR light. Seedlings were grown on media without sucrose for 4 d under FR light intensities as indicated. Data are presented as average hypocotyl length ± standard deviations (SD; n = 20). (A) RT–PCR analyses showing reduction of LAF1 transcript in LAF1RNAi/hfr1-201 (panel a) and reduction of HFR1 transcript in HFR1RNAi/laf1 (panel b). (B) Hypocotyl length of wild-type (Col; Columbia, Ler; Lansberg erecta), phyA mutants (phyA-211 and phyA-201), hfr1-201, laf1, LAF1RNAi/hfr1-201, and HFR1RNAi/laf1 seedlings after irradiation with FR (1.5 μmol m−2 sec−1). (Panel a) Measurement of hypocotyl lengths. (Panel b) Comparison of seedling morphology. (C) Responses of hfr1laf1 double mutants under different fluence rates (1, 3, 6, and 10 μmol m−2 sec−1) of FR. (Panel a) Measurement of hypocotyl lengths. (Panel b) Comparison of seedling morphology.
Altered phyA-dependent gene expression in hfr1laf1 double mutants
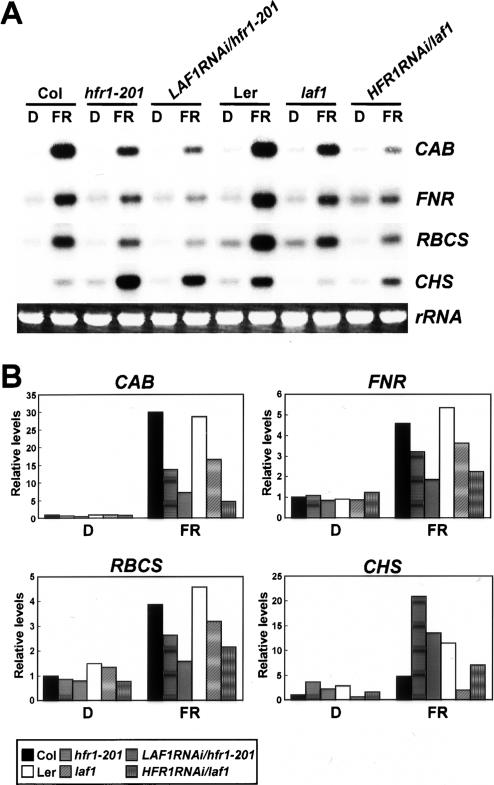
We performed Northern blot analyses with four phyA-dependent genes—CAB, FNR, RBCS, and CHS—using hfr1-201, laf1, and hfr1laf1 double mutant seedlings grown in darkness for 4 d followed by exposure to FR light for 18 h. Figure 2 shows that expression of these four genes was induced by FR in wild-type (Col and Ler) plants. FR-induced expression of CAB, FNR, and RBCS was reduced in hfr1-201 and laf1 mutants compared with those in wild type. Moreover, expression levels of CAB, FNR, and RBCS were lower in LAF1RNAi/hfr1-201 or HFR1RNAi/laf1 double mutants compared with single mutants (hfr1-201 and laf1). These results indicate that both LAF1 and HFR1 function as positive regulators for gene expression. In contrast to CAB, FNR, and RBCS, CHS expression was reduced in laf1 but increased in hfr1-201. This opposing pattern of CHS expression had been previously described (Soh et al. 2000; Ballesteros et al. 2001) and is consistent with the view that LAF1 functions as a positive regulator while HFR1 functions as a negative regulator. Northern blot analyses of CHS expression in the double mutants support this view (Fig. 2A,B).
Figure 2.
FR-dependent expression levels of CAB, FNR, RBCS, and CHS in wild type and various mutants. The seedlings were grown in darkness for 4 d and then transferred to FR light (3 μmol m−2 sec−1) or kept in darkness for 18 h. Each lane contained 5 μg of total RNA. (A) Representative Northern blot analyses. CAB, FNR, RBCS, and CHS indicate RNA levels detected with the respective probes. Ethidium bromide-stained 28S rRNA was used as a loading control. (B) Quantitative expression levels of CAB, FNR, RBCS, and CHS in wild type (Col and Ler), hfr1-201, laf1, LAF1RNAi/hfr1-201, and HFR1RNAi/laf1. Expression levels of each gene and rRNA shown in A were quantified using the program Image Gauge version 3.12 (Fuji) of a PhosphorImage analyzer and normalized to D in the Col sample.
LAF1 interacts with HFR1
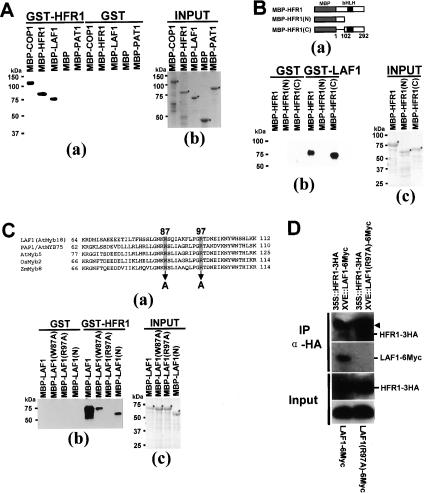
To investigate possible LAF1/HFR1 interaction, we used several MBP (maltose-binding protein) and GST (glutathione S-transferase) fusion proteins for in vitro pull-down assays. Figure 3A shows that GST-HFR1 interacted with MBP-COP1, MBP-HFR1, and MBP-LAF1 but not with MBP-PAT1, another phyA signaling component (Bolle et al. 2000), nor with MBP alone. GST alone did not pull down any of the MBP fusions mentioned above excluding nonspecific binding. These results indicate that HFR1 can self-associate as well as interact with LAF1. The interaction of HFR1 with COP1 confirms previous observations of in vitro ubiquitination by the E3 ligase (Jang et al. 2005; Yang et al. 2005). We also confirmed LAF1/HFR1 interaction using LAF1 synthesized by in vitro transcription/translation (Supplementary Fig. S1).
Figure 3.
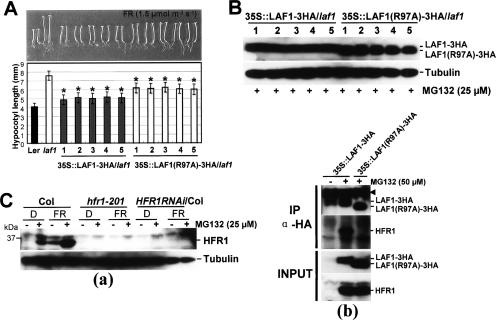
HFR1 and LAF1 interact in vitro and in vivo. (A) In vitro pull-down assay of full-length HFR1 with other proteins. (Panel a) Five-hundred nanograms of target proteins were pull downed with HFR1 protein (1 μg each) and detected by anti-MBP antibody. (Panel b) Purified target proteins used in pull-down assays were resolved on SDS-PAGE. Asterisks indicate the input proteins. (B) Definition of HFR1 interaction domain that binds to LAF1. (Panel a) Schematic diagrams of target proteins [MBP-HFR1, MBP-HFR1(N), and MBP-HFR1(C)]. (Panel b) In vitro pull-down assay to define the interaction domain of HFR1. (Panel c) Purified target proteins used in pull-down assays were resolved on SDS-PAGE. Asterisks indicate the input proteins. (C) Identification of noninteracting LAF1 mutant protein. (Panel a) The amino acid sequence of the LAF1 R3 domain (GenBank accession no. NM_118688) was aligned with those of PAP1/AtMYB75 (GenBank accession no. NM_104541), AtMyb5 (GenBank accession no. U26935), OsMyb2 (GenBank accession no. D88618), and ZmMyb8 (GenBank accession no. AM156905). Two amino acid residues, W87 and R97 (shaded), were mutated to Ala (A). The number of amino acids is indicated at the left and right sides. (Panel b) In vitro pull-down assays of LAF1(W87A) and LAF1(R97A). (Panel c) Purified target proteins used in pull-down assays were resolved on SDS-PAGE. (D) Coimmunoprecipitation of HFR1 with LAF1 protein. Protein extracts of double transgenic Arabidopsis seedlings [35S-HFR1-3HA and XVE-LAF1-6Myc or 35S-HFR1-3HA and XVE-LAF1(R97A)-6Myc] treated with MG132 (50 μM) plus β-estradiol (10 μM) were immunoprecipitated with anti-HA. Input proteins and the immunoprecipitates were separated on 10% SDS–polyacrylamide gels, blotted onto membranes, and detected with anti-HA and anti-Myc antibodies. Input refers to the starting protein amount in extracts used for immunoprecipitation reactions. The arrowhead indicates the cross-reaction with the heavy chain of the protein A-conjugated antibody.
Next, we generated deletion mutants of each protein to identify their interacting region. The C-terminal region of HFR1, designated HFR1(C), was able to interact with the N-terminal region of LAF1, designated LAF1(N) (Fig. 3B,C). Note that HFR1(C) is identical to the HFR1(ΔN) that we reported previously (Jang et al. 2005).
We also examined the subcellular location of HFR1 and LAF1 using onion epidermis cells. Coexpression of HFR1-YFP and LAF1-CFP showed that the two proteins localized to the same nuclear bodies (Supplementary Fig. S2).
LAF1, which is also known as AtMYB18 (Kranz et al. 1998), shares sequence homology in the R3 domain to those of AtMYB5 and AtMYB75, as well as of rice MYB2 and maize MYB8 (Fig. 3C, panel a). Using site-directed mutagenesis, we created two LAF1 mutants, W87A and R97A. Compared with wild type, LAF1 binding to HFR1 in vitro was much weaker in LAF1(W87A) and LAF1(N), and not detectable with LAF1(R97A) (Fig. 3C). We conclude that amino acid residues 87 and 97 of LAF1 are important for HFR1 association and the R97A mutation completely abolishes interaction in vitro.
We generated double transgenic plants coexpressing LAF1-6Myc or LAF1(R97A)-6Myc and HFR1-3HA. Both proteins were unstable in transgenic plants, but their expression levels can be increased by MG132, which blocks 26S proteasomes (Fig. 3D). Immunoprecipitation of HFR1-3HA pulled down LAF1-6Myc, indicating that the two factors associate in vivo. Consistent with the in vitro interaction results, this LAF1/HFR1 association was dependent on amino acid R97 of LAF1 (Fig. 3D). Interestingly, the R97 mutation did not affect LAF1 localization to nuclear bodies (Supplementary Fig. S2), indicating that HFR1 interaction is not needed for this process.
LAF1/HFR1 association reduces ubiquitination by COP1
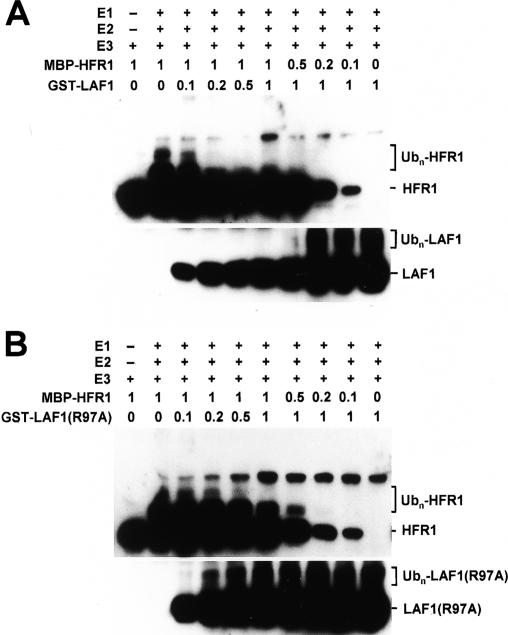
Both LAF1 (Seo et al. 2003) and HFR1 (Jang et al. 2005; Yang et al. 2005) have been shown to be COP1 substrates in vitro. We considered the possibility that interaction of the two substrates may somehow affect their ubiquitination by COP1. To test this possibility, we examined the effects of increasing concentrations of one factor on COP1-mediated ubiquitination of the other. Figure 4A shows that increasing concentrations of LAF1 attenuated polyubiquitination of HFR1 and vice versa. This attenuation was not seen with the noninteracting LAF1(R97A) mutant (Fig. 4B). Together, these results show that LAF1/HFR1 association reduces their polyubiquitination by COP1.
Figure 4.
LAF1 inhibits HFR1 ubiquitination in vitro. Epitope-tagged HFR1, LAF1, LAF1(R97A), and COP1 were expressed in E. coli, and in vitro ubiquitination assays were performed. COP1 (E3)-mediated ubiquitination of HFR1 was performed with increasing amounts of LAF1 (A) and LAF1(R97A) (B) and vice versa, and confirmed by Western blot analyses. Numbers indicate the relative amounts of proteins present in the reaction, where 1 represents 100 ng of MBP-HFR1-3HA, GST-LAF1, or GST-LAF1(R97A).
Complementation of laf1 mutation
Previous work showed that HFR1-3HA can complement the deficiency of the hfr1-201 mutant (Jang et al. 2005). To examine the activity of a similarly tagged LAF1 in transgenic plants, we transformed laf1 with 35S-LAF1-3HA and also with 35S-LAF1(R97A)-3HA, and five independent lines were analyzed for each construct. Figure 5 shows that the hypocotyl length of laf1 can be restored to near wild-type levels by expression of wild-type LAF1-3HA. Similar complementation was observed with the noninteracting mutant, LAF1(R97A)-3HA, expressed at a similar protein level, except that the mutant was less efficient than wild-type LAF1-3HA. In all cases, the protein products of the transgenes in all the five lines for each construct were expressed at comparable levels (Fig. 5B). These experiments show that appending an epitope at the C terminus of LAF1 does not compromise its biological activity. Moreover, LAF1(R97A), which does not interact with HFR1, was still able to complement the mutation, albeit with reduced efficiency.
Figure 5.
Phenotypes of transgenic Arabidopsis seedlings expressing LAF1 or LAF1(R97A) in laf1. (A) Complementation of the laf1 mutant by LAF1 or LAF1(R97A)-3HA overexpression under FR light. Five independent transgenic lines were analyzed for each construct. LAF1 #1 (n = 100), #2 (n = 100), #3 (n = 100), #4 (n = 80), and #5 (n = 80), and LAF1(R97A) #1 (n = 100), #2 (n = 100), #3 (n = 100), #4 (n = 100), and #5 (n = 100) seedlings were grown for 4 d under FR light (1.5 μmol m−2 sec−1) on media without sucrose. Data are presented as the average hypocotyl length ± standard deviation (SD). (*) LAF1-overexpressing transgenic seedlings are significantly shorter than LAF1(R97A) overexpression seedlings (P < 0.01, Student’s t-test). (B) Western blot analysis of transgenic Arabidopsis seedlings overexpressing LAF1-3HA (lanes 1–5) or LAF1(R97A)-3HA (lanes 1–5) under FR irradiation with MG132. After treatment with MG132 (25 μM), seedlings were incubated for 12 h under continuous FR light (1.5 μmol m−2 sec−1). Tubulin levels were used as loading controls. Note that LAF1(R97A)-3HA has a faster mobility than LAF1-3HA. As these samples were analyzed on the same blot, their signal strengths can be directly compared. (C) Endogenous HFR1 levels in wild type and laf1. (Panel a) Western blot analysis of wild type (Col), hfr1-201, HFR1RNAi/Col using anti-HFR1 antibody. Seedlings grown in darkness for 4 d were transferred to FR light (1.5 μmol m−2 sec−1) or darkness with or without MG132 (25 μM) for 12 h. Tubulin levels were used as loading controls. (Panel b) Coimmunoprecipitation of endogenous HFR1 with LAF1 protein. Protein extracts of transgenic Arabidopsis seedlings [35S∷LAF1-3HA or 35S∷LAF1(R97A)-3HA] treated with MG132 (50 μM) under FR light (1.5 μmol m−2 sec−1) for 12 h were immunoprecipitated with anti-HA. Four-day-old 35S∷LAF1-3HA seedlings grown in darkness were used as a control. Input proteins and the immunoprecipitates were separated on 10% SDS–polyacrylamide gels, blotted onto membranes, and detected with anti-HA and anti-HFR1 antibodies. Input refers to the starting protein amount in extracts used for immunoprecipitation reactions. The arrowhead indicates the cross-reaction with the heavy chain of the protein A-conjugated antibody.
Using specific HFR1 antibodies for in vivo pull-down assays, we showed that wild-type LAF1-3HA but not its R97 mutant interacted with the endogenous HFR1 (Fig. 5C).
LAF1/HFR1 interaction stabilizes HFR1 levels
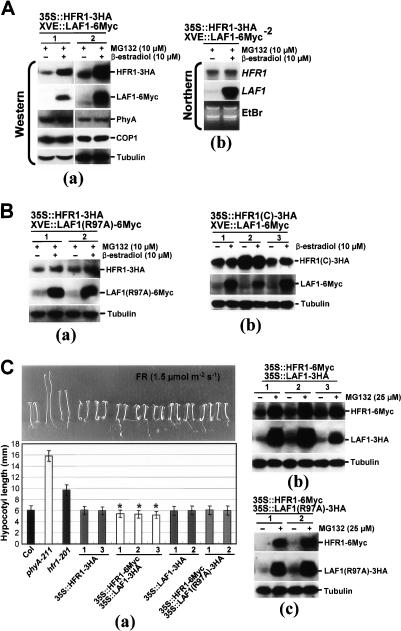
The observation that epitope-tagged HFR1 and LAF1 still retain their biological activities allowed us to use double transgenic plants expressing these two factors to investigate the consequence of their interaction in vivo. If LAF1/HFR1 association inhibits HFR1 ubiquitination by COP1, we would expect HFR1 to be stabilized in vivo by this association. To examine this issue with appropriate internal controls, we generated double transgenic plants expressing HFR1-3HA and inducible LAF1-6Myc and selected two lines for further analysis. We treated transgenic seedlings with low concentrations of MG132 to accumulate low levels of HFR1 to establish a baseline for comparison (Fig. 6A). Figure 6A, panel a, shows that induced expression of LAF1-6Myc increased HFR1 levels by two to five times depending on the transgenic line, and under this condition, there was no change in levels of COP1 nor phyA. Northern blot analysis confirmed that HFR1 transcript remained at similar levels with or without inducer treatment (Fig. 6A, panel b).
Figure 6.
Stabilization of HFR1 by LAF1. (A) HFR1 protein levels are increased by induced LAF1 expression. (Panel a) Two independent transgenic lines containing 35S-HFR1-3HA and XVE-LAF1-6Myc were treated with MG132 (10 μM) with and without induction of LAF1-6Myc expression by inducer (10 μM β-estradiol) for 12 h. HFR1 and LAF1 expression levels were detected by anti-HA and anti-Myc, respectively. These protein extracts were analyzed with anti-COP1 and anti-phyA antibodies. Tubulin expression was used to normalize loading. (Panel b) Northern blot analysis of transgenic plants containing 35S-HFR1-3HA and XVE-LAF1-6Myc-2. Total RNAs were isolated from transgenic plants treated as in panel a. Transgenic HFR1 transcript levels were detected using a full-length HFR1 cDNA probe. Ethidium bromide staining (EtBr) of total RNAs (3 μg per lane) was used to monitor RNA loading. (B) HFR1 protein levels are not affected by LAF1(R97A) expression. (Panel a) Two independent transgenic lines containing 35S-HFR1-3HA and XVE-LAF1(R97A)-6Myc were treated as in A. Antibodies for detection of HFR1-3HA, LAF1(R97A)-6Myc, and loading control were identical to those of A, except anti-COP1 and anti-phyA antibodies. (Panel b) Three independent transgenic lines containing 35S-HFR1(C)-3HA and XVE-LAF1-6Myc were treated with and without induction of LAF1-6Myc expression by inducer (10 μM β-estradiol) for 12 h. Antibodies for detection of HFR1(C)-3HA, LAF1-6Myc, and loading control were identical to those of A, except anti-COP1 and anti-phyA antibodies. (C) Phenotypes of HFR1/LAF1 double overexpressing transgenic seedlings under FR light. (Panel a) Hypersensitivity of HFR1/LAF1 double overexpressing transgenic seedlings under FR light. Seedlings were grown for 4 d under FR light (1.5 μmol m−2 sec−1) on media without sucrose. Data are presented as average hypocotyl length ± standard deviations (SD; n > 100). Seedlings are shown above the histograms. (*) HFR1/LAF1 double overexpressing transgenic seedlings are significantly shorter than HFR1 or LAF1 single overexpression seedlings (P < 0.01, Student’s t-test; n > 100). Western blot analysis of transgenic Arabidopsis seedlings overexpressing HFR1-6Myc and LAF1-3HA (panel b) or HFR1-6Myc and LAF1(R97A)-3HA (panel c) under FR irradiation with or without MG132. After treatment with 25 μM MG132 or mock treatment, seedlings were incubated for a further 12 h under continuous FR light (1.5 μmol m−2 sec−1). Tubulin levels were used as loading controls.
We performed control experiments using transgenic plants expressing HFR1-3HA and inducible LAF1(R97A)-6Myc, which was unable to interact with HFR1 (Fig. 3). In contrast to wild-type LAF1, induced expression of the noninteracting LAF1(R97A) did not significantly alter HFR1 levels (Fig. 6B, panel a). Additional control experiments showed that LAF1 expression had a small stabilization effect on HFR1(C)-3HA, which already accumulated to high levels in the absence of MG132 (Fig. 6B, panel b).
To investigate the phenotypic consequence of LAF1/HFR1 interaction, we compared seedling phenotypes of double overexpressors with single overexpressors. Figure 6C, panel a confirmed previous data that overexpression of neither HFR1 nor LAF1 alone produced FR hypersensitivity. In contrast, FR hypersensitivity was obtained in plants expressing the two factors, and this effect was seen in three independent transgenic lines. Western blot analysis showed that HFR1-6Myc and LAF1-3HA were present in the coexpressing plants, although the levels were substantially increased with MG132, indicating continued degradation (Fig. 6C, panel b). Control experiments showed that in plants expressing 35S-HFR1-6Myc, coexpression of LAF1(R97A)-3HA to comparable levels as the wild-type LAF1-3HA (Fig. b 6C, panel b,c; Supplementary Fig. S4) did not confer FR hypersensitivity, indicating the requirement of LAF1/HFR1 interaction for enhanced photoresponses.
Regulated post-translational degradation of LAF1 and HFR1
We have previously shown that HFR1 is degraded post-translationally with a half-life of ∼1 h (Jang et al. 2005). Figure 7A confirms this observation and, in addition, demonstrates that the half-life of HFR1 decay can be prolonged to 2 h upon induced LAF1 expression. Similar experiments using cycloheximide to block new protein synthesis showed that the LAF1 post-translational degradation half-life (∼1 h) was similarly prolonged to 2–3 h in the presence of coexpressed HFR1. As a negative control, we examined the effects of induced expression of the noninteracting mutant LAF1(R97A). Figure 7B, panel a, shows that the HFR1 decay rate was not significantly altered by induced expression of LAF1(R97A), indicating the requirement of LAF1/HFR1 interaction to delay HFR1 degradation. Note that the decay rate of LAF1(R97A) was ∼1–2 h, irrespective of the presence of HFR1 (Fig. 7B, panel b).
Figure 7.
Post-translational decay of HFR1. (A) HFR1 decay is delayed by induced LAF1 expression. (Panel a) Four-day-old seedlings (35S-HFR1-3HA and XVE-LAF1-6Myc) grown in darkness were incubated in liquid MS medium with MG132 (50 μM) or MG132 (50 μM) plus β-estradiol (10 μM) for 12 h with continuous light, washed, and then transferred to MS medium with 100 μM cycloheximide (CHX). Treated seedlings were then incubated under FR light (1.5 μmol m−2 sec−1). Proteins were extracted at the indicated times and analyzed by Western blotting using anti-HA or anti-Myc antibodies. Tubulin levels were used as loading controls. Expression levels of HFR1 and tubulin were measured using the program Image Gauge version 3.12 (Fuji), and the values were normalized to 0 time in the “−” inducer or “+” inducer sample of both panels. (Panel b) Post-translational decay of LAF1. Four-day-old seedlings (35S-LAF1-3HA) grown in darkness were treated with MG132 (50 μM). All conditions and loading control were identical to those of panel a. (B) HFR1 decay is not delayed by induced LAF1(R97A) expression. (Panel a) Four-day-old seedlings [35S-HFR1-3HA and XVE-LAF1(R97A)-6Myc] grown in darkness were used, and all conditions were identical to those of A. (Panel b) Post-translational decay of LAF1 (R97A). Four-day-old seedlings [35S-LAF1(R97A)-3HA] grown in darkness were treated with MG132 (50 μM). All conditions and loading control were identical to those of panel a in A. (C) Post-translational decay of endogenous HFR1 in LAF1RNAi/Col. Four-day-old seedlings (wild type and LAF1RNAi/Col) grown in darkness were incubated in liquid MS medium with MG132 (50 μM), and other conditions were identical to those of A, except anti-HFR1 antibody was used. (D) Post-translational decay of LAF1 in hfr1-201. 35S∷LAF1-3HA/Col- or 35S∷LAF1-3HA/hfr1-201-overexpressing seedlings were used. All conditions were identical to those of C except anti-HA antibody.
Using antibody to HFR1, we also investigated the post-translational fate of endogenous HFR1 in wild-type and transgenic plants deficient in LAF1. Figure 7C shows that the HFR1 half-life was reduced in LAF1RNAi plants compared with wild type, indicating that a deficiency in LAF1 destabilized the endogenous HFR1. Because we were unsuccessful in preparing antibody to LAF1, we compared the decay rate of transgenic LAF1-3HA, rather than the endogenous LAF1, in wild type and hfr1-201. Similar to the case of HFR1, the LAF1-3HA half-life was also reduced by the absence of HFR1. Taken together, these results provide additional evidence that the stability of one factor is decreased in the absence of its interacting partner in the endogenous setting.
Discussion
LAF1 and HFR1 regulate largely independent pathways
Genetic screens for phyA signaling mutants have been largely based on seedling morphology under FR light. Wild-type seedlings grown under FR are deetiolated and have expanded cotyledons and short hypocotyls, whereas seedlings of a phyA-null mutant are completely insensitive to FR light and remain etiolated with folded cotyledons and long hypocotyls. The use of these criteria for a block in phyA signal transduction—i.e., FR hyposensitivity—has resulted in the isolation and characterization of >10 mutants deficient, but not totally blocked, in FR responses. The phenotype of all of these mutants is not as severe as that of a phyA-null mutant. Although mutant hypocotyls are longer than wild-type hypocotyls under FR light, they are not as long as those of a phyA-null mutant. This result suggests that phyA signals may be distributed via several branches downstream from the photoreceptor, each of which contributes to an inhibition of hypocotyl elongation. This notion has received some support from the analysis of double mutants. Kim et al. (2002) showed that the hy5-1hfr1-201 double mutant has hypocotyls longer than those of the two single mutants hfr1-201 and hy5-1. Here, we have extended the analysis to laf1 and hfr1-201 and shown that the double mutant (hfr1laf1) also has an additive phenotype of the two single mutants, indicating that LAF1 and HFR1 regulate largely independent pathways. We note that seedling hypocotyls of either the hy5-1hfr1-201 (Kim et al. 2002) or the hfr1laf1 double mutant are still shorter than those of a phyA-null mutant (Fig. 1), suggesting the contribution of additional positive factors in FR-mediated inhibition of hypocotyl elongation. Whether LAF1 and HY5 act in the same or different pathway downstream from the phyA photoreceptor awaits future analysis of laf1hy5 double mutants.
Expression analysis of selected FR-responsive genes—CAB, RBCS, and FNR—confirmed the additive molecular phenotype of the hfr1laf1 double mutant (Fig. 2). Transcript levels of each of these three genes are lower in the double mutant compared with those in the single mutants. The regulation of CHS is clearly different from that of CAB, RBCS, and FNR. Soh et al. (2000) previously reported that CHS transcript levels are elevated in hfr1-201, and this observation is confirmed by our results. Together, they indicate that HFR1 acts as a negative regulator of CHS expression. On the other hand, LAF1 is a positive regulator for CHS expression since transcript levels of this gene are depressed in laf1.
Although our analysis indicates that LAF1 and HFR1 regulate largely independent pathways downstream from the photoreceptor, we cannot rule out the possibility that a portion of the two factors may also function in a shared pathway that contributes to the photoresponses. This is because the phenotype attributed to the shared pathway, which is nonadditive, will be masked in the double mutant by the phenotypes of the independent pathways, which are additive.
LAF1 interacts with HFR1
Previous reports of MYB/bHLH interaction in plant anthocyanin biosynthesis (Goff et al. 1992; Grotewold et al. 2000; Zimmermann et al. 2004; Quattrocchio et al. 2006) have prompted us to examine whether LAF1 (MYB) would interact with HFR1 (bHLH) in phyA signaling. In vitro studies showed that LAF1 indeed interacts with HFR1, and this interaction is dependent on the R97 residue in the LAF1 R3 domain (Fig. 3). This result confirms previous findings that the R3 domain of MYB is needed for bHLH interaction (Grotewold et al. 2000; Zimmermann et al. 2004). In vivo pull-down assays support the notion that the LAF1/HFR1 interaction observed in vitro has physiological relevance. LAF1, but not the noninteracting mutant LAF1(R97A), could be coimmunoprecipitated with endogenous HFR1 as well as transgenic HFR1-3HA (Figs. 3D, 5C [panel b]).
The Arabidopsis genome contains two myb factors, AtMyb5 and AtMyb75, with high sequence homology with LAF1 in the R2R3 region. AtMyb75 is also known as PAP1, which associates with the bHLH protein EGL3 in mediating trichome development (Zhang et al. 2003). In view of our results here, it is possible that PAP1/EGL3 association may also enhance factor stability. The function of AtMyb5 is unknown. Whether AtMyb5 might interact with HFR1 or other bHLH proteins in the context of other signaling pathways is a subject of future investigation.
Transient expression assays show that LAF1 and HFR1 colocalize in nuclear bodies, and the localization of LAF1 is not affected by the R97 mutation. These results indicate that LAF1/HFR1 interaction is not needed for nuclear body localization. Note that the colocalization of LAF1/COP1 and of HFR1/COP1 in such nuclear bodies has been previously reported (Seo et al. 2003; Jang et al. 2005).
In addition to the MYB/bHLH interaction for anthocyanin biosynthesis (Goff et al. 1992; Grotewold et al. 2000; Zimmermann et al. 2004; Quattrocchio et al. 2006) and the LAF1/HFR1 interaction for phyA responses described here, MYB/bHLH complexes have been implicated in the regulation of gene expression in other systems as well. Kaspar et al. (2005) reported that c-Myb directly interacts with MyoD, a bHLH factor, to repress differentiation of myoblasts, and Agarwal et al. (2006) found that the MYB15/ICE1(bHLH) complex binds to cis-elements upstream of CBF promoters to mediate cold responses of Arabidopsis. It should be pointed out that both LAF1 and HFR1 bind to the COP1 E3 ligase, which contains WD40 repeats (Seo et al. 2003; Jang et al. 2005), and with respect to this finding, a MYB/bHLH/WD40 protein complex has been shown to specify epidermal cell fate (Ramsay and Glover 2005).
HFR1/LAF1 association stabilizes both factors and confers FR hypersensitivity
The association of LAF1 with HFR1 raises the question of the physiological relevance of the protein complex in phyA signaling. These two positive regulators have been identified as substrates of the COP1 E3 ligase, and their degradation in FR likely operates to desensitize phyA signals (Seo et al. 2003; Jang et al. 2005; Yang et al. 2005). We examined the effects of LAF1 on the ubiquitination of HFR1 mediated by COP1 and vice versa and found that LAF1/HFR1 interaction depresses ubiquitination of both factors in vitro (Fig. 4). Consistent with the in vitro results, analysis of endogenous HFR1 in LAF1RNAi plants showed a faster post-translational degradation in plants deficient in LAF1 compared with wild type. Moreover, the stability of transgenic HFR1-3HA can be enhanced by the coexpression of wild-type LAF1 but not the noninteracting mutant LAF1(R97A) (Fig. 7), which nonetheless can still complement the laf1 mutation but with reduced efficiency (Fig. 5). Similarly, the post-translational degradation rate of LAF1 is increased in hfr1-201 compared with wild type but delayed in plants with coexpressed HFR1 (Fig. 7). These results are similar to those reported for the Myc oncoprotein, which forms a ternary complex with the zinc finger protein Miz1 and the E3 ligase HectH9. It was found that Miz1 inhibits ubiquitination of Myc by HectH9 E3 ligase through competition for binding to the latter (Adhikary et al. 2005).
To elucidate the physiological impact of LAF1/HFR1 association, we analyzed seedling phenotypes of transgenic plants. We found that double transgenic plants expressing LAF1 and HFR1 are hypersensitive to FR (shorter hypocotyls) compared with wild-type plants. In contrast, plants expressing LAF1(R97A) and HFR1 are like wild type, with no FR hypersensitivity. The increased photosensitivity can be explained by the stabilization of LAF1 and HFR1 resulting in higher factor expression levels to promote greater photoresponses. The reduced capacity of the noninteracting mutant LAF1(R97A) as compared with wild-type LAF1 to complement the laf1 mutation also supports this view (Fig. 5). Future work should address whether the LAF1/HFR1 complex may bind to and activate novel cis-elements upstream of FR-responsive promoters.
Materials and methods
Plant material, growth conditions, and production of transgenic plants
The wild-type Columbia (Col) and Landsberg erecta (Ler) ecotypes of Arabidopsis thaliana, hfr1-201 (in the Col background) (Soh et al. 2000), and laf1 (in the Ler background) (Ballesteros et al. 2001) were used in this study. Conditions for plant growth and FR irradiation were described by Bolle et al. (2000), and the following FR fluence rates were used: 1, 1.5, 3, 6, and 10 μmol/m2/sec. Plants were transformed by the Agrobacterium strain EHA105 using the floral dip method (Clough and Bent 1998).
Construction of LAF1RNAi/hfr1-201 and HFR1RNAi/laf1
The mutants hfr1-201 (Col) and laf1 (Ler) are in different ecotypes, and they both contain a kanamycin-resistance marker. To avoid potential issues with ecotypic differences in photoresponses, we used hairpin RNA to silence either HFR1 or LAF1 in the appropriate mutant background. The LAF1-RNAi contained ∼500 base pairs (bp), whereas the HFR1-RNAi construct contained ∼350 bp. DNA fragments were amplified by PCR using the following oligo sets: 5′-CACCGCTCAAGTCTCAGAGCTTAC-3′ and 5′-ACGTCGTTGTTGATGGAGAA-3′ for LAF1-RNAi, and 5′-CACCTGCGTAAGCTACAGCAACTC-3′ and 5′-CGTGAAGAGACTGAGGAGAA-3′ for HFR1-RNAi. PCR-amplified DNA fragments were cloned into pENTR/D, followed by LR reaction with pBA-DC-RNAi to generate pBA-RNAi-LAF1 and pBA-RNAi-HFR1, which confer Basta resistance. These RNAi constructs were transformed into wild-type (Col or Ler) or mutant (hfr1-201 or laf1) backgrounds. Homozygous T3 Basta- and kanamycin-resistant double mutants were selected and used for further analysis.
RNA extraction, RT–PCR, and Northern blot hybridization
Total RNA was extracted from Arabidopsis seedlings using Qiagen RNeasy Plant Mini Kits (Qiagen). Reverse transcription was performed using the SuperScript II RT kit (Invitrogen) according to the manufacturer’s instructions.
RT–PCR was performed as described (Hare et al. 2003a). The oligonucleotides were as follows: 5′-TTCTTGCTGGACCACT GTTC-3′ and 5′-ACGTCGTTGTTGATGGAGAA-3′ for LAF1 amplification, 5′-GTCGGATCACTTGCTGTGAA-3′ and 5′-CGTGAAGAGACTGAGGAGAA-3′ for HFR1 amplification, and 5′-GGACAAGCTGGGATCCAGG-3′ and 5′-CGTCTCCA CCTTCAGCACC-3′ for Tubulin (Tub) amplification.
Northern blot analysis was performed as described previously (Jang et al. 2005) using HFR1, CAB, FNR, RBCS, and CHS probes identical to those described elsewhere (Soh et al. 2000; Ballesteros et al. 2001; Møller et al. 2001).
Vector construction and site-directed mutagenesis of LAF1
Plasmids encoding MBP-COP1, MBP-HFR1, and MBP-HFR1(C) for protein expression in Escherichia coli and plasmids encoding 35S-HFR1-3HA, 35S-HFR1-6Myc, and 35S-HFR1(C)-3HA for protein expression in plants were described previously (Jang et al. 2005). Most constructs except GST-tagged vectors in this study were made using the Gateway system (Invitrogen). Construction of these destination vectors has been described elsewhere (Zhang et al. 2005).
HFR1(N) contains amino acids 1–101 of HFR1, and LAF1(N) contains amino acids 1–160 of LAF1. DNA sequences encoding these regions were amplified by PCR using full-length cDNAs of HFR1 (Jang et al. 2005) and LAF1 (Ballesteros et al. 2001). cDNA encoding full-length PAT1 was amplified by PCR to generate pMBP-DC-PAT1. All cDNA or DNA fragments were cloned in pENTR/D vector (Invitrogen) and then transferred into the appropriate vectors by recombination using the LR Clonase enzyme according to the manufacturer’s instructions (Invitrogen).
To generate LAF1 mutant genes (W87A or R97A), site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene), pENTR-LAF1 as a template, and the following primer sets: 5′-TCTTCCTTGGGTAA CAAGGCGTCGCAAATAGCTAAATTC-3′ and 5′-GAATTT AGCTATTTGCGACGCCTTGTTACCCAAGGAAGA-3′ for LAF1(W87A), and 5′-GCTAAATTCTTACCGGGAGCAACAG ACAATGAGATAAAG-3′ and 5′-CTTTATCTCATTGTCTGT TGCTCCCGGTAAGAATTTAGC-3′ for LAF1(R97A). After mutagenesis of LAF1, the mutant genes were used as a template for PCR reaction and cloned in a pENTR/D vector, followed by LR reaction to generate pMBP-DC-LAF1(W87A) and pMBP-DC-LAF1(R97A).
cDNAs encoding full-length HFR1, LAF1, and LAF1(R97A) were amplified by PCR and inserted into pGEX-4T-1 (Amersham Biosciences) to generate pGST-HFR1, pGST-LAF1, and pGST-LAF1(R97A), respectively. All constructs used in this study were verified by sequencing.
Preparation of recombinant proteins
Recombinant proteins were expressed in E. coli and purified from bacterial extracts as described (Jang et al. 2005).
In vitro pull-down, in vitro ubiquitination assays, and in vivo coimmunoprecipitation
Experimental procedures were essentially as described before (Jang et al. 2005). For in vitro pull-down assays, the reaction mix was incubated with glutathione Sepharose 4B for 2 h. After washing with buffer (50 mM Tris-HCl at pH 7.5, 100 mM NaCl, 0.6% Triton X-100), pulled-down proteins were separated on 10% SDS–polyacrylamide gels and were detected by Western blotting using anti-MBP antibody (Santa Cruz Biotechnology).
For in vitro ubiquitination assays, each reaction mixture (30 μL) contained ∼100 ng of protein substrate (MBP-HFR1-3HA), 20 ng of rabbit E1 (Boston Biochem), 20 ng of human E2 UbcH5b (Boston Biochem), 10 μg of His6-ubiquitin (Sigma), and 200 ng of E3 (MBP-COP1) with or without GST-LAF1 or GST-LAF1(R97A).
Reactions were carried out for 2 h at 30°C. Ten microliters of the reaction mixtures were separated on 8% SDS–polyacrylamide gels, and ubiquitinated MBP-HFR1-3HA and GST-LAF1 or GST-LAF1(R97A) were detected by Western blotting with anti-HA (Santa Cruz Biotechnology) and anti-GST (Sigma) antibodies, respectively.
For in vivo coimmunoprecipitation, 10-d-old double transgenic Arabidopsis seedlings [35S-HFR1-3HA and XVE-LAF1-6Myc or 35S-HFR1-3HA and XVE-LAF1(R97A)-6Myc] treated with MG132 (50 μM) plus β-estradiol (10 μM) for 16 h were used. Five micrograms of anti-HA monoclonal (Santa Cruz Biotechnology) antibody were used for coimmunoprecipitation, and eluted proteins were analyzed by Western blotting using anti-HA, and anti-Myc (Santa Cruz Biotechnology) antibodies.
HFR1 antibody production
We generated polyclonal HFR1 antibody using MBP-HFR1 protein. Specific HFR1 antibody was partially purified from rabbit serum using GST-HFR1 protein.
Subcellular localization experiment, protein extraction, and Western blotting
The subcellular localization experiment, protein extraction, and Western blot analysis were identical to those described by Jang et al. (2005).
Acknowledgments
I.-C.J. was supported by a Human Frontier Science Program post-doctoral fellowship (LT00309/2004-C). S.W.Y. was supported by a post-doctoral fellowship from the Korea Research Foundation. This work was supported by NIH grant GM44640.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1568207
References
- Adhikary S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., Hock A., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., Beier R., Bernard S., Quarto M., Capra M., Goettig S., Bernard S., Quarto M., Capra M., Goettig S., Quarto M., Capra M., Goettig S., Capra M., Goettig S., Goettig S., et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Agarwal M., Hao Y., Kapoor A., Dong C.H., Fujii H., Zheng X., Zhu J.K., Hao Y., Kapoor A., Dong C.H., Fujii H., Zheng X., Zhu J.K., Kapoor A., Dong C.H., Fujii H., Zheng X., Zhu J.K., Dong C.H., Fujii H., Zheng X., Zhu J.K., Fujii H., Zheng X., Zhu J.K., Zheng X., Zhu J.K., Zhu J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006;281:37636– 37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Ballesteros M.L., Bolle C., Lois L.M., Moore J.M., Vielle-Calzada J.P., Grossniklaus U., Chua N.H., Bolle C., Lois L.M., Moore J.M., Vielle-Calzada J.P., Grossniklaus U., Chua N.H., Lois L.M., Moore J.M., Vielle-Calzada J.P., Grossniklaus U., Chua N.H., Moore J.M., Vielle-Calzada J.P., Grossniklaus U., Chua N.H., Vielle-Calzada J.P., Grossniklaus U., Chua N.H., Grossniklaus U., Chua N.H., Chua N.H. LAF1, a MYB transcription activator for phytochrome A signaling. Genes & Dev. 2001;15:2613–2625. doi: 10.1101/gad.915001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C., Koncz C., Chua N.H., Koncz C., Chua N.H., Chua N.H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes & Dev. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Desnos T., Puente P., Whitelam G.C., Harberd N.P., Puente P., Whitelam G.C., Harberd N.P., Whitelam G.C., Harberd N.P., Harberd N.P. FHY1: A phytochrome A-specific signal transducer. Genes & Dev. 2001;15:2980–2990. doi: 10.1101/gad.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild C.D., Schumaker M.A., Quail P.H., Schumaker M.A., Quail P.H., Quail P.H. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes & Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Chory J., Chory J. RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol. 2000;124:39–45. doi: 10.1104/pp.124.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., Cone K.C., Chandler V.L., Cone K.C., Chandler V.L., Chandler V.L. Functional analysis of the transcriptional activator encoded by the maize B gene: Evidence for a direct functional interaction between two classes of regulatory proteins. Genes & Dev. 1992;6:864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- Grotewold E., Sainz M.B., Tagliani L., Hernandez J.M., Bowen B., Chandler V.L., Sainz M.B., Tagliani L., Hernandez J.M., Bowen B., Chandler V.L., Tagliani L., Hernandez J.M., Bowen B., Chandler V.L., Hernandez J.M., Bowen B., Chandler V.L., Bowen B., Chandler V.L., Chandler V.L. Identification of the residues in the MYB domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. 2000;97:13579–13584. doi: 10.1073/pnas.250379897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare P.D., Møller S.G., Huang L.F., Chua N.H., Møller S.G., Huang L.F., Chua N.H., Huang L.F., Chua N.H., Chua N.H. LAF3, a novel factor required for normal phytochrome A signaling. Plant Physiol. 2003a;133:1592–1604. doi: 10.1104/pp.103.028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare P.D., Seo H.S., Yang J.Y., Chua N.H., Seo H.S., Yang J.Y., Chua N.H., Yang J.Y., Chua N.H., Chua N.H. Modulation of sensitivity and selectivity in plant signaling by proteasomal destabilization. Curr. Opin. Plant Biol. 2003b;6:453–462. doi: 10.1016/s1369-5266(03)00080-3. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczian A., Bury E., Tscheuschler A., Kircher S., Toth R., Honsberger A., Nagy F., Fankhauser C., Schafer E., Viczian A., Bury E., Tscheuschler A., Kircher S., Toth R., Honsberger A., Nagy F., Fankhauser C., Schafer E., Bury E., Tscheuschler A., Kircher S., Toth R., Honsberger A., Nagy F., Fankhauser C., Schafer E., Tscheuschler A., Kircher S., Toth R., Honsberger A., Nagy F., Fankhauser C., Schafer E., Kircher S., Toth R., Honsberger A., Nagy F., Fankhauser C., Schafer E., Toth R., Honsberger A., Nagy F., Fankhauser C., Schafer E., Honsberger A., Nagy F., Fankhauser C., Schafer E., Nagy F., Fankhauser C., Schafer E., Fankhauser C., Schafer E., Schafer E. Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 2005;15:2125–2130. doi: 10.1016/j.cub.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Tscheuschler A., Viczian A., Kunkel T., Kircher S., Schafer E., Tscheuschler A., Viczian A., Kunkel T., Kircher S., Schafer E., Viczian A., Kunkel T., Kircher S., Schafer E., Kunkel T., Kircher S., Schafer E., Kircher S., Schafer E., Schafer E. FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 2006;47:1023–1034. doi: 10.1093/pcp/pcj087. [DOI] [PubMed] [Google Scholar]
- Hoecker U., Xu Y., Quail P.H., Xu Y., Quail P.H., Quail P.H. SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell. 1998;10:19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh H.L., Okamoto H., Wang M., Ang L.H., Matsui M., Goodman H., Deng X.W., Okamoto H., Wang M., Ang L.H., Matsui M., Goodman H., Deng X.W., Wang M., Ang L.H., Matsui M., Goodman H., Deng X.W., Ang L.H., Matsui M., Goodman H., Deng X.W., Matsui M., Goodman H., Deng X.W., Goodman H., Deng X.W., Deng X.W. FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes & Dev. 2000;14:1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Hudson M., Ringli C., Boylan M.T., Quail P.H., Ringli C., Boylan M.T., Quail P.H., Boylan M.T., Quail P.H., Quail P.H. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes & Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Yang J.Y., Seo H.S., Chua N.H., Yang J.Y., Seo H.S., Chua N.H., Seo H.S., Chua N.H., Chua N.H. HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes & Dev. 2005;19:593–602. doi: 10.1101/gad.1247205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar P., Pajer P., Sedlak D., Tamaoki T., Dvorak M., Pajer P., Sedlak D., Tamaoki T., Dvorak M., Sedlak D., Tamaoki T., Dvorak M., Tamaoki T., Dvorak M., Dvorak M. c-Myb inhibits myogenic differentiation through repression of MyoD. Exp. Cell Res. 2005;309:419–428. doi: 10.1016/j.yexcr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Kim Y.M., Woo J.C., Song P.S., Soh M.S., Woo J.C., Song P.S., Soh M.S., Song P.S., Soh M.S., Soh M.S. HFR1, a phytochrome A signaling component, acts in a separate pathway from HY5, downstream of COP1 in Arabidopsis thaliana. Plant J. 2002;30:711–719. doi: 10.1046/j.1365-313x.2002.01326.x. [DOI] [PubMed] [Google Scholar]
- Kranz H.D., Denekamp M., Greco R., Jin H., Leyva A., Meissner R.C., Petroni K., Urzainqui A., Bevan M., Martin C., Denekamp M., Greco R., Jin H., Leyva A., Meissner R.C., Petroni K., Urzainqui A., Bevan M., Martin C., Greco R., Jin H., Leyva A., Meissner R.C., Petroni K., Urzainqui A., Bevan M., Martin C., Jin H., Leyva A., Meissner R.C., Petroni K., Urzainqui A., Bevan M., Martin C., Leyva A., Meissner R.C., Petroni K., Urzainqui A., Bevan M., Martin C., Meissner R.C., Petroni K., Urzainqui A., Bevan M., Martin C., Petroni K., Urzainqui A., Bevan M., Martin C., Urzainqui A., Bevan M., Martin C., Bevan M., Martin C., Martin C., et al. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Misera S., Deng X.W., von Arnim A.G., Araki T., Komeda Y., Misera S., Deng X.W., Araki T., Komeda Y., Misera S., Deng X.W., Komeda Y., Misera S., Deng X.W., Misera S., Deng X.W., Deng X.W. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell. 1994;6:487–500. doi: 10.1105/tpc.6.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller S.G., Kunkel T., Chua N.H., Kunkel T., Chua N.H., Chua N.H. A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes & Dev. 2001;15:90–103. doi: 10.1101/gad.850101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.M., Frankhauser C., Chory J., Frankhauser C., Chory J., Chory J. Light: An indicator of time and place. Genes & Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K., Shimura Y., Okada K., Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes & Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H. Phytochrome photosensory signaling networks. Nat. Rev. Mol. Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F., Verweij W., Kroon A., Spelt C., Mol J., Koes R., Verweij W., Kroon A., Spelt C., Mol J., Koes R., Kroon A., Spelt C., Mol J., Koes R., Spelt C., Mol J., Koes R., Mol J., Koes R., Koes R. PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix–loop–helix transcription factors of the anthocyanin pathway. Plant Cell. 2006;18:1274–1291. doi: 10.1105/tpc.105.034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay N.A., Glover B.J., Glover B.J. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Riechmann L., Heard J., Martin G., Reuber L., Jiang C.-Z., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., Heard J., Martin G., Reuber L., Jiang C.-Z., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., Martin G., Reuber L., Jiang C.-Z., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., Reuber L., Jiang C.-Z., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., Jiang C.-Z., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., Pineda O., Ratcliffe O.J., Samaha R.R., Ratcliffe O.J., Samaha R.R., Samaha R.R., et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Saijo Y., Sullivan J.A., Wang H., Yang J., Shen Y., Rubio V., Ma L., Hoecker U., Deng X.W., Sullivan J.A., Wang H., Yang J., Shen Y., Rubio V., Ma L., Hoecker U., Deng X.W., Wang H., Yang J., Shen Y., Rubio V., Ma L., Hoecker U., Deng X.W., Yang J., Shen Y., Rubio V., Ma L., Hoecker U., Deng X.W., Shen Y., Rubio V., Ma L., Hoecker U., Deng X.W., Rubio V., Ma L., Hoecker U., Deng X.W., Ma L., Hoecker U., Deng X.W., Hoecker U., Deng X.W., Deng X.W. The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes & Dev. 2003;17:2642–2647. doi: 10.1101/gad.1122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H., Bolle C., Ballesteros M.L., Chua N.H., Ballesteros M.L., Chua N.H., Chua N.H. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature. 2003;26:995–999. doi: 10.1038/nature01696. [DOI] [PubMed] [Google Scholar]
- Soh M.S., Kim Y.M., Han S.J., Song P.S., Kim Y.M., Han S.J., Song P.S., Han S.J., Song P.S., Song P.S. REP1, a basic helix–loop–helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell. 2000;12:2061–2074. doi: 10.1105/tpc.12.11.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim A.G., Deng X.W., Deng X.W. Ring finger motif of Arabidopsis thaliana COP1 defines a new class of zinc-binding domain. J. Biol. Chem. 1993;268:19626–19631. [PubMed] [Google Scholar]
- Wang H., Deng X.W., Deng X.W. Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J. 2002;21:1339–1349. doi: 10.1093/emboj/21.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Deng X.W., Deng X.W. Dissecting the phytochrome A-dependent signaling network in higher plants. Trends Plant Sci. 2003;8:172–178. doi: 10.1016/S1360-1385(03)00049-9. [DOI] [PubMed] [Google Scholar]
- Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X.W., Wang H., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X.W., Wang H., Sullivan J., Hoecker U., Liu B., Xu L., Deng X.W., Wang H., Hoecker U., Liu B., Xu L., Deng X.W., Wang H., Liu B., Xu L., Deng X.W., Wang H., Xu L., Deng X.W., Wang H., Deng X.W., Wang H., Wang H. Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell. 2005;17:804–821. doi: 10.1105/tpc.104.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler M., Bolle C., Chua N.H., Bolle C., Chua N.H., Chua N.H. The phytochrome A specific signaling component PAT3 is a positive regulator of Arabidopsis photomorphogenesis. Plant Cell Physiol. 2001;42:1193–1200. doi: 10.1093/pcp/pce177. [DOI] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A., Gonzalez A., Zhao M., Payne C.T., Lloyd A., Zhao M., Payne C.T., Lloyd A., Payne C.T., Lloyd A., Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- Zhang X., Garreton V., Chua N.-H., Garreton V., Chua N.-H., Chua N.-H. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes & Dev. 2005;19:1532–1543. doi: 10.1101/gad.1318705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Hare P.D., Yang S.W., Zeidler M., Huang L.F., Chua N.-H., Hare P.D., Yang S.W., Zeidler M., Huang L.F., Chua N.-H., Yang S.W., Zeidler M., Huang L.F., Chua N.-H., Zeidler M., Huang L.F., Chua N.-H., Huang L.F., Chua N.-H., Chua N.-H. FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J. 2005;43:356–370. doi: 10.1111/j.1365-313X.2005.02453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F., Heim M.A., Weisshaar B., Uhrig J.F., Weisshaar B., Uhrig J.F., Uhrig J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]