Abstract
Activating mutations of the p110 α subunit of PI3K (PIK3CA) oncogene have been identified in a broad spectrum of malignant tumors. However, their role in benign or preneoplastic conditions is unknown. Activating FGF receptor 3 (FGFR3) mutations are common in benign skin lesions, either as embryonic mutations in epidermal nevi (EN) or as somatic mutations in seborrheic keratoses (SK). FGFR3 mutations are also common in low-grade malignant bladder tumors, where they often occur in association with PIK3CA mutations. Therefore, we examined exons 9 and 20 of PIK3CA and FGFR3 hotspot mutations in EN (n = 33) and SK (n = 62), two proliferative skin lesions lacking malignant potential. Nine of 33 (27%) EN harbored PIK3CA mutations; all cases showed the E545G substitution, which is uncommon in cancers. In EN, R248C was the only FGFR3 mutation identified. By contrast, 10 of 62 (16%) SK revealed the typical cancer-associated PIK3CA mutations E542K, E545K, and H1047R. The same lesions displayed a wide range of FGFR3 mutations. Corresponding unaffected tissue was available for four EN and two mutant SK: all control samples displayed a WT sequence, confirming the somatic nature of the mutations found in lesional tissue. Forty of 95 (42%) lesions showed at least one mutation in either gene. PIK3CA and FGFR3 mutations displayed an independent distribution; 5/95 lesions harbored mutations in both genes. Our findings suggest that, in addition to their role in cancer, oncogenic PIK3CA mutations contribute to the pathogenesis of skin tumors lacking malignant potential. The remarkable genotype–phenotype correlation as observed in this study points to a distinct etiopathogenesis of the mutations in keratinocytes occuring either during fetal development or in adult life.
Keywords: oncogene, senescence, skin, benign tumor
Phosphatidylinositol 3-kinases (PI3Ks) are involved in the generation of the second-messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3). Three classes of PI3Ks are known. Class I PI3Ks are composed of a regulatory (p85) and a catalytic (p110) subunit. The Class IA subgroup proteins are involved in signaling from receptor tyrosine kinases upon growth factor binding, whereas class IB proteins signal from G-coupled receptors (1). The levels of PIP3 are also controlled by phosphatase and tensin homologue, a classical tumor suppressor protein with lipid phosphatase activity. PIP3 generated by the p110 catalytic subunit of class I PI3Ks is anchored to the cell membrane, leading to the binding of PKD through its plekstrin homology domain, its subsequent activation, and the downstream phosphorylation and activation of the survival kinase AKT. The PI3K pathway has been shown to play major roles in cell growth, proliferation, survival, and malignant transformation (2).
Four different p110 subunits (α, β, γ, and δ) have been identified, and all of them have transforming potential in vitro. Overexpression of WT p110 β, γ, or δ is oncogenic; by contrast, p110 α is oncogenic only when constitutively activated through point mutation (3). Indeed, activating somatic mutations have only been reported in the gene coding for the α isoform, PIK3CA, in a wide variety of human cancers, such as breast, colon, ovarian, and gastric cancer (4, 5). Somatic PIK3CA point mutations cluster in hotspots of exons 9 (helical domain) and 20 (kinase domain). These mutations result in a constitutive gain of function and confer oncogenic properties to mutated cells by deregulation of transcription and translation (6). The in vivo oncogenicity of mutant p110α carrying the most common hotspot mutations E542K, E545K, and H1047R has been recently demonstrated in a chicken model (7).
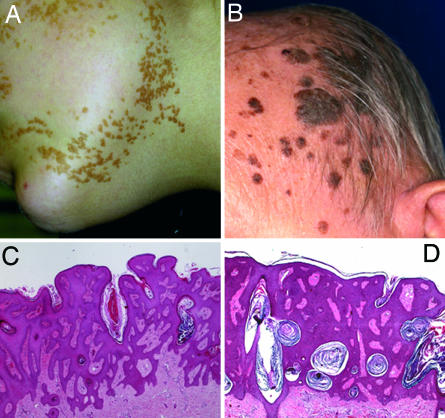
Epidermal nevi (EN) and seborrheic keratoses (SK) are keratinocyte-derived benign acanthotic skin tumors that never progress to acquire invasive features (Fig. 1). Both entities share clinical and histological characteristics. However, their natural history is very different. EN are congenital lesions that are present at birth or that develop early during childhood as localized epidermal thickening with hyperpigmentation (Fig. 1A). EN follow frequently the lines of Blaschko and may vary in their extent from a single linear lesion to widespread and systematized involvement. It has been proposed that EN result from mosaicism in the skin (8). By contrast, SK are rare in young people. Their prevalence increases with age, and they can be identified by age 50 in 80–100% of healthy individuals (9), although there is wide variation in the number of lesions. SK present as well demarcated brownish plaques with a verrucous surface predominantly localized in the head, neck, and trunk (Fig. 1B). EN and SK share many histological characteristics such as acanthosis, papillomatosis, and variable degrees of hyperkeratosis and hyperpigmentation (Fig. 1 C and D).
Fig. 1.
Morphological similarities of linear lesions of congenital EN from a child following Blaschko's lines (A) and SK from an elderly patient (B). At the microscopic level, both lesions are characterized by acanthosis, papillomatosis, and variable degrees of hyperkeratosis and hyperpigmentation (C, EN; D, SK).
Recently, activating mutations in the gene coding for FGF receptor 3 (FGFR3) were identified in both EN and SK (10, 11). The same mutations detected in these skin lesions have previously been identified as somatic alterations in several types of human cancer (12, 13) as well as in the germ-line in patients with short limb dysplasias (14). The reported FGFR3 mutations lead to a constitutive ligand-independent activation of the receptor tyrosine kinase activity, and a good correlation between the level of kinase activation and the severity of the skeletal phenotype has been observed (15). Because only ≈30% of EN (10, 16) and 25–85% of SK (11, 17, 18) harbor FGFR3 mutations, the involvement of additional genes has been postulated (16, 19). There is strong evidence indicating that the PI3K pathway is involved in FGFR signaling (20, 21). Therefore, genes in this pathway are potential candidates involved in the pathogenesis of EN and SK. Interestingly, the highest prevalence of FGFR3 mutations in cancer is found among superficial papillary bladder tumors (22–25). Activating PIK3CA mutations have also been reported in superficial papillary bladder tumors, often in association with FGFR3 mutations (26). These findings suggested that PIK3CA mutations might also occur in benign skin tumors. Therefore, we screened EN and SK for PIK3CA mutations in exons 9 and 20.
Here, we report that both types of benign acanthotic skin lesions indeed harbor somatic PIK3CA mutations in the hotspot codons as previously identified in malignant tumors. In addition, we demonstrate that the mutation spectrum of PIK3CA is different in EN and SK, indicating a remarkable genotype–phenotype correlation.
Results
Table 1 summarizes mutational analyses. Supporting information (SI) Tables 2 and 3 provide detailed information on the clinical characteristics of the patients and molecular findings.
Table 1.
PIK3CA mutations in EN and SK
| PIK3CA mutation | Base substitution | Amino acid substitution | n (EN = 33) | n (SK = 62) | Concomitant FGFR3 mutation |
|---|---|---|---|---|---|
| E542K | c.1624G > A | Glu > Lys | — | 7 | 3 (R248C, K652E) |
| E545K | c.1633G > A | Glu > Lys | — | 2 | — |
| E545G | c.1634A > G | Glu > Gly | 9 | — | 2 (R248C) |
| H1047R | c.3140A > G | His > Arg | — | 1 | — |
Activating PIK3CA Mutations Are Common in EN.
We sequenced exons 9 and 20 of PIK3CA in 33 EN. The E545G mutation was detected in nine cases (27%) (Fig. 2); no other alterations were found. In all cases, the WT sequence was also present, indicating heterozygosity. Codon 545 is a hotspot for activating mutations in a wide variety of tumors (5). However, in cancer, the E545K mutation is much more common than the E545G mutation, which has been reported only in a few carcinomas (4, 26) (www.sanger.ac.uk/genetics/CGP/cosmic). Mutations were not associated with sex, localization, or histological subtype. It has been proposed that EN result from somatic mutations occurring during development, leading to mosaicism, and evidence supporting this notion has been acquired in the case of FGFR3 mutations (10, 16). We analyzed six unaffected tissue samples from four patients with EN harboring mutant PIK3CA sequences and found that only the WT sequence was detectable. In an additional case, leukocyte DNA also displayed only the WT sequence. These findings provide evidence that PIK3CA mutations occur in mosaicism in the skin, analogous to FGFR3 mutations.
Fig. 2.
Mutational analysis of EN and SK. In all cases, the mutant sequence was accompanied by the presence of the WT allele, indicating heterozygosity.
Activating PIK3CA Mutations Occur in SK.
Because of the histological similarities between EN and SK and the recent demonstration that FGFR3 mutations occur in both types of lesions, we also analyzed exons 9 and 20 of PIK3CA in SK. Ten of 62 (16%) lesions analyzed were found to harbor PIK3CA mutations. Of them, eight displayed an acanthotic, one a hyperkeratotic, and one an adenoid histologic differentiation. The following amino acid substitutions were found: E542K (n = 7), E545K (n = 2), and H1047R (n = 1) (Table 1; Fig. 2). Analogous to the EN, all PIK3CA mutations detected in the SK were heterozygous. Corresponding normal skin, which was available from two patients with mutant SK, was found to be WT, confirming that PIK3CA mutations in SK are somatic events. Furthermore, multiple individual SK from three patients were analyzed (four, six, and eight SK per patient, respectively). In two patients, all SK were WT; in the third case, one of the SK was found to be mutated, whereas the other seven lesions from this patient displayed a WT sequence. PIK3CA mutations occurred preferentially in SK of the trunk (80%), but this association was not statistically significant (P = 0.29). Mutations were not associated with sex, age, or histologic subtype.
PIK3CA Mutations and FGFR3 Mutations in EN and SK.
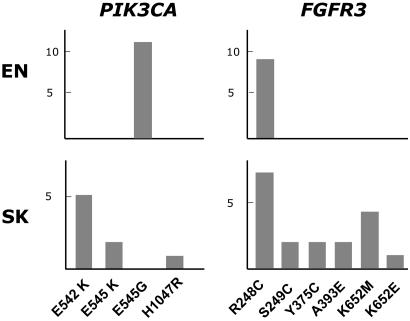
Fig. 3 summarizes the mutational spectrum findings. The presence of FGFR3 mutations in this series of EN, and in a proportion of the SK studied, has been reported (10, 16–18). In seven of nine (78%) EN cases with mutant PIK3CA sequences, no mutations in FGFR3 were identified. In the remaining two cases, the R248C mutation in FGFR3 was associated with the E545G mutation in PIK3CA. Five EN showed the R248C hotspot mutation in the FGFR3 gene in the absence of detectable PIK3CA mutations. Similar to EN, 7 of 10 (70%) SK with a PIK3CA mutation did not harbor an additional FGFR3 mutation at the known hotspots. Nineteen of 62 (31%) SK showed activating FGFR3 mutations: R248C (n = 9), S249C (n = 2), Y375C (n = 2), A393E (n = 2), K652E (n = 1), and K652M (n = 4), one of them with two mutations. Three SK revealed simultaneous PIK3CA and FGFR3 mutations (in two cases, the E542K/R248C and in one case, the E542K/K652E combination was present). In 16 of 62 (26%) SK, an FGFR3 mutation was found in the absence of PIK3CA alterations.
Fig. 3.
Spectrum of mutations found in benign skin lesions in PIK3CA and FGFR3. The number of lesions with each mutation is indicated in the ordinates. EN display a very homogenous mutational pattern in both genes, whereas, in SK, a wide spectrum of mutations as found in malignant cancers is observed.
Taken together, PIK3CA and/or FGFR3 mutations were detected in 14 of 33 (42%) EN and in 26 of 62 (42%) SK in this study. Mutations in both genes showed an independent distribution (P = 0.908). In 5 of 95 (5%) of the benign skin lesions analyzed, the simultaneous occurrence of mutations in these two oncogenes was demonstrated.
Discussion
An improved knowledge of the molecular basis of benign tumors is of fundamental importance to understand why benign tumors, unlike malignant ones, fail to progress and invade. Accordingly, a great deal of emphasis has been placed recently in investigating the cellular response to oncogenic stress as a defense mechanism against tumor progression.
Activating mutations in several oncogenes involved in malignant human tumors, such as K-ras or B-RAF, have been shown to occur frequently in benign lesions, such as colorectal adenomas or melanocytic nevi (27–29). These benign proliferative lesions carrying mutations show a variable risk for malignant transformation. In this report, we provide evidence that activating mutations in PIK3CA previously identified in a wide variety of human cancers also occur in two types of benign skin lesions that bear no relevant risk of progression to malignancy. These findings favor the notion that some benign lesions without malignant potential are initiated by oncogenic pathways shared with cancer. Why some types of benign lesions have the potential to progress to malignancy whereas others do not remains an important question (see below).
EN and SK are histologically similar but display a different natural history. EN are congenital lesions that have been proposed to result from mosaicism in the skin. Approximately 30% of EN display the R248C FGFR3 mutation in the absence of alterations in unaffected tissue (10, 16). However, there is no information on the genetic basis of the remaining cases. We show that ≈30% of EN display a specific mutation, E545G, in PIK3CA. This mutation has only exceptionally been reported in malignant tumors. By contrast, the E545K substitution in the same codon represents ≈25% of all PI3KCA mutations in cancers (4). Gymnopoulos et al. (30) have recently compared the activity of mutant PIK3CA proteins in vitro and have shown that the E545G substitution displays strong transforming activity on chicken embryo fibroblasts, although its effect is lower than that of the more common E542K and E545K substitutions.
The mutations identified in PIK3CA and FGFR3 in EN display a remarkable specificity: regarding the former, only the E545G amino acid substitution was present, and regarding the latter, the R248C mutation was almost exclusively found. This genetic homogeneity is in marked contrast with that reported among malignant tumors for both genes (4, 5, 12, 31). The E545G substitution results from an AT:GC transition, whereas the R248C substitution results from a CG:TA transition, suggesting that the biological nature of the mutation, rather than the mutagenic mechanism, plays a crucial role in the development of EN (32).
Unlike EN, SK display a wide spectrum of mutations in both genes (Fig. 3). Regarding PIK3CA, the three most common cancer-associated mutations (E542K, E545K, and H1047R) were found; as of FGFR3, the mutational spectrum showed even more diversity, including the R248C, S249C, G372C, S373C, Y375C, A393E, K652M, and K652E substitutions (19). Moreover, we have recently reported that, within a given individual with multiple SK, at least four different FGFR3 mutations were present in each case (18). The etiological and biological relevance of this genetic heterogeneity is currently unknown. However, it suggests that SK arise as a result of a wider range of mutagenic mechanisms than do EN. These observations may have implications for the understanding of the pathogenesis of other tumors in which genetic alterations have been proposed to occur during development.
Most of the mutations in PIK3CA present in SK were GC:AT transitions. The diversity of FGFR3 nucleotide substitutions reported in these lesions points to the selection of variants because of their biological properties rather than to a pivotal role of a carcinogen fingerprint associated with UV exposure in the skin (32).
This is a comprehensive study of PIK3CA mutations in human skin and, more generally, of point mutations in an oncogene of the PI3K pathway in benign lesions without malignant potential. The PI3K pathway is thought to play a predominant role in cell survival; mutations in genes coding for proteins acting upstream of PIKs, such as Ras genes or tyrosine kinase growth factor receptors, are thought to affect more directly cell proliferation. Pathologic activation of the FGFR3/PI3K/AKT pathway seems to be a frequent event in the pathogenesis of human EN and SK. That the majority of EN and SK in this study revealed a WT status in both PIK3CA and FGFR3 suggests that additional genes of this or of other pathways contribute to the pathogenesis of acanthotic skin tumors.
In addition, we report previously undescribed oncogenic mutations in two different genes occurring concomitantly in a completely benign lesion, whether congenital or acquired. That the majority of SK appear to be clonal (33) lends support to the notion that both mutations may occur in the same cell, although more work is required to conclusively establish this fact. Oncogenic germ-line FGFR3 mutations cause short limb dysplasia through the activation of Stat1 and the up-regulation of p21, thus leading to premature cell cycle exit in chondrocytes (34). We have proposed that this mechanism may, at least in part, be related to the association of FGFR3 mutations with good prognosis in patients with superficial papillary bladder tumors (25). In addition, we have proposed that, in bladder tumors, PIK3CA mutations may bypass a putative growth disadvantage effect provided by FGFR3 mutations (26). The findings in benign skin lesions reported here clearly indicate that, at least in keratinocytes, the presence of mutations in these two genes does not imply the malignant phenotype, supporting the notion that other genetic alterations, possibly in tumor suppressor genes, must occur to acquire a “progressor” phenotype. Furthermore, our findings suggest that the accumulation of mutations in oncogenes may effectively contribute to activate protective cellular mechanisms that act as a barrier for tumor development (35). Sarkisian et al. (36) have recently demonstrated the relevance of “the level of signaling” to the activation of senescence in a study using transgenic mice. In these mice, an oncogenic mutated Ras gene was expressed in the mammary gland under the control of a doxycycline-inducible promoter. Near-physiologic levels of oncogenic Ras led to hyperproliferation of epithelial cells, whereas high levels of Ras expression led to the activation of a senescence program. Importantly, high levels of oncogenic Ras failed to activate the senescence program in mice in which the Ink4a-Arf locus had been inactivated (36). Multiple mutations occurring in either a single oncogene, as is the case for FGFR3 or PIK3CA in benign skin tumors, or in multiple oncogenes may provide sufficient signals to activate a senescence program in humans as well. Interestingly, inhibition of the PI3K pathway has recently been shown to be required for Ras-induced senescence in vitro (37).
Our findings also bear evidence to the ability of abnormal PI3K signaling in the activation of senescence. Until now, genes located mainly upstream of PI3K have been reported to activate senescence (i.e., receptor tyrosine kinases, Ras, B-RAF, and NF1) (38, 39). This observation is in agreement with the activation of senescence upon acute Pten inactivation in the mouse (40). Mallette et al. (41) have reported that the DNA damage response (DDR) is involved in the induction of senescence mediated by oncogenic Ras and Stat5, and it will be important to determine whether aberrant PI3K signaling is also able to activate the DDR (41).
Overall, our findings extend the spectrum of proliferative lesions harboring PIK3CA mutations and underline the relevance of understanding the mechanisms involved in early steps of tumor development in human carcinogenesis. The skin provides a potent model for such studies given its ready accessibility for visual, pathological, and genetic examination.
Materials and Methods
Patients and Samples.
The files of the Department of Pathology of Hospital del Mar and Hospital de Sant Pau, the Department of Dermatology of the University of Regensburg, and the Department of Dermatohistopathology, Friedrichshafen, were searched to retrieve diagnostic slides corresponding to cases labeled as “epidermal nevus” (n = 33) and “seborrheic keratosis” (n = 62). Cases diagnosed as nonorganoid nonepidermolytic EN showing the “common” histopathological features (acanthosis, papillomatosis, and a variable degree of hyperkeratosis), as well as cases showing an acrokeratosis verruciformis-like findings, or an SK-like pattern were included in the study. For SK, the three major histologic subtypes (acanthotic, hyperkeratotic, and adenoid) were retrieved. H&E-stained sections were reviewed by an experienced dermatopathologist, and cases fulfilling criteria for the appropriate diagnoses were selected for study. The study was approved by the Ethics Committees of all participating institutions and was performed according to the Declaration of Helsinki.
PIK3CA and FGFR3 Mutation Analysis.
Microdissection, DNA extraction from formalin-fixed paraffin-embedded tissue, control samples, primers, and PCR conditions to amplify exons 9 and 20 of PIK3CA and exons 7, 10, and 15 of FGFR3 have been reported (16, 26). DNA from unaffected tissue was used to determine the somatic nature of the variants.
PCRs were done by using 10–50 ng of DNA/0.2 μmol/liter of each primer/200 μmol/liter deoxynucleotide triphosphates/3.5 mmol/liter MgCl2/1× PCR II buffer/1.5 units of Amplitaq Gold DNA Polymerase (Applied Biosystems, Foster City, CA). PCR conditions were as follows: 94°C (10 min) for one cycle; 94°C (40 sec), 60°C (40 sec), and 72°C (40 sec) for 42 cycles; and a final extension step of 72°C (10 min). In all experiments, several reactions were included in the absence of DNA to rule out contamination by PCR products. All mutations were confirmed by analyzing the products of a second independent PCR as well as by sequencing of both strands of PCR products. In addition, FGFR3 mutations for all SK in this study and 9 of 33 EN were analyzed by using a SNaPshot multiplex assay, as described (10, 42).
Statistical Analyses.
Independence of PIK3CA and FGFR3 mutational status was tested by applying χ2 Mantel–Haenszel test. The trend of PIK3CA mutational prevalence according to sex, age, localization, and histologic subtype was estimated by using Fisher's exact and Mann–Whitney tests.
Supplementary Material
Acknowledgments
We thank Manuel Serrano, Xavier Mayol, and Núria Malats for critical reading of the manuscript and helpful discussions; the Department of Pathology, Hospital del Mar, for valuable contributions; and Tania Lobato, Lydia Kuenzel, and Andrea Mueller for excellent technical support. This work was supported, in part, by Grants C03/010 and G03/174 from Instituto de Salud Carlos III, Ministerio de Sanidad, Spain; Grant SAF2004-01137 from Plan Nacional de I+D, Ministerio de Ciencia y Tecnología; and Grant SGR-00410 from Comissió Interdepartamental per la Recerca i Innovació Tecnològica–Generalitat de Catalunya. C.H. was supported by Deutsche Dermatologische Gesellschaft/Arbeitsgemeinschaft Dermatologische Forschung Research Grant 2007 and the ReForM-B grant of the University of Regensburg. E.L.-K. was supported by a Predoctoral Fellowship from Fundación Ramón Areces, Madrid, Spain. N.M.L. was supported by a Predoctoral Fellowship from Fundaçao para a Ciência e a Tecnologia, Portugal.
Abbreviations
- EN
epidermal nevus
- FGFR3
FGF receptor 3
- PI3K
phosphatidylinositol 3-kinase
- PIK3CA
p110 α subunit of PI3K
- SK
seborrheic keratosis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705218104/DC1.
References
- 1.Cantley LC. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 2.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 3.Kang S, Denley A, Vanhaesebroeck B, Vogt PK. Proc Natl Acad Sci USA. 2006;103:1289–1294. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 5.Karakas B, Bachman KE, Park BH. Br J Cancer. 2006;94:455–459. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bader AG, Kang S, Zhao L, Vogt PK. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 7.Bader AG, Kang S, Vogt PK. Proc Natl Acad Sci USA. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Happle R, Rogers M. Adv Dermatol. 2002;18:175–201. [PubMed] [Google Scholar]
- 9.Yeatman JM, Kilkenny M, Marks R. Br J Dermatol. 1997;137:411–414. [PubMed] [Google Scholar]
- 10.Hafner C, van Oers JM, Vogt T, Landthaler M, Stoehr R, Blaszyk H, Hofstaedter F, Zwarthoff EC, Hartmann A. J Clin Invest. 2006;116:2201–2207. doi: 10.1172/JCI28163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Logie A, Dunois-Larde C, Rosty C, Levrel O, Blanche M, Ribeiro A, Gasc JM, Jorcano J, Werner S, Sastre-Garau X, et al. Hum Mol Genet. 2005;14:1153–1160. doi: 10.1093/hmg/ddi127. [DOI] [PubMed] [Google Scholar]
- 12.Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, Chopin D, Thiery JP, Radvanyi F. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 13.Chesi M, Nardini E, Brents LA, Schrock E, Ried T, Kuehl WM, Bergsagel PL. Nat Genet. 1997;16:260–264. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vajo Z, Francomano CA, Wilkin DJ. Endocr Rev. 2000;21:23–39. doi: 10.1210/edrv.21.1.0387. [DOI] [PubMed] [Google Scholar]
- 15.Naski MC, Wang Q, Xu J, Ornitz DM. Nat Genet. 1996;13:233–237. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez S, Toll A, Baselga E, Ribe A, Azua-Romeo J, Pujol RM, Real FX. J Invest Dermatol. 2007;12:1064–1066. doi: 10.1038/sj.jid.5700705. [DOI] [PubMed] [Google Scholar]
- 17.Hafner C, van Oers JM, Hartmann A, Landthaler M, Stoehr R, Blaszyk H, Hofstaedter F, Zwarthoff EC, Vogt T. J Invest Dermatol. 2006;126:2404–2407. doi: 10.1038/sj.jid.5700422. [DOI] [PubMed] [Google Scholar]
- 18.Hafner C, Hartmann A, Real FX, Hofstaedter F, Landthaler M, Vogt T. J Invest Dermatol. 2007;127:1883–1885. doi: 10.1038/sj.jid.5700804. [DOI] [PubMed] [Google Scholar]
- 19.Hafner C, Vogt T, Hartmann A. Cell Cycle. 2006;5:2723–2728. doi: 10.4161/cc.5.23.3509. [DOI] [PubMed] [Google Scholar]
- 20.Schlessinger J. Science. 2004;306:1506–1507. doi: 10.1126/science.1105396. [DOI] [PubMed] [Google Scholar]
- 21.Eswarakumar VP, Lax I, Schlessinger J. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Billerey C, Chopin D, Aubriot-Lorton MH, Ricol D, Gil Diez de Medina S, Van Rhijn B, Bralet MP, Lefrere-Belda MA, Lahaye JB, Abbou CC, et al. Am J Pathol. 2001;158:1955–1959. doi: 10.1016/S0002-9440(10)64665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Rhijn BW, van der Kwast TH, Vis AN, Kirkels WJ, Boeve ER, Jobsis AC, Zwarthoff EC. Cancer Res. 2004;64:1911–1914. doi: 10.1158/0008-5472.can-03-2421. [DOI] [PubMed] [Google Scholar]
- 24.Jebar AH, Hurst CD, Tomlinson DC, Johnston C, Taylor CF, Knowles MA. Oncogene. 2005;24:5218–5225. doi: 10.1038/sj.onc.1208705. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez S, Lopez-Knowles E, Lloreta J, Kogevinas M, Amoros A, Tardon A, Carrato A, Serra C, Malats N, Real FX. J Clin Oncol. 2006;24:3664–3671. doi: 10.1200/JCO.2005.05.1771. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Knowles E, Hernandez S, Malats N, Kogevinas M, Lloreta J, Carrato A, Tardon A, Serra C, Real FX. Cancer Res. 2006;66:7401–7404. doi: 10.1158/0008-5472.CAN-06-1182. [DOI] [PubMed] [Google Scholar]
- 27.Barry EL, Baron JA, Grau MV, Wallace K, Haile RW. Cancer. 2006;106:1036–1040. doi: 10.1002/cncr.21721. [DOI] [PubMed] [Google Scholar]
- 28.Spring KJ, Zhao ZZ, Karamatic R, Walsh MD, Whitehall VL, Pike T, Simms LA, Young J, James M, Montgomery GW, et al. Gastroenterology. 2006;131:1400–1407. doi: 10.1053/j.gastro.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 29.Thomas NE. Melanoma Res. 2006;16:97–103. doi: 10.1097/01.cmr.0000215035.38436.87. [DOI] [PubMed] [Google Scholar]
- 30.Gymnopoulos M, Elsliger MA, Vogt PK. Proc Natl Acad Sci USA. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Rhijn BW, van Tilborg AA, Lurkin I, Bonaventure J, de Vries A, Thiery JP, van der Kwast TH, Zwarthoff EC, Radvanyi F. Eur J Hum Genet. 2002;10:819–824. doi: 10.1038/sj.ejhg.5200883. [DOI] [PubMed] [Google Scholar]
- 32.Vineis P, Malats N, Porta M, Real FX. Mutat Res. 1999;436:185–194. [PubMed] [Google Scholar]
- 33.Nakamura H, Hirota S, Adachi S, Ozaki K, Asada H, Kitamura Y. J Invest Dermatol. 2001;116:506–510. doi: 10.1046/j.1523-1747.2001.01289.x. [DOI] [PubMed] [Google Scholar]
- 34.Legeai-Mallet L, Benoist-Lasselin C, Munnich A, Bonaventure J. Bone. 2004;34:26–36. doi: 10.1016/j.bone.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Collado M, Serrano M. Cell Cycle. 2005;4:1722–1724. doi: 10.4161/cc.4.12.2260. [DOI] [PubMed] [Google Scholar]
- 36.Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA. Nat Cell Biol. 2007;9:493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 37.Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 39.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 40.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Oers JM, Lurkin I, van Exsel AJ, Nijsen Y, van Rhijn BW, van der Aa MN, Zwarthoff EC. Clin Cancer Res. 2005;11:7743–7748. doi: 10.1158/1078-0432.CCR-05-1045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.