Abstract
In Drosophila melanogaster, p53 (Dmp53) is an important mediator of longevity. Expression of dominant-negative (DN) forms of Dmp53 in adult neurons, but not in muscle or fat body cells, extends lifespan. The lifespan of calorie-restricted flies is not further extended by simultaneously expressing DN-Dmp53 in the nervous system, indicating that a decrease in Dmp53 activity may be a part of the CR lifespan-extending pathway in flies. In this report, we show that selective expression of DN-Dmp53 in only the 14 insulin-producing cells (IPCs) in the brain extends lifespan to the same extent as expression in all neurons and this lifespan extension is not additive with CR. DN-Dmp53-dependent lifespan extension is accompanied by reduction of Drosophila insulin-like peptide 2 (dILP2) mRNA levels and reduced insulin signaling (IIS) in the fat body, which suggests that Dmp53 may affect lifespan by modulating insulin signaling in the fly.
Keywords: Drosophila melanogaster, lifespan extension, p53
Calorie restriction (CR) has long been known to increase lifespan of mammals and in recent years been shown to increase life span of organisms as diverse as yeast, nematodes and flies (1). Another well studied pathway of lifespan extension is the insulin/insulin-like growth factor signaling pathway (IIS), down regulation of which has been shown to increase lifespan of nematodes, flies and mammals (2).
IIS is mediated through the insulin receptor (InR) and the InR substrate (CHICO in flies). Binding of insulin or, in Drosophila, insulin-like peptides (dILP) to the InR leads to activation of phosphoinositide-3-kinase (PI3K) and protein kinase B/Akt (3). This kinase cascade eventually phosphorylates the forkhead transcription factor FoxO, resulting in FoxOs retention in the cytoplasm via binding to 14-3-3 proteins (4, 5). The IIS signaling pathway is negatively regulated by the protein phosphatase PTEN. Down regulation of IIS results in increased FoxO dephosphorylation and its accumulation in the nucleus (6).
Inactivation of InR or CHICO (7, 8) and over-expression of dFoxO or dPTEN (9, 10) have been shown to increase Drosophila lifespan. Interestingly, the lifespan-extending effects of down-regulated IIS appear to be active in the fat body, Drosophila's major metabolic organ, because dFoxO or dPTEN can extend lifespan only when over-expressed in the fat body, but not in neurons (10). This is in contrast to what is seen with other regulators of Drosophila lifespan, like dSir2 (11) or dominant-negative (DN) Dmp53 (12), where lifespan is extended when over-expressed in neurons, but not in the fat body. The picture to date suggests that in flies lifespan is regulated by discreet, tissue-specific pathways. CR dependent lifespan extension appears to be mediated through alterations in the nervous system, whereas IIS-dependent lifespan extension seems to be mediated by changes in the fat body.
We showed (12) that expression of two different DN versions of Dmp53 in the Drosophila nervous system extends lifespan. This lifespan extension is not additive to the effects of CR. Because inhibition of caspase-dependent neuronal apoptosis does not appear to account for the lifespan-extending effects of DN-Dmp53 (12), we investigated other down stream mechanisms for DN-Dmp53-mediated lifespan regulation. We show that this mechanism is related to IIS, as IIS in the fat body is down-regulated in DN-Dmp53 long lived flies. Our results therefore suggest, that at least in flies, Dmp53 may be a modulator of IIS.
Results
Expression of DN-Dmp53 in Insulin Producing Neurons Is Sufficient to Extend Lifespan.
We showed (12) that expression of two structurally different DN versions of Drosophila p53 (Dmp53) in neurons leads to significant lifespan extension. However, it remains unclear how DN-Dmp53 expression extends lifespan. Inhibition of caspase-dependent apoptosis in the adult brain does not extend lifespan, and can therefore be ruled out as a mechanism for DN-Dmp53-dependent lifespan extension (12). Other effector pathways have thus to be explored that may explain the lifespan-extending effects of DN-Dmp53. Because the phenotypes of these two different long-lived flies are virtually identical, we concentrated on using the 259H mutant that carries a point mutation in the DNA-binding domain, rather than the Ct mutant, which consists of only the Dmp53 C-terminal domain.
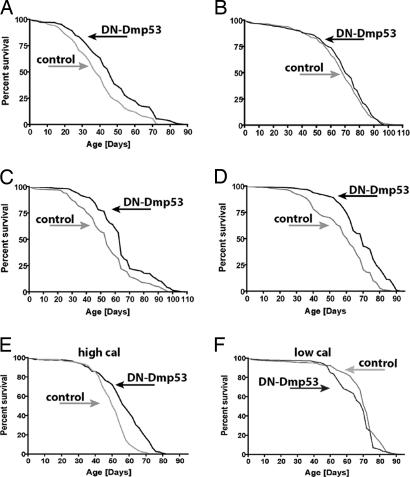
Neuronal expression of DN-Dmp53, using the widely expressing neuronal ELAV driver (henceforth referred to as “pan-neuronal”), extends lifespan by ≈20% (Fig. 1A), whereas expression with another widely expressing neuronal driver specific for cholinergic neurons leads to a smaller increase in lifespan (data not shown). Because expression of DN-Dmp53 in some other tissues (fat body or muscle) does not extend lifespan (12), we reasoned that identification of the smallest subset of neurons still capable of lifespan extension might yield clues as to the longevity mechanism engaged by Dmp53 reduction. We therefore made use of a variety of cell specific drivers that target subsets of cells in the nervous system to selectively express DN-Dmp53 in particular neurons. Expression of DN-Dmp53 specifically in dopaminergic or serotonergic neurons (Fig. 1B and data not shown) does not lead to lifespan extension. In addition, a variety of other brain specific drivers, including drivers expressing in the mushroom body, the corpora cardiaca and in glia, do not show lifespan extension [female survivorship data in Table 1, male data in supporting information (SI) Table 2]. The 14 insulin-producing cells (IPCs), the functional equivalent of the mammalian pancreatic β-cells, were the only cells of the brain in which expression of DN-Dmp53 was found to extend lifespan to a degree comparable to that observed with the pan-neuronal driver. Expression of DN-Dmp53 in the IPC, using two different GAL4 drivers derived from the promoter region of the Drosophila insulin-like peptide-2 (dILP2) gene, leads to lifespan extension of up to 19% (Fig. 1 C and D). In contrast, expression of WT-Dmp53 in these cells does not extend lifespan (Table 1). These data suggest that most, if not all, of the neuronal DN-Dmp53-dependent lifespan extension may be caused by reduction of Dmp53 activity in these 14 IPCs.
Fig. 1.
Expression of DN-Dmp53 in IPCs extends lifespan and is related to CR. (A–D) Survivorship curves of female DN-Dmp53 (black) or isogenic control flies (gray) crossed to various constitutively active neuron-specific drivers are shown. Pan-neuronal (A) (ELAV driver) expression of DN-Dmp53 extends mean lifespan up to 19%. (B) No lifespan extension is seen when a driver specific for dopaminergic neurons is used. When expressed in IPCs [dILP2 -Shen driver (C); dILP2 -Rulifson driver (D)] lifespan extension similar to ELAV driver is observed (18% and 19% extension of mean lifespan for Shen and Rulifson driver, respectively). (E and F) Survivorship curves of dILP2-DN-Dmp53 flies under CR conditions, using the Shen driver. Expression of DN-Dmp53 (black) in IPCs on high calorie food leads to a 14% increase in mean lifespan (E) over controls (gray). When flies are raised on lifespan-extending low calorie food (gray) (F), 37% extension of mean lifespan is observed. Simultaneous expression of DN-Dmp53 (black) does not lead to additional lifespan extension. All experiments shown were performed by using >200 flies each. Mean, median, and maximum lifespan data can be found in Table 1.
Table 1.
Effect of various proteins expressed in different expression patterns on female lifespan
| Driver | Mean LS vs. ctrl | Mean LS extension, % | Median LS vs. ctrl | Median LS extension, % | Max LS vs. ctrl | Max LS extension, % | Flies, no. |
χ2 | P | |
|---|---|---|---|---|---|---|---|---|---|---|
| Ctrl | Experimental | |||||||||
| ELAV (pan-neuronal) | 47/38 | 24 | 44/40 | 11 | 82/52 | 58 | 168 | 185 | 42.02 | <0.0001 |
| ELAV (pan-neuronal) | 46/39 | 19 | 46/38 | 21 | 72/70 | 3 | 210 | 281 | 22.86 | <0.0001 |
| Dopaminergic neurons | 68/66 | 2.5 | 71/68 | 4 | 94/94 | 0 | 255 | 258 | 1.005 | 0.3161 |
| Glia (repo driver) | 46/51 | −11 | 44/46 | −5 | 80/92 | −13 | 263 | 251 | 9.919 | 0.0016 |
| Brain driver c739 | 52/56 | −7 | 54/59 | −8 | 92/92 | 0 | 282 | 272 | 9.114 | 0.0025 |
| Brain driver c309 | 62/60 | 3 | 64/64 | 0 | 88/88 | 0 | 270 | 269 | 0.1567 | 0.6922 |
| Corpora cardiaca | 51/46 | −10 | 44/54 | −19 | 76/78 | −3 | 241 | 237 | 12.00 | 0.0005 |
| IPC (Rulifson driver) | 69/58 | 19 | 70/62 | 13 | 90/80 | 12.5 | 245 | 197 | 50.76 | <0.0001 |
| IPC (Rulifson driver) | 66/59 | 12 | 64/58 | 10 | 90/86 | 5 | 235 | 325 | 26.16 | <0.0001 |
| IPC (Shen driver) | 64/54 | 18 | 64/54 | 19 | 96/89 | 8 | 213 | 246 | 28.39 | <0.0001 |
| IPC (Shen driver) high cal | 57/50 | 14 | 58/52 | 12 | 75/66 | 14 | 245 | 236 | 71.36 | <0.0001 |
| IPC (Shen driver) low cal | 65/68 | −4 | 70/72 | −3 | 76/84 | −10 | 218 | 212 | 8.175 | 0.0042 |
| IPC (Rulifson driver) with WT Dmp53 | 54/51 | −6 | 58/56 | 4 | 76/74 | 3 | 239 | 234 | 0.09368 | 0.7595 |
The indicated drivers were crossed to UAS-Dmp53–259H flies. Offspring were collected and lifespan was determined on standard cornmeal food (except where indicated). Only females are shown. Mean, median, and maximum lifespan; log rank analysis; P value, percent change in mean, median, and maximum lifespan as compared with controls (w1118 for constitutive expression, without RU486 for GeneSwitch experiments); χ2; and P values derived from survivorship curves of each protein and driver are shown. Maximum lifespan was calculated as the median lifespan of the longest surviving 10% of the population. ctrl, control; cal, calorie; LS, lifespan.
Lifespan extension is usually greater in females than in males (Table 1 and SI Table 2), which likely reflects the differential effects of food nutrient conditions on male and female lifespan. In most of these studies, a cornmeal-based food was used, which is not a high calorie food. These food conditions may already be close to the maximum achievable nutrient dependent lifespan extension for males. When raised on higher-calorie food (1.5 N), males show a robust lifespan extension when DN-Dmp53 is expressed (SI Table 2). These data indicate a possible relationship between calorie restriction and DN-Dmp53 related lifespan extension.
Lifespan Extension by DN-Dmp53 Expression in IPCs Is Related to CR.
We showed (12) that lifespan extension by pan-neuronal DN-Dmp53 expression is not additive with CR lifespan extension, suggesting that a decrease in neuronal Dmp53 activity may be part of the CR lifespan-extending pathway. We therefore tested whether lifespan extension by DN-Dmp53 expression only in IPCs may also be related to CR. When control flies are subjected to CR conditions, robust lifespan extension of 37% is observed (Fig. 1E). However, additional expression of the DN-Dmp53 constructs in IPCs does not further extend the lifespan of these long-lived, calorie-restricted flies (Fig. 1F). This is consistent with our earlier observation with pan-neuronal DN-Dmp53 expression showing a lack of additivity to CR and suggests that IPC-specific DN-Dmp53-dependent lifespan extension may be related to CR.
Lifespan Extension of Flies Expressing DN-Dmp53 in Insulin-Producing Neurons Is Not Due to a Reduction in Female Fertility.
We reported (12) that pan-neuronally expressing DN-Dmp53 long-lived flies do not have a defect in female fertility. To verify that the dILP2-driven DN-Dmp53-dependent lifespan extension is also not due to a decrease in fertility, we measured daily egg production. As shown in SI Fig. 5, no difference in daily egg laying is observed over the first 17 days of life between control flies and DN-Dmp53 expressing flies. Therefore, as with pan-neuronal expression, longevity extension through expression of DN-Dmp53 in IPCs is not simply due to decreased fertility.
DN-Dmp53 Flies Demonstrate Slower Weight Gain.
Because expression of DN-Dmp53 in IPCs is sufficient to extend fly lifespan, DN-Dmp53-dependent lifespan extension might be connected to IIS. Flies with markedly reduced insulin signaling are visibly smaller and weigh less than their controls (7, 8, 13, 14). If lifespan extension through expression of DN-Dmp53 in the IPCs or pan-neuronally is indeed related to a change in insulin signaling, these long-lived flies might also be smaller or weigh less. As can be seen in SI Fig. 6, flies expressing DN-Dmp53 specifically in the IPCs, or pan-neuronally, weigh less and gain weight more slowly within the first 10 days of adult life, although they do not appear visibly smaller.
DN-Dmp53 Flies Have Lowered dILP2 mRNA Levels.
Ablation of the IPCs (13) leads to reduced mRNA levels of the Drosophila insulin-like peptides (dILP) 2, 3, and 5, whereas over-expression of the downstream insulin-signaling effector dFoxO in fat body cells (10) leads to reduction of dILP2 mRNA levels only. One consequence of the alteration of Dmp53 activity in the IPCs could therefore be a decrease in dILP levels. To test whether dILPs are decreased in the DN-Dmp53 long-lived flies, we performed quantitative PCR (QPCR) analysis of the three dILPs known to be secreted from the IPCs: dILP2, dILP3, and dILP5 (15). We found a specific reduction in the amount of the dILP2 transcript only but not of dILP3 or dILP5 transcripts. The mRNA levels of dILP2 in the heads of the long-lived flies are reduced by ≈60%, whereas mRNA levels of dILPs 3 and 5 are unchanged (SI Fig. 7).
Flies Expressing DN-Dmp53 in IPCs Have Lowered PI3K Activity.
Reduction of dILP2 mRNA levels does not directly demonstrate a change in systemic biological IIS activity. We therefore evaluated the functional state of the IIS system in long-lived DN-Dmp53 expressing flies and normal-lived controls by directly measuring the state of mediators of IIS in the fat body, one of the major sites for IIS activity in the fly.
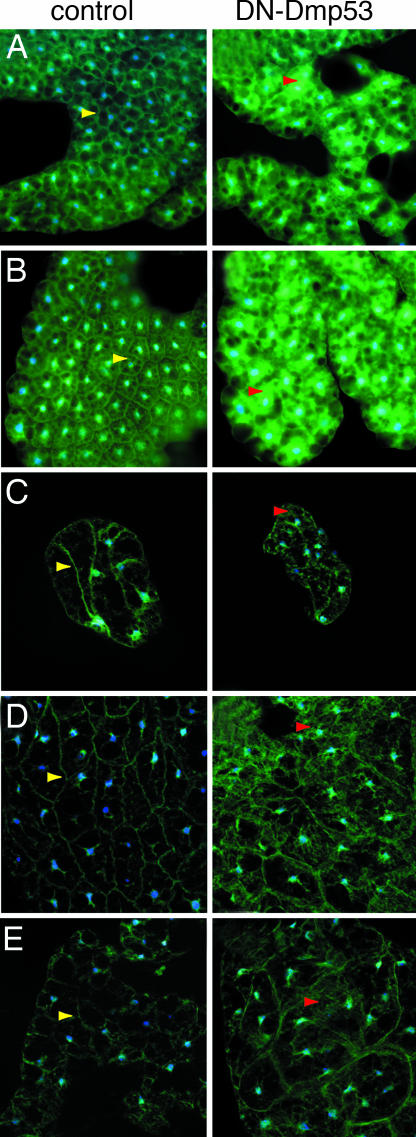
One consequence of IIS is the activation of PI3K and the generation of the second messenger phosphatidylinositol-3,4,5-trisphosphate (3). Phosphatidylinositol-3,4,5-trisphosphate in turn binds to proteins containing a pleckstrin homology (PH) domain, thereby sequestering them to the plasma membrane. Subcellular localization of a GFP fused to a PH domain can therefore be used to assess PI3K activity (16). DN-Dmp53 was expressed in the IPCs of a fly strain expressing GFP fused to the PH domain of Drosophila GRP1 under control of the tubulin promoter (tGPH) (16). If the lifespan-extending effect of expression of DN-Dmp53 is associated with lowered IIS activity, a redistribution of the tGPH reporter from the plasma membrane to the cytosol is expected to occur. In the larval fat body of control flies the tGPH reporter showed almost exclusive localization to the plasma membrane. In contrast, the fat body of larvae expressing DN-Dmp53 in the IPCs shows a marked reduction of plasma membrane bound tGPH and an increase in cytosolic staining (Fig. 2A). The same is seen with larvae expressing DN-Dmp53 pan-neuronally. Larvae induced to express DN-Dmp53 pan-neuronally show strong cytoplasmic tGPH localization, whereas in control larvae tGPH is localized almost exclusively at the plasma membrane (Fig. 2B).
Fig. 2.
PI3K activity is reduced in long-lived flies expressing DN-Dmp53 in the IPCs or pan-neuronally. The tGPH reporter construct was engineered into the control and the UAS-DN-Dmp53 strain. The resulting lines were then crossed to the dILP2 or ELAV-Switch driver. Fat bodies were isolated from the indicated tissues and analyzed for subcellular localization of the tGPH reporter (blue: DAPI; green: GFP). (A) Fat bodies were isolated from late third-instar of dILP2-DN-Dmp53 larvae. In control flies (Left), plasma membrane staining and no cytoplasic staining is observed. In DN-Dmp53-expressing flies (Right), plasma membrane staining is less pronounced and there is diffuse, cytoplasmic staining. (B) Similar results are observed when fat body cells are isolated from ELAV-Switch-DN-Dmp53 larvae. (C–E) When fat body is examined from heads (C) or abdomen (D) adult dILP2-DN-Dmp53 flies, or the abdomen (E) adult ELAV-Switch-DN-Dmp53 flies, control flies show plasma membrane staining with little cytoplasmic staining, whereas there is diffuse cytoplasmic staining in DN-Dmp53 expressing flies. Yellow arrows depict tGPH at the plasma membrane, red arrows cytosolic tGPH. Shown are representative images of at least five independent experiments using three to five flies per experiment.
We then investigated IIS in the fat body of the adult fly. Fat bodies from the head and the abdomen of adult flies were isolated and tGPH localization determined. In long-lived flies expressing DN-Dmp53 in IPCs, an increase in cytosolic tGPH is observed in the head (Fig. 2C) and abdominal (Fig. 2D) fat bodies, as compared with control flies. In flies expressing DN-Dmp53 pan-neuronally, tGPH is mostly cytoplasmic, whereas in control flies tGPH is localized at the plasma membrane (Fig. 2E) in the abdominal fat body. This suggests that DN-Dmp53 expression, specifically in the IPCs leads to reduction in the steady state of PI3K activity and IIS signaling in the fat body.
The Function of IPCs Is Not Negatively Impacted by DN-Dmp53 Expression.
Ablation of IPCs can extend lifespan, presumably through a reduction in IIS (13). Our results show reduction of dILP2 levels, but normal levels of dILP3 and dILP5 mRNA in the long-lived flies expressing DN-Dmp53 in the IPCs, suggesting that the IPCs are still present and functional. We further demonstrate that IPCs are not lost in long-lived DN-Dmp53 expressing flies by concomitant expression of GFP together with DN-Dmp53 in IPCs. Our results show no loss of GFP signal compared with control flies, suggesting that DN-Dmp53 expression does not lead to loss of the IPCs (SI Fig. 8). However, DN-Dmp53 expression might lead to a loss of cellular functionality short of cell death. We therefore tested the ability of the IPCs in the long-lived DN-Dmp53 expressing flies to respond to a functional (sugar) challenge.
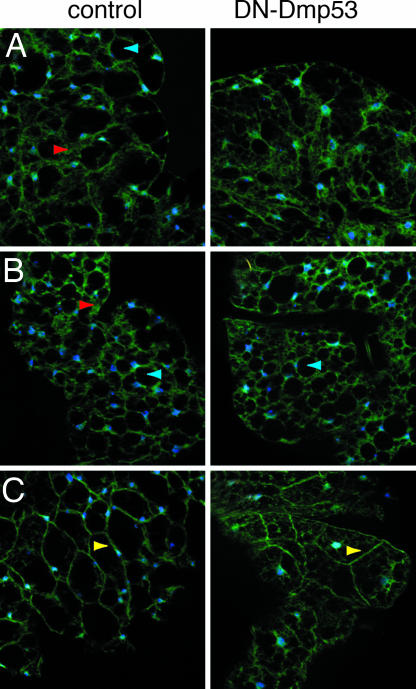
Brief (5-h) starvation treatment of flies expressing DN-Dmp53 in IPCs sensitizes the DN-Dmp53 expressing flies and leads to even stronger cytosolic tGPH staining (Fig. 3A). After 20 h of starvation, both control and DN-Dmp53 expressing flies show almost exclusive cytoplasmic tGPH localization (Fig. 3B). Prolonged starvation is also marked by the appearance of larger vacuoles inside the cells. To test whether IIS can be restored after starvation in these flies, adult flies were starved for 5 h and then refed for 3 h on 5% sucrose solution. This treatment leads to strong activation of IIS in the fat body, as demonstrated by the relocalization of tGPH back to the plasma membrane (Fig. 3C; compare with Fig. 3A). This relocalization is observed in both control and DN-Dmp53 expressing flies, albeit not quite as pronounced in the DN-Dmp53 expressing flies. These data suggest that when challenged by acute ingestion of a sugar load long-lived flies expressing DN-Dmp53 show a nearly normal functional response in their fat body cells.
Fig. 3.
The fat body cells of DN-Dmp53-expressing flies still respond to a starvation/sugar refeeding challenge. (A) Adult abdominal fat body from dILP2-DN-Dmp53 flies starved for 5 h shows a general decrease in membrane-associated tGPH in control (Left) and DN-Dmp53-expressing flies (Right) that is greater in DN-Dmp53 expressing flies. (B) After 20 h of starvation, membrane-associated tGPH is completely lost in both control (Left) and DN-Dmp53- expressing flies. (Right) (C) Three-hour feeding of a 5% sucrose solution after a 5-h period of water only starvation leads to almost exclusive plasma membrane staining in both control flies (Left) and flies expressing DN-Dmp53 in the IPCs. (Right) Yellow arrows depict tGPH bound at the plasma membrane, red arrows cytosolic tGPH, and blue arrows show starvation induced cytosolic vacuoles. Shown are representative images of at least five independent experiments using three to five flies each.
DN-Dmp53 Expressing Flies Show Nuclear Accumulation of dFoxO.
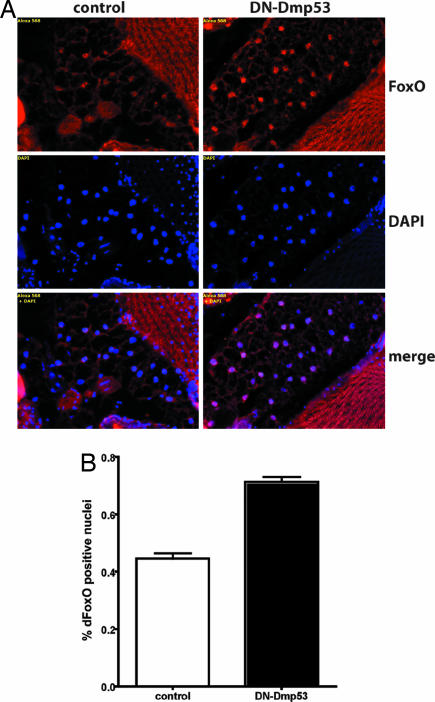
The downstream consequence of activated PI3K is the phosphorylation of the forkhead transcription factor FoxO (6). Phosphorylated FoxO is retarded in the cytosol, presumably through interaction with the scaffolding protein 14-3-3 (4, 5, 17). Inactivation of IIS thus leads to dephosphorylation and significant accumulation of dFoxO in the nucleus. The subcellular localization of dFoxO can consequently be used to assess the activity of IIS. DN-Dmp53 was expressed in the IPCs and dFoxO subcellular localization was measured in the pericerebral fat body of adult flies. Normal-lived control flies show very little nuclear localization of dFoxO in the pericerebral fat body, whereas long-lived flies expressing DN-Dmp53 in the IPCs show a marked increase in nuclear dFoxO staining (Fig. 4A). Quantification of the number of cells with nuclear dFoxO versus cells with only cytoplasmic dFoxO, shows that the percentage of fat body cells with nuclear dFoxO localization is increased by at least 30% in DN-Dmp53 expressing flies compared with controls (Fig. 4B).
Fig. 4.
dFoxO accumulates in the nucleus of fat body cells in long-lived dILP2 driven DN-Dmp53 expressing flies. (A) Heads of 10-day-old dILP2-DN-Dmp53 flies were cryosectioned and stained with an α-dFoxO antibody. In control flies (Left), only a small percentage of fat body cells show dFoxO staining. In contrast, in DN-Dmp53 expressing flies (Right) almost all fat body nuclei are positive for dFoxO antibody staining (red, α-dFoxO; blue, DAPI; purple, merge). (B) Quantification of dFoxO nuclear translocation in fat body cells. A total of ≈1,300 individual nuclei were scored for dFoxO staining. Compared with control flies, 30% more nuclei in DN-Dmp53 expressing flies also stain positive for dFoxO. A representative of two independent experiments using at least 10 animals each is shown. Error bars represent the SEM; P < 0.0001.
Taken together, these data suggest that reduction of Dmp53 activity leads to a decrease in IIS in a relevant target tissue, the fat body. These observations provide a possible mechanism by which DN-Dmp53 expression leads to lifespan extension.
Discussion
We recently demonstrated that the expression of DN-Dmp53 constructs in the adult nervous system extends lifespan. This lifespan extension is not additive to CR (12), suggesting that Dmp53 may be part of the CR pathway of lifespan extension. To understand more about the mechanisms by which neuronal reduction of Dmp53 extends lifespan, we examined the expression of DN-Dmp53 in subsets of neuronal cells. General expression of DN-Dmp53 in the nervous system, using two different, broadly expressing neuronal specific promoters, ELAV (pan-neuronal) or Cha (cholinergic neurons), both lead to significant lifespan extension. When expression of DN-Dmp53 was restricted to smaller subsets of neurons, including dopaminergic neurons, serotoninergic neurons, neurons of the mushroom body, or to IPCs, only expression in the 14 IPCs cells led to lifespan extension (Fig. 1 C and D). Thus, expression of DN-Dmp53 in only the 14 IPCs is sufficient to extend Drosophila lifespan. When combined with CR, IPC-specific DN-Dmp53 expression did not further extend lifespan (Fig. 1F), suggesting that reduction of Dmp53 activity in the IPCs may be a component of CR-dependent lifespan extension. Interestingly, IPCs are the functional equivalent of mammalian pancreatic β-cells, but reside in the fly brain. Our data suggests that Dmp53 controls insulin secretion from the IPCs. A consequence of inhibition of Dmp53 activity is reduced dILP2 mRNA and subsequent down regulation of IIS in the fat body, Drosophila's major insulin responsive metabolic organ. In this relevant target tissue, two different assays, tGPH and dFoxO subcellular localization, show that IIS is diminished. These data suggest that DN-Dmp53 expression might extend lifespan through modulation of IIS. Interestingly, p53 has been linked to insulin regulation in mammals. Mice over-expressing the shorter p44 isoform of p53, that is thought to resemble Dmp53 more than p53 itself, have elevated levels of IGF-1and IGF-1R (18). Furthermore, p53 null mice have 75% reduced levels of IGF-1 (19).
Loss or destruction of IPCs has been postulated to extend lifespan in flies (13, 14). A loss of the IPCs does not explain the lifespan extension seen with the long-lived flies expressing DN-Dmp53 specifically in IPCs as we are able visualize the presence of the IPCs, at least up until day 44, as measured by simultaneous expression of GFP (SI Fig. 8). We have also presented evidence suggesting that the IPCs remain functionally active. Of the three dILPs produced in the IPCs, only dILP2 mRNA levels are lowered, whereas dILP3 and dILP5 levels remain unchanged (SI Fig. 7). Furthermore, the fat body cells exhibit a nearly normal insulin response to a short period of starvation and sucrose re-feeding in the long-lived DN-Dmp53- expressing flies (Fig. 3). Thus, the lifespan extension induced by expression of DN-Dmp53 in IPCs is not due to loss or damage of the IPCs.
IIS related lifespan extension in the fly is thought to be mediated through alterations in the InR, CHICO (InR substrate), dPTEN and perhaps dFoxO. Interestingly, increasing levels of dPTEN or dFoxO in the fat body, but not the nervous system, extends lifespan (9, 10). Experiments on another nutrient sensing system, the TOR signaling pathway, also supports the importance of the fat body in lifespan extension as down regulation of the TOR pathway in the fat body, but not the nervous system leads, to lifespan extension (20). IIS can modulate TOR signaling via phosphorylation of Tsc2 by protein kinase B/Akt (21). Thus, down-regulated IIS may lead to suppression of TOR signaling.
The fly thus appears to have at least two different tissues that can influence longevity: one, neuronal in nature, of which Dmp53 is part; the other as part of a larger nutrient-sensing signaling network active in the fat body. Are these two separate systems or is there cross-talk between them? One possible means of connecting these two systems could be through the neuroendocrine system, where alterations in the nervous system, through control of hormonal secretion, could affect the physiology of the fat body. It is thus of considerable interest that the site of production and secretion for three of the major insulin-like peptides, dILP2, dILP3, and dILP5, is the 14 ICPs located in the brain of the fly. Our finding that expression of DN-Dmp53 specifically in these IPCs affects IIS in fat body cells and extends lifespan is intriguing. It remains to be determined whether this is one of the cellular sites linking the longevity determining effects of the brain with those associated with the fat body.
It is tempting to speculate even further: Our data suggest that the CR and DN-Dmp53 lifespan-extending pathways are related. It may therefore be that the lifespan-extending effects of CR are also accompanied by a down regulation of IIS. Dmp53 might thus be a point of convergence for these two lifespan-extending pathways, but further experiments are needed to clarify this point.
Materials and Methods
Fly Culture and Strains.
All flies were kept in a humidified, temperature-controlled incubator with 12 h on/off light cycle at 25°C in vials containing standard cornmeal medium (11). The ELAV-GeneSwitch line was from H. Keshishian (Yale University, New Haven, CT), and tGPH was from B. Edgar (Fred Hutchinson Cancer Research Center, Seattle, WA). One dILP2 driver line was from P. Shen (University of Georgia, Athens, GA) and the other dILP2 driver and the AKH driver were from E. Rulifson (Stanford University, Stanford, CA). The driver made by the Rulifson lab contains an 859-bp fragment of the dILP2 promoter (14), whereas the Shen driver contains a 2-kb fragment (22). Cha (23) and Ddc (24) drivers were from R. Reenan (Brown University). All other driver lines were from the Drosophila Stockcenter (Bloomington, IN). The following stocks were used: 458 (ELAV), 7362 (c739) (25), 6906 (c309) (25), 7415 (repo) (26). All Dmp53 lines (UAS-Dmp53 and UAS-Dmp53-259H) were originally generated in a w1118 background (27).
Lifespan Analysis.
Flies were collected under light anesthesia, randomly divided into treatment groups and housed at a density of 25 males and 25 females each per vial. At least 10 such vials were used per treatment as per (11). Flies were passed every other day, and the number of dead flies was recorded.
All lifespans were performed on regular cornmeal food [2% yeast/10% sucrose/5% cornmeal (all wt/vol)] without added live yeast, except where indicated. High- and low-calorie food contained 5% (0.5 N) and 15% (1.5 N), respectively, each yeast and sucrose (wt/vol), but no cornmeal (28). For induction with the GeneSwitch system, RU486 (Sigma–Aldrich, St. Louis, MO) was added directly to the food to a final concentration of 200 μM. The same concentration of diluent was added to control food. For expression with constitutive drivers, all UAS-lines were backcrossed to w1118 for 10 generations, and isogenic controls were generated from the last backcross.
Fertility and Weight Measurements.
Fertility was examined by using at least 15 individual mating pairs. Egg laying was stimulated by culture on higher-caloric food (1.5 N). Flies were passed daily and eggs counted over a 17-day period. For weight measurements, flies were collected within 8 h of eclosion and aged for the indicated days. Groups of 10 flies were weighed in three biological replicates each on an AB-S fine balance (Mettler-Toledo, Columbus, OH).
QPCR.
Total RNA was isolated from at least 75 heads of 10-day old females, using TRIzol (Invitrogen, Carlsbad, CA) and further purified by using the RNeasy kit (Qiagen, Valencia, CA). cDNA was generated with 0.5 μg of total RNA, using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) in a 10-μl reaction volume. iScript reaction (0.8 μl) was used as QPCR template. QPCR was performed on an ABI 7500 Real-Time PCR machine, using the ABI SYBR-Green PCR master mix following the manufacturers instructions. Each QPCR was performed by using four biological replicates in triplicate each. The following primers were used: dilp2-F, AGC AAG CCT TTG TCC TTC ATC TC; dilp2-R, ACA CCA TAC TCA GCA CCT CGT TG; dilp3-F, AGA GAA CTT TGG ACC CCG TGA A; dilp3-R, TGA ACC GAA CTA TCA CTC AAC AGT CT; dilp5-F, GAG GCA CCT TGG GCC TAT TC; dilp5-R, CAT GTG GTG AGA TTC GGA GCA A; GAPDH-F, GAC GAA ATC AAG GCT AAG GTC G; GAPDH-R, AAT GGG TGT CGC TGA AGA AGT C; Tubulin-F, ACA TCC CGC CCC GTG GTC; Tubulin-R, AGA AAG CCT TGC GCC TGA ACA TAG.
Fluorescence Microscopy.
For dFoxO subcellular localization, 10-day old adult fly heads were fixed in fresh 4% paraformaldehyde, embedded into Tissue Freezing Medium (TFM, Triangle Biomedical), frozen and cryosectioned to 10 μM. Cryosections were washed to remove TFM and stained with α-dFoxO antibody [1:500, rabbit antiserum, kind gift from O. Puig (University of Helsinki, Helsinki, Finland)], followed by Alex 568-conjugated secondary antibody (1:2,000; Molecular Probes, Eugene, OR). Nuclei were costained with DAPI. All images were taken by using a Zeiss (Thornwood, NY) Axiovision Z1 fluorescent microscope. For quantification of dFoxO nuclear localization, individual nuclei were scored for fluorescent activity and compared with cytoplasmic dFoxO staining, using the Axiovision software suite, Version 4.5. Only nuclei that showed 1.5-fold greater than cytoplasmic dFoxO background staining were considered dFoxO positive.
For tGPH subcellular localization, fat body tissue was dissected into PBS from third-instar larvae and stained with DAPI (Sigma). Adult fat body was dissected from the head or the abdomen, respectively, of 10-day-old adults. Adult fat body was visualized by using ApoTome optics (Zeiss).
Statistics.
Statistical analyses, including log rank tests, were performed by using the Prism suite of biostatistical software (GraphPad, San Diego, CA). Maximum lifespan was calculated as the median lifespan of the longest surviving 10% of the population.
Supplementary Material
Acknowledgments
We thank M. Tatar (Brown University), R. Reenan, H. Keshishian, B. Edgar, P. Shen, and E. Rulifson for the kind gift of fly stocks; O. Puig for the dFoxO antibody; and Will Lightfoot and Zen Canady for technical assistance. This work was supported by National Institute on Aging Grants AG16667, AG24353, and AG25277; the Donaghue Foundation; and the Ellison Medical Foundation (to S.L.H.). S.L.H. is an Ellison Medical Research Foundation Senior Investigator. This research was conducted while J.H.B. was an Ellison Medical Foundation/AFAR Senior Postdoctoral Fellow.
Abbreviations
- CR
calorie restriction
- DN
dominant-negative
- dILP2
Drosophila insulin-like peptide 2
- IIS
Insulin/insulin-like growth factor signaling
- InR
insulin receptor
- IPC
insulin-producing cells
- QPCR
quantitative PCR
- PI3K
phosphoinositide-3-kinase
- tGPH
tubulin promoter-driven GFP fused to pleckstrin homology domain.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706121104/DC1.
References
- 1.Masoro EJ. Sci Aging Knowledge Environ. 2003:RE2. doi: 10.1126/sageke.2003.8.re2. [DOI] [PubMed] [Google Scholar]
- 2.Tatar M, Bartke A, Antebi A. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 3.Goberdhan DC, Wilson C. Differentiation. 2003;71:375–397. doi: 10.1046/j.1432-0436.2003.7107001.x. [DOI] [PubMed] [Google Scholar]
- 4.Obsil T, Ghirlando R, Anderson DE, Hickman AB, Dyda F. Biochemistry. 2003;42:15264–15272. doi: 10.1021/bi0352724. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Oh SW, Deplancke B, Luo J, Walhout AJ, Tissenbaum HA. Mech Ageing Dev. 2006;127:741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Van Der Heide LP, Hoekman MF, Smidt MP. Biochem J. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 8.Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 9.Giannakou ME, Goss M, Junger MA, Hafen E, Leevers SJ, Partridge L. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- 10.Hwangbo DS, Gersham B, Tu MP, Palmer M, Tatar M. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 11.Rogina B, Helfand SL. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauer JH, Poon PC, Glatt-Deeley H, Abrams JM, Helfand SL. Curr Biol. 2005;15:2063–2068. doi: 10.1016/j.cub.2005.10.051. [DOI] [PubMed] [Google Scholar]
- 13.Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, Partridge L. Proc Natl Acad Sci USA. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rulifson EJ, Kim SK, Nusse R. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 15.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- 16.Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Dev Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- 17.Obsilova V, Vecer J, Herman P, Pabianova A, Sulc M, Teisinger J, Boura E, Obsil T. Biochemistry. 2005;44:11608–11617. doi: 10.1021/bi050618r. [DOI] [PubMed] [Google Scholar]
- 18.Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, Sutherland A, Thorner M, Scrable H. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hursting SD, Lavigne JA, Berrigan D, Donehower LA, Davis BJ, Phang JM, Barrett JC, Perkins SN. J Nutr. 2004;134:2482S–2486S. doi: 10.1093/jn/134.9.2482S. [DOI] [PubMed] [Google Scholar]
- 20.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hafen E. Swiss Med Wkly. 2004;134:711–719. doi: 10.4414/smw.2004.09885. [DOI] [PubMed] [Google Scholar]
- 22.Wu Q, Zhao Z, Shen P. Nat Neurosci. 2005;8:1350–1355. doi: 10.1038/nn1540. [DOI] [PubMed] [Google Scholar]
- 23.Kitamoto T, Ikeda K, Salvaterra PM. J Neurosci. 1992;12:1628–1639. doi: 10.1523/JNEUROSCI.12-05-01628.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Auluck PK, Bonini NM. Nat Med. 2002;8:1185–1186. doi: 10.1038/nm1102-1185. [DOI] [PubMed] [Google Scholar]
- 25.Rodan AR, Kiger JA, Jr, Heberlein U. J Neurosci. 2002;22:9490–501. doi: 10.1523/JNEUROSCI.22-21-09490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sepp KJ, Schulte J, Auld VJ. Dev Biol. 2001;238:47–63. doi: 10.1006/dbio.2001.0411. [DOI] [PubMed] [Google Scholar]
- 27.Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 28.Chapman T, Partridge L. Proc Biol Sci. 1996;263:755–759. doi: 10.1098/rspb.1996.0113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.