Abstract
The perinucleolar compartment (PNC) is a nuclear substructure present in transformed cells. The PNC is defined by high concentrations of certain RNA binding proteins and a subset of small RNAs transcribed by RNA polymerase III (pol III), including the signal recognition particle RNA and an Alu RNA as reported here. To determine if the PNC is dependent on pol III transcription, HeLa cells were microinjected with the selective pol III inhibitor, Tagetin. This resulted in disassembly of the PNC, whereas inhibition of pol I by cycloheximide or pol II by α-amanitin did not significantly affect the PNC. However, overexpression of one of the PNC-associated RNAs from a pol II promoter followed by injection of Tagetin blocked the Tagetin-induced PNC disassembly, demonstrating that it is the RNA rather than pol III activity that is important for the PNC integrity. To elucidate the role of the PNC-associated protein PTB, its synthesis was inhibited by siRNA. This resulted in a reduction of the number of PNC-containing cells and the PNC size. Together, these findings suggest, as a working model, that PNCs may be involved in the metabolism of specific pol III transcripts in the transformed state and that PTB is one of the key elements mediating this process.
INTRODUCTION
The perinucleolar compartment (PNC) was first described during characterization of the polypyrimidine tract binding (PTB) protein (Ghetti et al., 1992). The PTB protein binds to the 3′ end of pre-mRNA introns (Garcia-Blanco et al., 1989; Wang and Pederson, 1990) and is localized throughout the nucleoplasm and cytoplasm in addition to being concentrated in the PNC (Ghetti et al., 1992). Subsequently, several small RNAs transcribed by RNA polymerase III (RNase MRP RNA, RNase P RNA, and some of the Ro autoantigen-associated hY RNAs; Matera et al., 1995; Lee et al., 1996), and other RNA binding proteins, CUG-BP/hNab50 (Timchenko et al., 1996; Huang et al., 1998), and Raver 1 (Huttelmaier et al., 2001) have also been identified in the PNC. However, not all pol III transcripts are detected in the PNC. In situ hybridization experiments failed to show other pol III transcripts, including tRNA, U6, or 5S rRNA, in the PNC (Matera et al., 1995, and C. Wang and S. Huang, unpublished data). The PNC is an irregularly shaped structure ranging from 0.25 to 1 μm in diameter. Electron microscopic analyses show that the PNC is structurally distinct from the nucleolus and is composed of multiple electron-dense strands ∼80–180 nm in diameter (Huang et al., 1998; Huang, 2000).
The most engaging feature of PNCs is that they are present predominantly in transformed cells and are rarely found in normal cells (Huang et al., 1997). Analyses of breast cancer tissue have indicated that PNC prevalence (the percentage of cells that contains at least one PNC) positively correlates with the progression of the disease (R.V. Kamath, A.D. Thor, C. Wang, S.M. Edgerton, J. Wang, E.L. Wiley, B. Jovanovic, Q. Wu, R. Nayar, and S. Huang, unpublished data). PNCs persist through interphase, disassemble at mitosis, and reform at early G1. They are dynamic structures through which the PTB protein shuttles in and out rapidly (Huang et al., 1997). The localization of the PTB protein in the PNC is dependent on its RNA binding activity, and the structural integrity of the PNC is related to transcription (Huang et al., 1998). Furthermore, the PNC incorporates newly synthesized RNA after a brief (5 min) pulse label (Huang et al., 1998). These findings suggest a fundamental relationship between the PNC and RNA metabolism. Because the genes for some of the known PNC-associated RNAs are not physically near PNCs in the nucleus (Matera et al., 1995; C. Wang and S. Huang unpublished data), the PNC is unlikely to be the transcription site of these RNAs.
In the present investigation we have further explored the relationship between the PNC, the PTB protein, and nuclear RNA metabolism. We find that two additional small RNAs transcribed by RNA polymerase III, signal recognition particle RNA, and an Alu RNA are present in the PNC. In addition, the presence of the PNC is dependent on the continuous production of PNC-associated transcripts, not the pol III activity itself. Furthermore, inhibition of PTB protein expression by siRNA resulted in a decrease in the number and size of PNCs. Taken together, our results indicate that active on-going syntheses of pol III transcripts and the PTB protein are essential for the integrity of the PNC.
MATERIALS AND METHODS
Cell Culture
HeLa cells were grown on 22 × 22-mm glass coverslips in 35-mm Petri dishes in Dulbecco modified Eagle's minimum essential medium (DMEM) containing 10% fetal bovine serum (FBS). Inhibition of RNA polymerase II transcription was achieved by addition of α-amanitin (75 μg/ml for 5 h; Kedinger et al., 1970; Lindell et al., 1970) or 5,6-dichloro-1-d-ribofuranosylbenzimidazole (DRB; 25 μg/ml for 3 h; Sehgal et al., 1976; Fraser et al., 1978). Inhibition of RNA polymerase II and III transcription was achieved by addition of α-amanitin (300 μg/ml for 5 h) to the culture medium (Kedinger et al., 1970; Lindell et al., 1970).
Tagetin Microinjection
HeLa cells were plated on coverslips 24 h before injection. A 0.6 mM solution of Tagetin (Epicentre Technology, Madison, WI) containing fixable, Texas Red– conjugated 70 kDa dextran (Molecular Probes, Eugene, OR) at 1 μg/ml was loaded into microinjection needles and cells were injected using an Eppendorf Microinjector (Brinkman Instruments, Westbury, NY) system paired with a Zeiss Axiovert microscope (Thornwood, NY). Following microinjection, the coverslips were gently rinsed three times with DMEM supplemented with 10% FBS and then cultured at 37°C for varying periods of time before immunostaining or in situ hybridization.
In Situ Hybridization
In situ hybridization was carried out using rhodamine-labeled peptide nucleic acid probes complementary to human SRP RNA, as detailed elsewhere (Politz et al., 2002). The biotinylated oligonucleotide probe to an Alu RNA was 5′-bio/CAGGCGCCCGCCACCACGCCCGGCTAATTTTTTGTATTTTTAGTAGAG. Cells grown on glass coverslips were fixed in 4% formaldehyde in phosphate-buffered saline (PBS) and then stored in 100% ethanol at 4°C over-night. Before in situ hybridization, cells were permeabilized in 100% acetone at -20°C for 10 min. In situ hybridization was carried out in 40% (vol/vol) formamide, 40% dextran sulfate, and 4× SSC at 37°C for 3 h (1× SSC is 0.15 M NaCl, 0.015 M sodium citrate). Cells were washed with 40% formamide, 2× SSC and then 20% formamide, 1× SSC, for 30 min each at 37°C, followed by three 15-min washes in 1× SSC at room temperature. For sequential in situ hybridization and immunostaining, cells were refixed in 2% formaldehyde in PBS for 5 min after in situ hybridization and were then incubated with antibodies as detailed below in Immunostaining.
In situ hybridization to RNase MRP RNA using a biotin-labeled 2′-O-Me oligonucleotide probe (Matera et al., 1995) was carried out after cells were fixed as described above and permeabilized with 0.5% Triton X-100 (vol/vol) for 5 min. The hybridization buffer contained 4× SSC, 10% dextran sulfate, 1 μg/μl tRNA, and the oligo probe at 1 pmol/μl. After 1 h of hybridization at 37°C, the cells were washed with 4× SSC containing 0.1% (vol/vol) Tween-20 three times, each for 10 min at room temperature. The cells were then incubated for 20 min in 4× SSC containing 10% bovine serum albumin as a blocking step. The biotin-labeled probe was then detected by incubating the cells for 1 h at room temperature with Texas Red–labeled avidin.
Immunostaining
Cells were fixed with 2% or 4% formaldehyde in PBS for 10 min followed by 5 min permeabilization with 0.5% (vol/vol) Triton X-100 at room temperature. Primary antibody was applied for 1 h, and cells were washed with PBS three times before incubations with secondary antibodies that were conjugated with FITC, Texas Red, or Amca (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Signal was visualized using a Nikon Eclipse E800 microscope equipped with a SenSys cooled CCD camera (Photometrics, Tucson, AZ). Images were captured using Metamorph image acquisition software (Universal Imaging, Downingtown, PA). The primary antibodies used include those against PTB (Huang et al., 1997), CUG-BP (Timchenko et al., 1996), fibrillarin (Sigma, St. Louis, MO), Sam68 (Chen et al., 1999), SC35 (Fu and Maniatis, 1990), and UBF (kindly provided by Edward Chan and Eng Tan, Scripps Research Institute, La Jolla, CA).
Transfection
Plasmids encoding either a GFP fusion of the human PTB protein (Huang et al., 1997), GFP itself or RNase MRP RNA from a CMV promoter (constructed by inserting RNase MRP RNA coding DNA into the pcDNA vector (Invitrogen, Carlsbad, CA), were transfected into HeLa cells by electroporation. Cells in 100-mm dishes were collected by trypsinization, resuspended in DMEM containing 10% FBS, and mixed with 4 μg of the desired plasmid DNA and 16 μg of sheared salmon sperm DNA. The mixture was electroporated using a Bio-Rad GENE PULSER II electroporator (Bio-Rad, Hercules, CA) at 250 V and 950 μF. The cells were subsequently seeded onto glass coverslips in 35-mm Petri dishes and were cultured for 24–48 h.
RNA Interference
Twenty-one nucleotide-pair double-stranded RNA was chemically synthesized, deprotected, and purified by Dharmacon Research Inc. (Lafayette, CO). One strand of the dsRNA was homologous to the PTB mRNA sequence: 5′-UGACAAGAGCCGUGACUAC(dTdT)-3′. The control dsRNA was a 21 nucleotide-pair RNA with a sequence homologous to a region of the mRNA of the pol I specific transcription factor UBF: 5′-CAGGAGUUCGAGCGAAACC(dTdT)-3′. This dsRNA of UBF does not behave as a siRNA in that it has no effect on the expression of UBF or on overall cell nuclear morphology up to 72 h after transfection (C. Wang and S. Huang, unpublished data). Transfection of siRNA duplexes was performed using Oligofectamine (Invitrogen, Carlsbad, CA). HeLa cells were plated in 24-well plates at a density that reached 30–50% confluence in 24 h. Two microliters of Oligofectamine and 3 μl of siRNA containing 60 pmol oligos were mixed and added to each well (containing 200 μl of medium). Cells were then incubated in DMEM without serum or antibiotics at 37°C for 5 h followed by replacement with DMEM containing 10% FBS and antibiotics. Seventy-two hours after transfection, cells were fixed and immunostained with various antibodies.
RESULTS
SRP and Alu RNAs Are Localized in the PNC
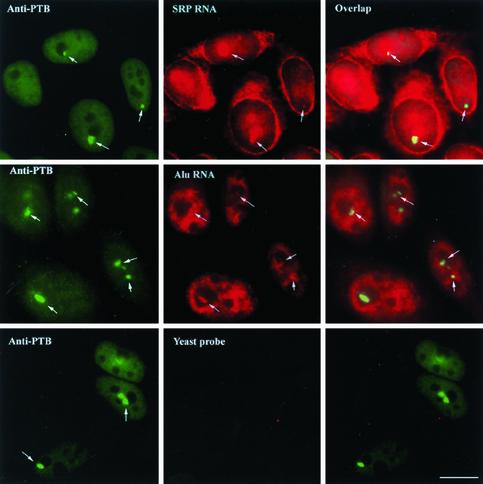
Previous studies showed that all PNC-associated RNAs identified so far are transcribed by RNA polymerase III, including RNase P RNA, RNase MRP RNA, and several of the Ro autoantigen-associated hY RNAs (Matera et al., 1995; Lee et al., 1996). We were interested in investigating whether other pol III RNA transcripts are also present in the PNC. The localization of RNAs in the PNC was examined using in situ hybridization followed by immunostaining cells for the PTB protein. As shown in Figure 1, top panels, SRP RNA was detected in PNCs (perinucleolar high concentrations of PTB protein [arrows]) as indicated by the yellow merged signals (arrows) of both SRP RNA and PTB protein. The specificity of the SRP RNA in situ hybridization was confirmed by the lack of detectable signal when the probe was complementary to a region of S. pombe SRP RNA that diverges from the comparable human SRP RNA sequence (Figure 1, bottom row, center panel). In addition to being present in the PNC, SRP RNA was also localized throughout the nucleoli, as well as in the cytoplasm, in confirmation of previous observations (Politz et al., 2000, 2002). A similar PNC colocalization of the PTB protein and an Alu RNA species was also observed (Figure 1, second row, right panel).
Figure 1.
SRP RNA and Alu RNA are localized with the PTB protein in the PNC (arrows) in addition to their other cellular distributions. The PNC is marked by immunolabeling using SH54, a mAb against the PTB protein. The right panels are the computational merger of the RNA and PTB localizations. Bar, 10 μm.
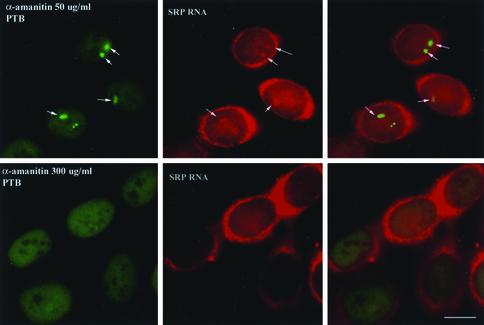
To determine whether the PNC localization of these RNAs is related to transcriptional activity, cells were treated with α-amanitin at both a concentration (50 μg/ml) that selectively inhibits RNA polymerase II activity and a concentration (300 μg/ml) that inhibits both pol II and pol III transcription (Kedinger et al., 1970; Lindell et al., 1970). When pol II was inhibited, the structure of the PNC was slightly altered, becoming somewhat more extended (Figure 2, top left panel, arrows), but SRP RNA was still detected in these slightly altered PNCs (Figure 2, top right panel). However, when cells were treated with α-amanitin at the concentration that inhibits both pol II and pol III, PNCs were no longer detectable in most cells (Figure 2, bottom left panel) and SRP RNA was not observed to be colocalized with the PTB protein at any distinct nuclear sites (Figure 2, bottom right panel). These results indicate that the structure of the PNC depends on the activity of RNA polymerase III. However, because the use of α-amanitin does not conclusively discriminate between the activities of pol II vs. pol III, we undertook a more direct approach to analyze the relationship between pol III transcription and the maintenance of the PNC.
Figure 2.
SRP RNA localization to the PNCs is sensitive to treatment with a concentration of α-amanitin (bottom panels) that inhibits both pol II and pol III transcription. However, SRP RNA remains detectable in PNCs when cells are treated with a lower concentration of α-amanitin that selectively inhibits pol II transcription (top panels, arrows). Bar, 10 μm.
PNC Structural Integrity Is Dependent on pol III Transcription
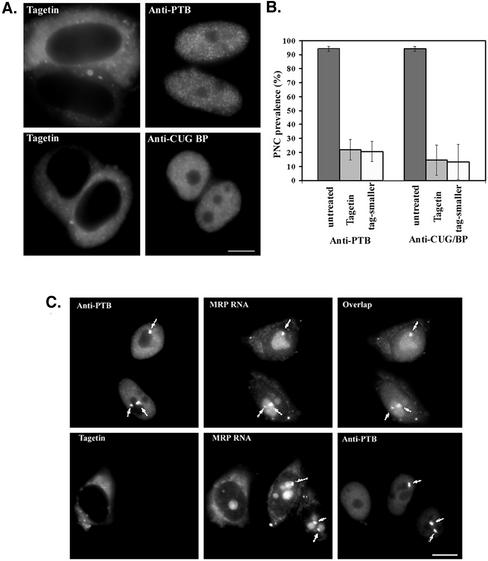
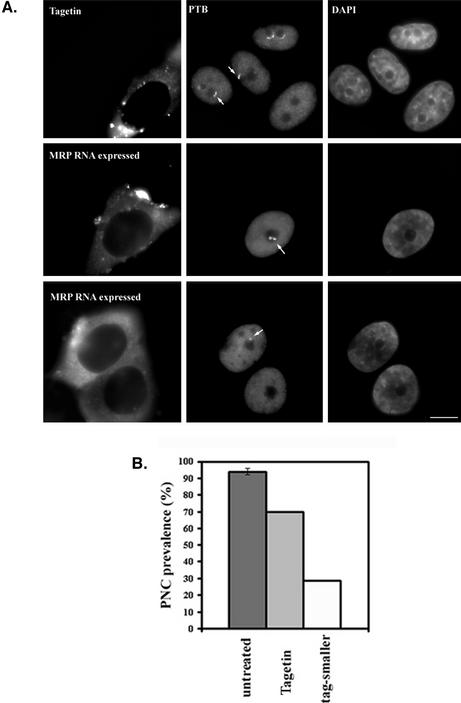
Tagetin is a bacterial toxin that has been shown to preferentially inhibit eukaryotic RNA polymerase III elongation without significant effects on the transcriptional activities of other polymerases (Steinberg et al., 1990; Steinberg and Burgess, 1992). However, cultured cells do not take up Tagetin directly from the medium. We therefore microinjected 600 μM Tagetin into HeLa cells and evaluated its effect on PNCs. Tagetin was injected into the cytoplasm in most cases but into nuclei in some experiments. Microinjected cells were identified on the coverslips by the presence of coinjected Texas Red– conjugated dextran (Figure 3A, left panels). Four hours after injection, distinct perinucleolar structures were no longer detectable in the majority of injected cells based on immunostaining of PTB and a second PNC-associated protein, CUG-BP (Figure 3A, right panels). More than 400 injected cells were counted for PNC prevalence, and the results showed a significant reduction of PNC prevalence in Tagetin injected cells, from 95 to 15–20% (Figure 3B, dark vs. light gray bars). Most of the remaining PNCs were reduced in size to a dot-like appearance (white bars in Figure 3B).
Figure 3.
Microinjection of Tagetin, a selective inhibitor for pol III polymerase, into HeLa cell cytoplasm (as marked by Texas Red–labeled, 70 kDa dextran, A, left panels) significantly reduces PNC prevalence as evaluated by immunolabeling with either anti-PTB or anti-CUG-BP antibodies (A, right panels, and B). The light gray bars in B show the PNC prevalence after Tagetin injection, and the white bars in B show the percentage of PNCs with significantly reduced size. RNase MRP RNA, which normally shows a high concentration in PNCs (top panels in C, arrows), is no longer detected as highly concentrated dots at the nucleolar periphery (bottom middle panel, the left cell, in C) along with the disappearance of PNCs marked by immunolabeling of PTB (bottom right panel in C). Bar, 10 μm.
To address whether PNC-associated RNA components behave similarly as the RNA binding proteins during the inhibition of pol III transcription, we examined the localization of a known PNC-associated RNA, RNase MRP RNA, after Tagetin microinjection. RNase MRP RNA, as previously shown, is prominently concentrated in PNCs and is less intensely detected in nucleoli (Figure 3C, top panels; Matera et al., 1995). After Tagetin injection, the prominent perinucleolar MRP RNA signals that are coincident with PTB were no longer detected, as shown in the bottom panels of Figure 3C). The loss of the distinct perinucleolar signals for the PTB protein, CUG-BP, and RNase MRP RNA were observed as early as 2 h after Tagetin microinjection (unpublished data), indicating a relative immediacy of the requirement for on-going pol III transcription for their presence in the PNC and for the structural integrity of the PNC. These findings together demonstrate that PNCs, as defined by the perinucleolar concentration of the PTB and CUG-BP proteins and small pol III RNAs, are dependent on on-going pol III transcription.
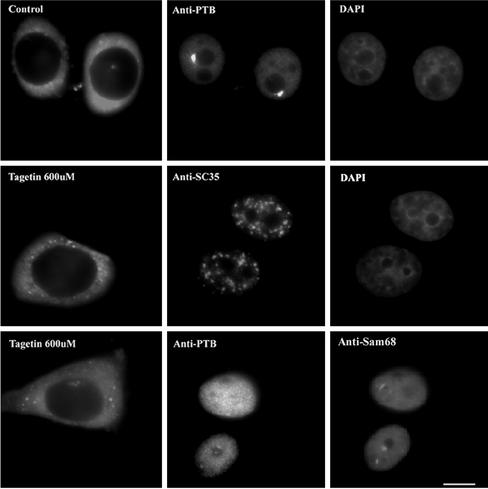
To control for the possibility that PNC disassembly might have been due to the injection process, cells were injected with Texas Red– conjugated dextran alone. No change in PNC size, shape, or prevalence was observed 2– 4 h after injection (Figure 4, upper middle panel). To evaluate whether Tagetin injection might, despite its known selectivity for pol III, also significantly influence overall intranuclear structural organization, we examined the distribution of SC35, an essential pre-mRNA splicing factor, whose “speckled” distribution in the nucleoplasm is highly sensitive to pol II transcription inhibition (Spector, 1993). We found that injection of Tagetin did not significantly affect the distribution of SC35 (Figure 4), suggesting that Tagetin does not have a global effect on pol II transcription or overall nuclear structure in these experiments.
Figure 4.
Injection of Texas-red dextran alone has no effects on the PNC (top panels). Tagetin does not appear to significantly change the overall nuclear organization. The distributions of SC-35 (C, middle panels) and Sam68 bodies (C, bottom panels, arrowheads) remain similar compared with those in noninjected cells. Bar, 10 μm.
In addition to PNCs, a recently described population of intranuclear structures termed Sam68 bodies has also been shown to have a primarily perinucleolar location (Chen et al., 1999; Huang, 2000). Sam68 bodies are also enriched with RNA and RNA binding proteins and are eliminated by treatment of actinomycin D (Chen et al., 1999; Huang, 2000). We were interested in whether selective inhibition of pol III transcription affects Sam68 bodies. Cells injected with Tagetin were simultaneously immunolabeled for PTB and for Sam68, a protein that is enriched in the Sam68 bodies. In contrast to PNCs, Tagetin injection did not detectably change Sam68 bodies (Figure 4, bottom row, right panel, arrowheads), indicating that Sam68 bodies, unlike PNCs, are not dependent on pol III transcription and further confirming, as the SC35 results, that Tagetin is not inducing overall alterations of nuclear organization beyond its specific effect on the pol III transcription and the PNCs.
PNCs Depend on the Specific pol III Transcripts Irrespective of the Polymerase That Generates Them
We next asked whether the activity of pol III itself or the specific newly synthesized RNAs are important for maintaining the structural integrity of PNCs. To address this question, we constructed an expression vector through which RNase MRP RNA was transcribed by pol II polymerase under the pol II-specific cytomegalovirus (CMV) promoter. We asked whether overexpression of this PNC-associated RNA could prevent the Tagetin-induced PNC disassembly. The CMV-RNase MRP RNA plasmid and a GFP plasmid were cotransfected into HeLa cells at a 10:1 ratio, so that the great majority of GFP-expressing cells would also be expressing MRP RNA. Tagetin was injected into GFP-expressing cells. As can be seen in Figure 5, PNC prevalence was reduced by only ∼25% in MRP RNA-expressing cells (Figure 5B), compared with the 80–85% reduction observed in untransfected cells (Figure 3B). Although a proportion of the cells had smaller and dot-like PNCs (Figure 5, A, bottom panels and B), the overexpression of MRP RNA prevented the Tagetin-induced PNC-disassembly in a large proportion of cells (Figure 5, A and B). This finding strongly supports the notion that the structural integrity of PNCs depends on a continuous supply of certain pol III transcripts, irrespective of the actual polymerase that synthesizes them.
Figure 5.
When cells were injected with Tagetin (injection dye as shown in left panels in A), a loss of perinucleolar labeling of the PTB protein is observed (top panels in A). Overexpression of RNase MRP RNA directed by a pol II promoter partially blocks the loss of PNCs in Tagetin-injected cells. The middle panel in the second row (A) shows that a cell expressing MRP RNA maintains a large and irregularly shaped PNC, and the bottom panels show one of the injected cells with a smaller and dot-like PNC (arrow). (B) Quantitative analyses show a smaller percentage of cells losses PNCs compared with those without expression of MRP RNA from a pol II promoter (Figure 3). Bar, 10 μm.
The PTB Protein Is Essential for Maintaining the Structural Integrity of PNCs
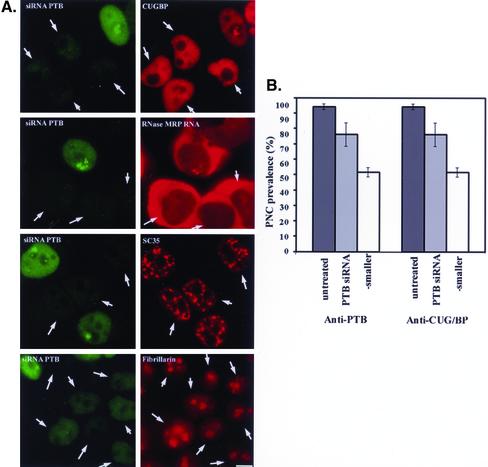
Although the PTB protein was the original defining feature of the PNC, its function in the PNC remains unknown. To analyze the role of PTB in the maintenance of PNCs, we performed RNA interference (RNAi) experiments targeting the mRNA of the PTB protein. A 21 nucleotide-pair, double-stranded RNA of appropriate sequence (see MATERIALS AND METHODS) was introduced into HeLa cells, and the levels of PTB protein expression and the presence of PNCs were determined after various periods of time. When evaluated by immunostaining, cells that were transfected with the siRNA showed a significant reduction in the nuclear level of PTB protein compared with cells that were not transfected (Figure 6A, arrows indicate the transfected cells). The decreases in the nuclear level of PTB protein could be detected 48 h after transfection, and the maximum inhibition was reached at around 72 h. Although immunoblots of whole cell extracts showed up to 95% inhibition of PTB expression in siRNA-transfected cells (unpublished data), a complete loss of PTB labeling was rarely observed in transfected cells when they were immunostained with an anti-PTB antibody. Cells that displayed a level of PTB immunostaining ≤25% of that of untransfected cells (measured by fluorescence densitometry) were analyzed for the presence of PNCs using either anti-PTB or anti-CUG-BP antibody. As shown in Figure 6A, left panels, PNCs were not detected in some of the cells (arrows) that displayed very low PTB protein levels. The loss of PNCs, evaluated with an anti-PTB antibody, in PTB protein expression-suppressed cells coincided with the loss of the nucleolar periphery concentration of a second PNC-associated protein, CUG-BP (Figure 6A, top right panel, arrows). In addition, in situ hybridization to a PNC-associated RNA, RNase MRP RNA, also demonstrated the loss of the perinucleolar concentrated labeling in PTB protein expression-suppressed cells (Figure 6A, second panels, arrows), confirming that PNCs are no longer present in these cells.
Figure 6.
siRNA against PTB protein mRNA significantly reduces the nuclear level of PTB protein (left panels in A, arrows). The decrease in PTB protein expression reduces PNC prevalence (gray bar in B) and two thirds of the remaining PNCs shrink into dot-like structures (white bar in B). However, the suppression of PTB expression does not significantly affect either the nuclear speckled distribution of a splicing factor or the nucleolar distribution of fibrillarin (bottom two panels in A). Bar, 10 μm.
Quantitative analyses of >500 cells showed an ∼20% lower PNC prevalence when PTB expression was significantly suppressed (Figure 6, A and B, light gray bars). Approximately two thirds of the remaining PNC-containing cells showed a significant reduction in the size of PNCs and these smaller PNCs became dot-like structures (Figure 6B, white bars). Although the reduction of PNC prevalence in PTB protein expression–suppressed cells was small, it was statistically significant (p <0.01, Student's t test), indicating that the level of PTB expression affects the structural integrity of PNCs. The lack of more extensive reduction in PNC prevalence in PTB protein expression-suppressed cells could be due to the inability of the siRNA to completely curtail PTB protein expression. Because more than half of the cells with suppressed PTB protein expression showed either loss of PNCs or alterations in their shape and size, these results suggest that the availability of PTB protein is a limiting factor for PNC integrity in many of the cells.
To control for possible nonspecific effect that siRNA oligos might have on PNCs, we transfected a control siRNA (MATERIALS AND METHODS) into HeLa cells under the same conditions. The introduction of this siRNA had no effect on the PNC structure and overall nuclear morphology (C. Wang and S. Huang, unpublished data). Furthermore, to evaluate whether the suppression of PTB protein expression might cause overall nuclear reorganization, we examined the distribution of several nuclear proteins in cells with significantly suppressed PTB expression 72 h after siRNA transfection. As shown in Figure 6A, right panels, the lack of PTB protein expression did not affect the diffuse nucleoplasmic distribution of CUG-BP (top right panel), the speckled nucleoplasmic distribution of the SC35 splicing factor (middle lower right panel), or the predominantly nucleolar localization of fibrillarin (bottom right panel). Thus, the effect of PTB protein suppression on PNCs cannot be attributed to a global reorganization of nuclear structure. Altogether, these findings suggest that the formation or the maintenance of PNCs require a certain essential level of PTB protein expression.
DISCUSSION
The Integrity of PNCs Is Dependent on Ongoing Pol III Transcription
Previous work demonstrated that the PNC is enriched in newly synthesized RNA as determined by Br-UTP incorporation after a brief (5 min) label, suggesting the involvement of transcription and/or RNA metabolism in PNC formation and maintenance (Huang et al., 1998). Actinomycin D at a concentration selective for pol I inhibition was found to eliminate PNCs, suggesting at the time that pol I transcription may be involved in PNC formation and possibly its functions (Huang et al., 1998). However, treatment of cells with cycloheximide for up to 5 h does not affect the structure of PNC (C. Wang and S. Huang, unpublished data), which argues against an involvement of pol I transcription because inhibition of translation by cycloheximide inhibits pol I transcription within 2 h (Tower and Sollner-Webb, 1987; O'Keefe et al., 1994). Thus, the sensitivity of the PNC to a low concentration of actinomycin D is not due to inhibition of pol I transcription, but instead may reflect a yet to be understood connection between the ribosomal DNA and PNCs. Although selective inhibition of pol II using 50 μg/ml α-amanitin alters, but does not eliminate PNCs (Huang et al., 1998), treatment of cells with a concentration of α-amanitin that inhibits both pol II and pol III transcription abolishes PNCs. These results thus raised the question of the effect on PNCs if pol III alone were inhibited. In this study, we injected cells with a highly specific pol III inhibitor, Tagetin, and the results clearly demonstrated that the structural integrity of the PNC requires on-going pol III transcription. Although we could not directly determine that Tagetin is selectively and extensively inhibiting pol III transcription in our experiments, the facts that α-amanatin does not affect PNCs until at an concentration known to block pol III transcription and that Tagetin does not affect the speckle distribution of splicing factors that are sensitive to pol II transcription leave little room for doubt that the Tagetin selectively affects pol III transcription. In addition, overexpression of a typical PNC-associated RNA, RNase MRP RNA, directed by a pol II promoter, partially blocked the PNC disassembly in the presence of Tagetin. This finding indicates that continuous production of the specific PNC-associated RNAs rather than the pol III polymerase activity is responsible for PNC maintenance. This finding also demonstrates that overexpression of only one of the PNC-associated RNAs is sufficient to maintain the PNC structure even through several other pol III small RNAs are observed in the PNC. In addition, this observation is an interesting contrast to previous studies showing that the intranuclear behavior of a given RNA can be profoundly affected when transcribed by a polymerase other than its usual one (e.g., Sisodia et al., 1987).
What is the biological relevance of these newly synthesized small RNAs accumulating in the PNC? The formation of PNCs is associated with the malignant phenotype (Huang et al., 1997), and they are not detected in normal cells or tissues in vivo, for example, breast or colon epithelium (R.V. Kamath and S. Huang, unpublished data). Thus, the presence of PNCs is likely to be a consequence, either directly or indirectly, of malignant transformation. There is a large body of literature describing structural and functional alterations of nucleoli during malignant transformation (Busch and Smetana, 1970; Derenzini et al., 1998; Smetana, 2002). Clinicians have long used changes in nucleolar shape, size, and enhanced silver staining of NORs (reflecting increased nucleolar levels of certain proteins) as phenotypic markers for judging the degree of malignancy (Ruschoff et al., 1990; Rzymowska, 1997). Although these changes may not always correlate with the levels of on-going ribosome synthesis, because some cancer cells are not particularly fast growing, they may represent alterations in steps of ribosome synthesis or nucleolar functions other than ribosome synthesis such as cell cycle regulation, p53 metabolism, telomerase activities, and macromolecule trafficking (Pederson, 1998). Because PNCs are spatially close to nucleoli, it is possible that the PNC formation is related to nucleolar changes during malignant transformation. The accumulation of some pol III transcripts in the PNC may reflect alternative pathways for the processing and metabolism of these RNAs, which could be changed in their expression levels during transformation. Further studies will be needed to understand the mechanistic basis for the connections between PNCs and nucleolus at the malignant state.
The newly synthesized pol III transcripts that are found in the PNC are unlikely to be transcribed in or nearby the structure because the genes of some known PNC RNAs are not detected in or near the PNCs (Matera et al., 1995; C. Wang and S. Huang, unpublished data). In addition, the PNCs do not appear to have a statistically significant association with any particular chromosome in HeLa cells (C. Wang and S. Huang, unpublished data). Thus, it is more likely that PNC-associated RNAs transit into PNCs shortly after their transcription elsewhere. One of the earliest proteins to associate with certain pol III transcripts is La, a nuclear phosphoprotein that is thought to chaperone pol III transcripts through posttranscriptional modifications (Maraia, 2001; Maraia and Intine, 2001, 2002). However, we have not detected a high concentration of La in PNCs either by immunostaining or by expression of GFP- or HA- tagged La protein (C. Wang and S. Huang, unpublished data), which is in agreement with earlier observations using immunostaining (Matera et al., 1995). Although RNase MRP RNA, which is an La-binding RNA (Yuan and Reddy, 1991), is highly concentrated in the PNC, two other well-known, La-associated pol III transcripts, 5S rRNA and U6 small nuclear RNA were not detected in PNCs using in situ hybridization (Matera et al., 1995). The lack of enrichment of La protein in the PNC suggests that the PNC-associated pol III RNAs may represent ones that are on a different processing pathway or have already released their La protein.
One of the open questions about some of the most abundant pol III transcripts in PNCs is how their abundance there relates to their levels throughout the nucleoli. Different pol III transcripts can have different PNC:nucleolus ratios, as determined by the signal intensity of in situ hybridization. For example, in cells with PNCs, RNase P RNA and RNase MRP RNA are found in both PNCs and nucleoli, with the labeling signal higher in PNCs than in nucleoli (Matera et al., 1995; Lee et al., 1996; and the present report), allowing the PNC to be unmistakably identified by the RNA signals alone. In contrast, the level of SRP RNA and Alu RNA in the PNC detected in the present study is not particularly higher than it appears to be throughout the nucleolus. Although the PNC localization of these RNAs can be seen in some cases by direct examination of the in situ hybridization pattern, it becomes most evident when the PNC is simultaneously identified by immunostaining (Figure 1). The significance of these differences in the relative levels of a given RNA in PNCs vs. the nucleolus is not known.
Another intriguing fact is that the PNC-associated RNAs do not appear to be in complexes with the usual protein subunits that form their functional RNPs. The Ro autoantigen that is known to be complexed with hY RNAs was not detected in PNCs (Matera et al., 1995; C. Wang and S. Huang, unpublished data), nor were the protein subunits of RNase MRP or RNase P (C. Wang and S. Huang, unpublished data). Similar observations seem to hold as well for SRP RNA, in that the three GFP-SRP proteins that have been observed in the nucleolus (Politz et al., 2000) were not detected in PNCs under the same transfection and expression conditions (C. Wang, J.C. Politz, T. Pederson, and S. Huang, unpublished data). This raises again the question of how the PNC-associated RNAs are related to these same RNA species that are normally present in nucleoli or in the cytoplasm. PNC-associated RNAs could transit through PNCs and then become assembled into functional ribonucleoproteins elsewhere in a way similar to their nucleolar populations. Alternatively, the PNC RNAs might not be on a productive pathway to functional ribonucleoproteins altogether. As a hypothetical example, one could imagine that if cells have a feedback pathway operating on the transcription of pol III genes, it might function by sensing the nucleoplasmic steady-state concentration of these transcripts. The PNC might, by sequestering a constant amount of, or a constant proportion of these transcripts, determine the set point for such a feedback circuit. Furthermore, transit through PNCs may represent a pathway of processing of these RNAs during malignancy that is different from that in normal cells. These are intriguing possibilities that will require further investigation.
The PTB Protein Is an Essential Component of the PNC
Although the PTB protein was the basis of the PNC's discovery and for much of its structural characterization (Huang, 2000), it had not been clear, until the present study, to what extent the PNC is actually dependent on this protein. The results of the present siRNA experiments indicate that the PTB protein is indeed an essential component of the PNC. Previous findings based on the expression of mutant PTB proteins indicated that the protein's RNA binding capacity is necessary for its PNC localization, further suggesting a stabilizing role of the PTB protein in holding the PNC-associated RNAs (Huang et al., 1997). However, polypyrimidine tracts of the length and composition known to be required for PTB protein binding are not present in human RNase MRP RNA, RNase P RNA, or SRP RNA. Indeed, in vitro RNA binding using recombinant PTB protein did not show interactions with either MRP RNA or SRP RNA (R.V. Kamath and S. Huang, unpublished data). Although MRP RNA and SRP RNA lack long stretches of pyrimidines, it remains possible that the PTB protein interacts with these RNAs directly or indirectly in vivo. This possibility is supported by our findings that anti-PTB antibody prevented a gel supershift of32P-labeled SRP RNA or MRP RNA when they were incubated with HeLa nuclear extracts (R.V. Kamath and S. Huang, unpublished data). In addition, incubation of PNC RNAs (but not non-PNC RNAs) in nuclear extracts changed the electrophoretic mobility of the PTB protein in native gels (R.V. Kamath and S. Huang, unpublished data). Together, these observations suggest that PTB may be involved in translocation of RNA-protein complexes into or through the PNC and that this activity is essential for the structural integrity of PNCs.
In summary, we have shown that the PNC is dependent on ongoing pol III transcription in HeLa cells. Overexpression of one of the PNC RNAs from a pol II promoter partially prevented the PNC disassembly that occurs upon inhibition of pol III transcription, indicating that it is the production of distinctive pol III transcripts that dictates the structural integrity of the PNC, not the transcriptional machinery or chromosomal locations that produce them. In addition, we find that reducing the nuclear levels of the PTB protein causes a decrease in PNC prevalence, indicating that the PTB protein is essential for the maintenance of the PNC. These findings together lead to a working model that PNC RNAs are complexed, directly or indirectly, with the PTB protein and possibly other proteins such as CUG-BP. These complexes may facilitate an alternative pathway of processing or degradation of certain pol III transcripts during transformation.
Acknowledgments
We are indebted to Rajesh Kamath in the Huang laboratory for his constructive discussion during this study, particularly his ideas and biochemical information on RNA-PTB protein interactions. We are also appreciative of the generous gifts of various reagents from many colleagues including Sidney Altman, Gregory Matera, Edward K. Chan, Stéphane Richard, Maurice Swanson, and Eng Tan. This investigation was supported by National Institutes of Health Grant GM-21595 to T.P. and National Cancer Institute Grant R01 CA77560 to S.H.
Abbreviations used: CMV, cytomegalovirus; CUG-BP, CUG-binding protein; PNC, perinucleolar compartment; PTB, polypyrimidine tract binding; SRP, signal recognition particle; UBF, upstream binding factor.
References
- Busch, H., and Smetana, K. (1970). Nucleoli of tumor cells. In: The Nucleolus. ed. H. Busch, New York: Academic Press.
- Chen, T., Boisvert, F.M., Bazett-Jones, D.P., and Richard, S. (1999). A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol. Biol. Cell 10, 3015-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini, M., Trere, D., Pession, A., Montanaro, L., Sirri, V., and Ochs, R.L. (1998). Nucleolar function and size in cancer cells. Am. J. Pathol. 152, 1291-1297. [PMC free article] [PubMed] [Google Scholar]
- Fraser, N.W., Sehgal, P.B., and Darnell, J.E. (1978). DRB-induced premature termination of late adenovirus transcription. Nature 272, 590-593. [DOI] [PubMed] [Google Scholar]
- Fu, X.D., and Maniatis, T. (1990). Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 343, 437-441. [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco, M.A., Jamison, S.F., and Sharp, P.A. (1989). Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 3, 1874-1886. [DOI] [PubMed] [Google Scholar]
- Ghetti, A., Pinol-Roma, S., Michael, W.M., Morandi, C., and Dreyfuss, G. (1992). hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 20, 3671-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. (2000). Review: perinucleolar structures. J. Struct. Biol. 129, 233-240. [DOI] [PubMed] [Google Scholar]
- Huang, S., Deerinck, T.J., Ellisman, M.H., and Spector, D.L. (1997). The dynamic organization of the perinucleolar compartment in the cell nucleus. J. Cell Biol. 137, 965-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., Deerinck, T.J., Ellisman, M.H., and Spector, D.L. (1998). The perinucleolar compartment and transcription. J. Cell Biol. 143, 35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier, S., Illenberger, S., Grosheva, I., Rudiger, M., Singer, R.H., and Jockusch, B.M. (2001). Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J. Cell Biol. 155, 775-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedinger, C., Gniazdowski, M., Mandel, J.L., Jr., Gissinger, F., and Chambon, P. (1970). Alpha-amanitin: a specific inhibitor of one of two DNA-pendent RNA polymerase activities from calf thymus. Biochem. Biophys. Res. Commun. 38, 165-171. [DOI] [PubMed] [Google Scholar]
- Lee, B., Matera, A.G., Ward, D.C., and Craft, J. (1996). Association of RNase mitochondrial RNA processing enzyme with ribonuclease P in higher ordered structures in the nucleolus: a possible coordinate role in ribosome biogenesis. Proc. Natl. Acad. Sci. USA 93, 11471-11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell, T.J., Weinberg, F., Morris, P.W., Roeder, R.G., and Rutter, W.J. (1970). Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science 170, 447-449. [DOI] [PubMed] [Google Scholar]
- Maraia, R.J. (2001). La protein and the trafficking of nascent RNA polymerase iii transcripts. J. Cell Biol. 153, F13-F18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia, R.J., and Intine, R.V. (2001). Recognition of nascent RNA by the human La antigen: conserved and divergent features of structure and function. Mol. Cell. Biol. 21, 367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia, R.J., and Intine, R.V. (2002). La protein and its associated small nuclear and nucleolar precursor RNAs. Gene Expr. 10, 41-57. [PMC free article] [PubMed] [Google Scholar]
- Matera, A.G., Frey, M.R., Margelot, K., and Wolin, S.L. (1995). A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J. Cell Biol. 129, 1181-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe, R.T., Mayeda, A., Sadowski, C.L., Krainer, A.R., and Spector, D.L. (1994). Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124, 249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson, T. (1998). The plurifunctional nucleolus. Nucleic Acids Res. 26, 3871-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J.C., Yarovoi, S., Kilroy, S.M., Gowda, K., Zwieb, C., and Pederson, T. (2000). Signal recognition particle components in the nucleolus. Proc. Natl. Acad. Sci. USA 97, 55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J.C., Lewandowski, L.B., and Pederson, T. (2002). Signal recognition particle RNA localization within the nucleolus differs from the classical sites of ribosome synthesis. J. Cell Biol. 159, 411-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruschoff, J., Bittinger, A., Neumann, K., and Schmitz-Moormann, P. (1990). Prognostic significance of nucleolar organizing regions (NORs) in carcinomas of the sigmoid colon and rectum. Pathol. Res. Pract. 186, 85-91. [DOI] [PubMed] [Google Scholar]
- Rzymowska, J. (1997). AgNOR counts and their combination with flow cytometric analyses and clinical parameters as a prognostic indicator in breast carcinoma. Tumori 83, 938-942. [DOI] [PubMed] [Google Scholar]
- Sehgal, P.B., Derman, E., Molloy, G.R., Tamm, I., and Darnell, J.E. (1976). 5,6-Dichloro-1-Beta-D-ribofuranosylbenzimidazole inhibits initiation of nuclear heterogeneous RNA chains in HeLa cells. Science 194, 431-433. [DOI] [PubMed] [Google Scholar]
- Sisodia, S.S., Sollner-Webb, B., and Cleveland, D.W. (1987). Specificity of RNA maturation pathways: RNAs transcribed by RNA polymerase III are not substrates for splicing or polyadenylation. Mol. Cell. Biol. 7, 3602-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetana, K. (2002). Structural features of nucleoli in blood, leukemic, lymphoma and myeloma cells. Eur. J. Histochem. 46, 125-132. [DOI] [PubMed] [Google Scholar]
- Spector, D.L. (1993). Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9, 265-315. [DOI] [PubMed] [Google Scholar]
- Steinberg, T.H., and Burgess, R.R. (1992). Tagetitoxin inhibition of RNA polymerase III transcription results from enhanced pausing at discrete sites and is template-dependent. J. Biol. Chem. 267, 20204-20211. [PubMed] [Google Scholar]
- Steinberg, T.H., Mathews, D.E., Durbin, R.D., and Burgess, R.R. (1990). Tagetitoxin: a new inhibitor of eukaryotic transcription by RNA polymerase III. J. Biol. Chem. 265, 499-505. [PubMed] [Google Scholar]
- Timchenko, L.T., Miller, J.W., Timchenko, N.A., DeVore, D.R., Datar, K.V., Lin, L., Roberts, R., Caskey, C.T., and Swanson, M.S. (1996). Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 24, 4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower, J., and Sollner-Webb, B. (1987). Transcription of mouse rDNA is regulated by an activated subform of RNA polymerase I. Cell 50, 873-883. [DOI] [PubMed] [Google Scholar]
- Wang, J., and Pederson, T. (1990). A 62,000 molecular weight spliceosome protein crosslinks to the intron polypyrimidine tract. Nucleic Acids Res. 18, 5995-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y., and Reddy, R. (1991). 5′ flanking sequences of human MRP/7–2 RNA gene are required and sufficient for the transcription by RNA polymerase III. Biochim. Biophys. Acta 1089, 33-39. [DOI] [PubMed] [Google Scholar]