Insights into virus–host cell interactions as uncovered by Randall et al. (1) in a recent issue of PNAS further our understanding of the hepatitis C virus (HCV) life cycle, persistence, and pathogenesis and might lead to the identification of new therapeutic targets. HCV persistently infects 180 million individuals worldwide, causing chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. The only approved treatment, combination therapy with IFN-α and ribavirin, targets cellular pathways (2); however, a sustained virologic response is achieved only in approximately half of the patients treated. Therefore, there is a pressing need for the identification of novel drugs against hepatitis C. Although most research focuses on the development of HCV-specific antivirals, such as protease and polymerase inhibitors (3), cellular targets could be pursued and might allow the development of broad-spectrum antivirals (2).
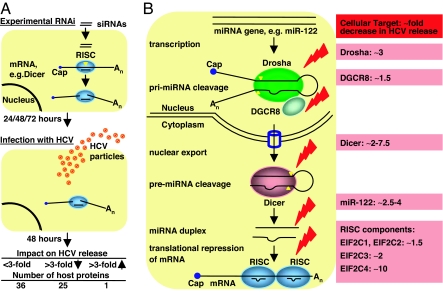
Because of the lack of HCV cell culture models, initial studies on HCV–host cell interactions used expressed or purified proteins. The impact of host cell proteins on HCV replication and entry could be studied after the development of the HCV replicon system (reviewed in ref. 1; see also ref. 4) and pseudoviral particles (5), respectively. Both systems were originally developed in the human hepatocellular carcinoma cell line Huh7. Randall et al. (1) investigated for the first time the biological relevance of a panel of putative cellular HCV interacting factors, identified in the described experimental systems and the current study, for the complete viral life cycle using a recently developed HCV genotype 2a (J6/JFH1) cell culture system (6), which uses the Huh7-derived highly permissible cell line Huh7.5 (7). After silencing of individual cellular proteins by using experimental RNA interference (RNAi) (Fig. 1A), Huh7.5 cells were infected with J6/JFH1, and the resulting HCV infectious titers were compared (1). Silencing of a total of 26 cellular host proteins modulated production of infectious HCV by at least 3-fold; silencing of 25 of these proteins decreased HCV infectious titers by as much as 42-fold, indicating their supportive role for HCV replication, assembly, release, or viral entry. Although the antiviral effect of silencing HCV interaction partners appeared to be moderate compared with direct silencing of HCV (>230-fold decrease in infectious titer), targeting of the same interaction partners by, e.g., small-molecule inhibitors or specific antibodies might result in increased antiviral efficacy. Thus, antibody-mediated blocking of the HCV coreceptor CD81 was shown to prevent infection with HCV (8) compared with an only 11-fold decrease in infectious titer after silencing (1). Such differences could be explained by the fact that in the experimental setup used (1), host protein expression is influenced not only by silencing efficacy but also by translation levels and turnover of these proteins, and possibly by regulation caused by HCV infection. On the other hand, moderate impacts on the HCV infectious titer after silencing of presumably important HCV interacting partners, such as the cellular peptidases involved in HCV processing (SEC11L1 and HM13), could be caused by redundancy of cellular proteins used during the HCV life cycle. Such phenomena could also explain why 36 putative HCV interacting partners did not substantially influence the release of infectious HCV particles.
Fig. 1.
Impact of host cell factors on production of infectious HCV particles: study design and results reported by Randall et al. (1). (A) Silencing of 62 host cell proteins by experimental RNAi. Small interfering RNAs (siRNAs) were transfected into Huh7.5 cells. Target mRNA is cleaved by cellular RNA-induced silencing complex RISC due to perfect sequence match to bound siRNA (9). Huh7.5 cells were infected with J6/JFH1 HCV viruses at 24, 48, or 72 h after transfection. HCV infectious titers in cell culture supernatants and intracellular HCV RNA levels were determined 48 h after infection. (B) Silencing of seven components of the miRNA biogenesis/RNAi pathway by experimental RNAi and depletion of miR-122. miRNAs such as miR-122 are transcribed as primary miRNAs (pri-miRNAs) and cleaved by Drosha (cofactor DGCR-8) in the nucleus. After nuclear export, the resulting pre-miRNA is processed by Dicer to form an miRNA duplex. Mature single-stranded miRNAs commonly bind their target mRNAs at several sites close to their 3′ end with imperfect match, resulting in assembly of RISCs and translational repression (14). HCV RNA contains two putative miR-122 binding sites in the untranslated regions (15); however, the mode and implications of miR-122 binding are unclear. Components targeted by Randall et al. are indicated by red flashes. Red boxes show the approximate resulting decrease in HCV infectious titers.
Excitingly, Randall et al. (1) discovered that the RNAi machinery exerted a proviral effect on the HCV life cycle because silencing of Dicer and components of the RNA-induced silencing complex RISC (Argonaute proteins EIF2C1–4) decreased HCV infectious titers (Fig. 1B). This adds strong evidence to the hypothesis that mammals, in contrast to plants and invertebrates, might not use RNAi as an innate anti viral defense mechanism (9–11). Previous findings supporting this hypothesis, specifically the absence of siRNAs and RNAi inhibitors during viral infections of mammalian cells (12), were also confirmed in the work of Randall et al. A likely explanation for this proviral effect of RNAi components could be their use in microRNA (miRNA) biogenesis (Fig. 1B) (11, 13, 14), given the demonstrated dependence of HCV on the liver-specific microRNA miR-122 (1, 15). This hypothesis is supported by the finding that silencing of Drosha or DCRG-8, specific for miRNA biogenesis (Fig. 1B), resulted in a similar decrease in the production of infectious HCV particles as silencing of the downstream components common for miRNA biogenesis and RNAi (1).
miRNAs are conserved 21- to 23-nt noncoding RNA molecules regulating gene expression at the posttranscription level in plants and animals (14). Their sheer abundance suggests that interactions between viruses and miRNAs might have evolved. Particularly, herpes viruses express miRNAs to regulate their own and possibly also host gene expression (11, 12). As an RNA virus with a cytoplasmatic life cycle, HCV apparently does not encode miRNAs (1, 12); however, HCV is so far the only virus for which a dependence on an endogenous miRNA has been demonstrated (1, 11, 15). While a direct interaction of miR-122 with the 5′ UTR of HCV seems to be important for HCV genotype 1 replication (15), Randall et al. (1) show the importance of miR-122 for the entire viral life cycle of genotype 2 (Fig. 1B). The mechanism of this interaction remains unknown. Alternatively to a direct role for HCV replication, miR-122 could be sequestered from its targets by physical interaction with HCV genomes; indeed, HCV replication has been shown to increase the expression of genes involved in cholesterol biosynthesis, which are modulated by miR-122 and thought to be of importance for the HCV life cycle (reviewed in ref. 1).
There is a pressing need for identification of drugs against hepatitis C.
Liver-specific factors necessary for the HCV life cycle could be determinants of HCV liver tropism. By detailed miRNA profiling, Randall et al. (1) show the liver-specific miR-122 to be the most predominant miRNA in human liver and in cultured Huh7.5 cells, as previously suggested by studies of human, rat, and mouse liver and Huh7 cells (15–17). Facing the differences observed between miRNA profiles of human liver and Huh7.5 cells (1), it should be emphasized that the currently available HCV cell culture models use a tumor cell line differing from normal hepatocytes. Thus, the biological relevance of results generated in these systems should ultimately be verified in primary hepatocytes and in vivo. Interestingly, analysis of Huh7.5 cells containing an HCV genotype 1b replicon showed that HCV replication differentially regulated the cellular miRNA environment (1), as already demonstrated for the mRNA and protein environment in replicon-containing cells and in vivo (18–23). Microarray and proteomics studies of HCV-infected Huh7.5 cells would complete the picture of this aspect of complex virus–host interactions. Finally, as soon as cell culture systems for the six major HCV genotypes become available, it would be interesting to study genotype-specific interaction with host cell proteins.
Footnotes
The authors declare no conflict of interest.
See companion article on page 12884 in issue 31 of volume 104.
References
- 1.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, et al. Proc Natl Acad Sci USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He Y, Duan W, Tan SL. Drug Discovery Today. 2007;12:209–217. doi: 10.1016/j.drudis.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Pawlotsky JM, Chevaliez S, McHutchison JG. Gastroenterology. 2007;132:1979–1998. doi: 10.1053/j.gastro.2007.03.116. [DOI] [PubMed] [Google Scholar]
- 4.Ng TI, Mo H, Pilot-Matias T, He Y, Koev G, Krishnan P, Mondal R, Pithawalla R, He W, Dekhtyar T, et al. Hepatology. 2007;45:1413–1421. doi: 10.1002/hep.21608. [DOI] [PubMed] [Google Scholar]
- 5.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset FL. J Biol Chem. 2003;278:41624–41630. doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- 6.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, et al. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 7.Blight KJ, McKeating JA, Rice CM. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen BR. Nat Immunol. 2002;3:597–599. doi: 10.1038/ni0702-597. [DOI] [PubMed] [Google Scholar]
- 10.Voinnet O. Nat Rev Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- 11.Cullen BR. Nat Genet. 2006;38(Suppl):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 12.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 13.Cullen BR. Nat Immunol. 2006;7:563–567. doi: 10.1038/ni1352. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 16.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 17.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 18.Fang C, Yi Z, Liu F, Lan S, Wang J, Lu H, Yang P, Yuan Z. Proteomics. 2006;6:519–527. doi: 10.1002/pmic.200500233. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs JM, Diamond DL, Chan EY, Gritsenko MA, Qian W, Stastna M, Baas T, Camp DG, Carithers RL, Jr, Smith RD, et al. J Virol. 2005;79:7558–7569. doi: 10.1128/JVI.79.12.7558-7569.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abe K, Ikeda M, Dansako H, Naka K, Shimotohno K, Kato N. Virus Res. 2005;107:73–81. doi: 10.1016/j.virusres.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Bigger CB, Brasky KM, Lanford RE. J Virol. 2001;75:7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bigger CB, Guerra B, Brasky KM, Hubbard G, Beard MR, Luxon BA, Lemon SM, Lanford RE. J Virol. 2004;78:13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, et al. Proc Natl Acad Sci USA. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]