Abstract
MiaE catalyzes the posttranscriptional allylic hydroxylation of 2-methylthio-N-6-isopentenyl adenosine in tRNAs. The Salmonella typhimurium enzyme was heterologously expressed in Escherichia coli. The purified enzyme is a monomer with two iron atoms and displays activity in in vitro assays. The type and properties of the iron center were investigated by using a combination of UV-visible absorption, EPR, HYSCORE, and Mössbauer spectroscopies which demonstrated that the MiaE enzyme contains a nonheme dinuclear iron cluster, similar to that found in the hydroxylase component of methane monooxygenase. This is the first example of an enzyme from this important class of diiron monooxygenases to be involved in the hydroxylation of a biological macromolecule and the second example of a redox metalloenzyme participating in tRNA modification.
Keywords: EPR spectroscopy, Mössbauer spectroscopy, nonheme dinuclear iron cluster, tRNA modification enzyme
Transfer RNAs (tRNAs) from all organisms contain modified nucleosides that are formed from the four normal nucleosides, adenosine (A), guanosine (G), uridine (U) and cytidine (C). At present, >100 different modified nucleosides have been characterized in tRNAs from various organisms (http://medstat.med.utah.edu/RNAmods). The wobble position (position 34) and the position immediately on the 3′ side of the anticodon (position 37) are frequently modified. Modifications are suggested to introduce conformational variations of the tRNA and to provide specific recognition sites for proteins or nucleic acids (1). They improve the fidelity and efficiency of tRNA in decoding the genome (2). The influence of tRNA modification on reading frame maintenance, on central and intermediary metabolism and on bacterial virulence has been reviewed (3, 4).
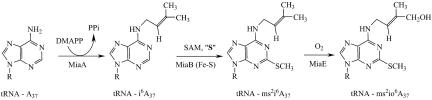
In general, modification reactions are catalyzed by enzymes acting posttranscriptionally on tRNA substrates and are thus an integral part of the tRNA maturation process. The genetic and regulatory properties of the tRNA modifying enzymes as well as the physiological consequences of modification defects have recently been reviewed (5, 6). tRNA modification is thus a major source of fascinating enzymes catalyzing a variety of interesting reactions with very high specificity because the chemical modification is introduced at a single site within a complex substrate (tRNA). Only a small part of these naturally occurring enzymes have been isolated and characterized so far. In particular, redox modifications of tRNAs have been surprisingly scarcely studied as compared with nonredox reactions such as methylations for example. Recently, we discovered and characterized the first, and so far the only, iron-sulfur enzyme involved in tRNA modification (7–9). This enzyme, the product of the miaB gene (Scheme 1), catalyzes a difficult C–H to C–S bond conversion during the thiomethylation of i6A-37 (N-6-isopentenyl adenosine) to ms2i6A-37 (2-methylthio-N-6-isopentenyl adenosine). This nucleoside is found at position 37, next to the anticodon at the 3′ position in almost all eukaryotic and bacterial tRNAs that read codons beginning with uridine except tRNAI.VSer (10). In Salmonella typhimurium, but not in Escherichia coli, for example, it is further modified by hydroxylation during an oxygen-dependent reaction catalyzed by the product of the miaE gene (Scheme 1) (11). The miaE mutant strain has several important phenotypes because it is unable to grow aerobically on succinate, fumarate or malate, whereas a miaA mutant can, suggesting that the bacteria are able to specifically sense the hydroxylation status of the tRNA–isopentenyl group and are growing on the dicarboxylic acids of the citric acid cycle only if this group is hydroxylated (12, 13).
Scheme 1.
Biosynthetic pathway for ms2io6A in S. typhimurium. The enzymes involved in this pathway are: MiaA, MiaB, and MiaE. DMAPP, dimethylallyl diphosphate; SAM, S-adenosylmethionine; S, sulfur atom from MiaB enzyme; PPi, pyrophosphate.
The highly selective incorporation of a single oxygen atom into such a complex macromolecule is a fascinating chemical issue. Whereas the gene (miaE), encoding the hydroxylase responsible for this reaction, was identified in the beginning of the 1990s, the protein has not been investigated yet and is thus the subject of this study. Here, we demonstrate that the MiaE protein is indeed a hydroxylase enzyme containing a carboxylate-bridged nonheme diiron center, as shown by a combination of UV-visible absorption, EPR, Mössbauer and HYSCORE spectroscopies coupled with iron analyses and in vitro enzyme activity assays. Thus, the MiaE protein is a new member of the carboxylate-bridged nonheme diiron protein family, which also includes soluble methane monooxygenase as a prototype (14). This is the first example of an enzyme from this important class of monooxygenases shown to participate in tRNA modification.
Results
Cloning, Expression, and Purification of MiaE from S. typhimurium.
The miaE gene is found as the second gene of a dicistronic operon with two possible translational start points for the gene product. The first AUG start codon is located 11 bp downstream of the stop codon of the first gene, ORF 15.6, of the operon, whereas the second one is located 49 bp downstream of the same stop codon. In between, a stem-loop terminator-like structure is present (12). Even though this feature suggested that translation would be allowed only from the second AUG, we made two constructs to verify whether only a 29,057-Da protein is overexpressed (if the second AUG is used) or a 31,140-Da protein is also overexpressed (if the first AUG is used). The two possible ORFs for the miaE gene were amplified from the genomic DNA of S. typhimurium by PCR and cloned into a pT7-7 vector as previously described (7, 9). The resulting plasmids are named pT7-miaE1 for the first AUG start codon and pT7-miaE2 for the second AUG start codon. The plasmids were then used to transform E. coli BL21(DE3) cells and protein expression was monitored by SDS/gel electrophoresis. It was clear that the observed level of MiaE expression in pT7-miaE1-transformed cells was much lower than that in pT7-miaE2-transformed ones (data not shown). pT7-miaE2 was used to construct a plasmid for expression of an N-terminal His-tagged protein as described (7). This plasmid was named pT7-miaE2H and led to the protein MiaE2H that was further studied here.
The E. coli strain BL21(DE3) was transformed by using the expression vector pT7-miaE2H. Isopropyl-1-thio-β-d-galactopyranoside induction of the transformed E. coli cells resulted in the overproduction of a protein that migrates at ≈30,000 Da on SDS gels and was found in the soluble fraction of cell-free extracts. After the final step of purification, the purity was evaluated by SDS/PAGE to be >95% [see supporting information (SI) Fig. 5]. The apparent molecular mass of MiaE2H determined by analytical gel filtration chromatography (Superdex 75 HR10/30; GE Healthcare, Vélizy, France) is ≈27,000 Da, which indicates that the protein behaves as a monomer in solution (data not shown).
An E. coli Strain Transformed with pT7-miaE2 or pT7-miaE2H Plasmids Produces ms2io6A.
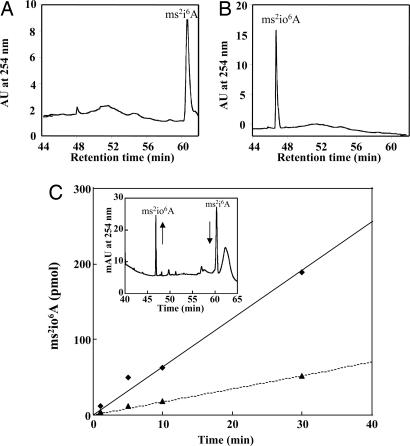
Knowing that E. coli has only the unhydroxylated form of ms2i6A in its tRNAs, we used it as a naturally occurring hydroxylase-deficient microorganism (12). The functionality of the two proteins (MiaE2 and MiaE2H) was assayed in vivo by using E. coli DH5α strain that was transformed with the two plasmids (pT7-miaE2 and pT7-miaE2H). Cells were grown at 37°C in LB medium, and tRNAs were then isolated, digested and their modified nucleosides were analyzed by HPLC, as described (15). Under these conditions and as expected, tRNAs from the control strain (DH5α), which was transformed with the parental pT7-7 lacking miaE plasmid, showed an accumulation of ms2i6A which elutes at ≈60 min with no evidence for the presence of ms2io6A (Fig. 1A). However, all tRNAs isolated from DH5α transformed with pT7-miaE2 or pT7-miaE2H plasmids showed the presence of ms2io6A, which elutes at ≈47 min (Fig. 1B). The identity of ms2i6A and ms2io6A was confirmed by their retention time and UV-visible spectra as described (15). These results demonstrated that MiaE2 and MiaE2H are functional in vivo during the ms2i6A–ms2io6A conversion. Because the introduction of a His tag at the N terminus in MiaE2H had no effect on enzymatic activity in vivo, we decided to work with the His-tagged MiaE2H enzyme in further in vitro experiments.
Fig. 1.
HPLC chromatograms of tRNA hydrolysates. (A) tRNAs were obtained from E. coli DH5α strain. (B) tRNAs were obtained from an in vivo complementation of DH5α strain transformed with pT7-miaE2 or pT7-miaE2H. The identification of ms2i6A and ms2io6A was based on UV-visible spectra (data not shown) and retention times (ms2i6A eluted at ≈60 min and ms2io6A at ≈47 min). (C) ms2io6A production as a function of reaction time and quantity of purified MiaE2H enzyme (♦, 20 μM; ▴, 5 μM). The assay mixture contained 50–100 μg of bulk tRNAs and 0.6 mg of cell-free extracts in 100 mM Tris·HCl (pH 7.5). Reactions were carried out at 37°C. (Inset) The HPLC detection of ms2i6A substrate (elution at 60 min) and ms2io6A, product of the reaction (elution at 47 min) the arrows indicate the decrease of the substrate and the increase of the product.
Metal Content and Amino Acid Analysis.
UV-Visible absorption bands in the optical spectrum of pure MiaE2H (see below) suggested the presence of a metal ion. Metal analysis revealed that MiaE2H is an iron-containing protein. Fe quantitation based on Fe assays and quantitative amino acid analysis of pure MiaE2H led to the value of ≈2.1 ± 0.2 mol of iron per mol of MiaE2H. No other metals were detected by atomic absorption. The same value was obtained by using MiaE2 protein. We noted that the Bradford protein assay overestimates the concentration of MiaE2H by a factor of ≈1.16.
Conversion of ms2i6A to ms2io6A Catalyzed by MiaE2H Protein in Vitro.
To investigate the enzymatic conversion of ms2i6A to ms2io6A in vitro, we established standard reaction conditions for assaying the purified MiaE2H protein. Typically the reaction mixture contained MiaE2H, total tRNAs, and cell free extracts obtained from DH5α strain, in a volume of 100 μl of 50 mM Tris·HCl (pH 7.5). The reaction was carried out at 37°C for 60 min in air. The tRNA substrate was total tRNAs obtained from E. coli DH5α, which contains ms2i6A, the reaction substrate, but no ms2io6A, the reaction product. After 60 min incubation in vitro, tRNAs were recovered by phenol extraction and ethanol precipitation and then completely hydrolyzed by nuclease P1 and alkaline phosphatase. The resulting hydrolysate was analyzed by HPLC and the results are summarized in Table 1. From the reaction mixtures containing the tRNA substrate, the purified MiaE2H enzyme and cell-free extracts altogether, the formation of the ms2io6A product was observed, thus reflecting selective in vitro tRNA hydroxylation. No ms2io6A could be detected when the MiaE2H protein or cell-free extracts were omitted from the reaction mixture. No ms2io6A could be detected either when cell-free extracts were used after extensive dialysis. In this case, the activity of MiaE2H protein was partially restored when NADPH was added to the reaction mixture. Substitution of hydrogen peroxide (H2O2) for cell free extracts in the reaction mixture allowed the conversion of ms2i6A to ms2io6A. Under the assay conditions described above and as shown in Fig. 1C, the production of ms2io6A was linear for at least half an hour, and the rate of reaction was proportional to MiaE2H concentration.
Table 1.
Assay conditions for in vitro conversion of ms2i6A to ms2io6A catalyzed by MiaE2H
| Experiments | tRNA | MiaE2H | Cell-free extracts | Dialyzed cell-free extracts | NADPH | H2O2 | ms2io6A |
|---|---|---|---|---|---|---|---|
| 1 | + | + | − | ||||
| 2 | + | + | − | ||||
| 3 | + | + | + | + | |||
| 4 | + | + | + | − | |||
| 5 | + | + | + | + | + | ||
| 6 | + | + | + | + |
The hydroxylase activity was assayed by incubating ≈50–100 μg of tRNAs with (+) or without (blank) 50 μM purified MiaE2H and 30 μl of cell-free extracts at 20 mg/ml in aerated buffer (Experiments 1–3). In Experiments 4 and 5, the cell-free extracts were dialyzed and assayed without (Experiment 4) or with (Experiment 5) 1 mM NADPH. In Experiment 6, 20 mM H2O2 was used in place of cell-free extracts. The last column indicates whether the experiment resulted (+) or not (−) in the production of ms2io6A as a probe for tRNA hydroxylation.
Spectroscopic Characterization of MiaE2H.
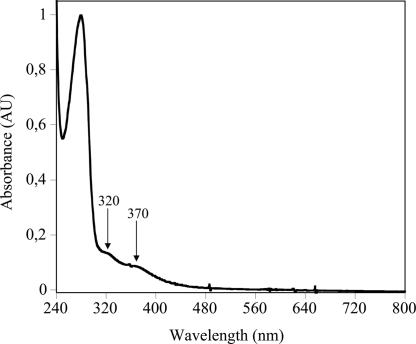
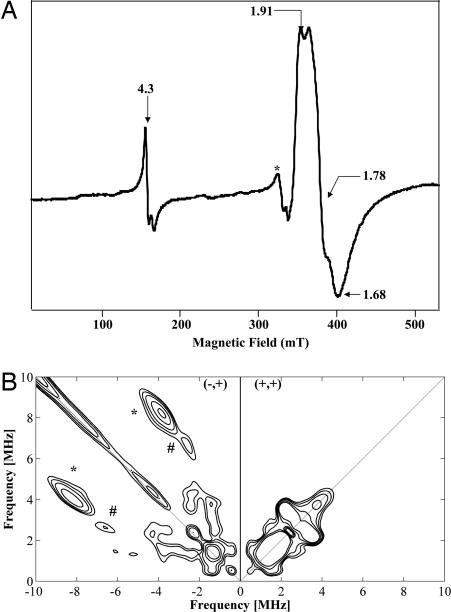
The light absorption spectrum of the as-isolated MiaE2H protein is shown in Fig. 2. In addition to the band at 280 nm (ε280 ≈ 60,000 M−1 cm−1) corresponding to protein absorption, the spectrum displays two additional bands at 320 and 370 nm associated with the presence of iron in the protein. The optical properties described here are comparable with those of the active site of the R2 subunit of aerobic ribonucleotide reductase from E. coli, phenol hydroxylase from Pseudomonas sp strain CF 600 and toluene-2-monooxygenase from Burkholderia cepacia G4 proteins known to contain μ-oxo-bridged diiron clusters with primary ligation sphere consisting of oxygen and nitrogen ligands (16–18). Similarity to these enzymes is further supported by EPR spectroscopy. In the low-field region of Fig. 3A the EPR spectrum displays a weak g = 4.3 signal associated with mononuclear adventitiously bound ferric ions in a rhombic environment. The intensity of the g = 4.3 signal varies from one preparation to another. In the high-field region of the spectrum of Fig. 3A, all g values of the signal are <g = 2 (g = 1.91, 1.78, 1.68) with gav = 1.83. This rhombic EPR spectrum is characteristic of a Zeeman split S = 1/2 ground state of an antiferromagnetically coupled mixed-valent [FeII–FeIII] center with an S = 5/2 FeIII and an S = 2 FeII. This type of mixed-valent cluster accounted for 27 ± 5% of the total cluster based on double integration of the gav = 1.83 signal. Chemical reduction of MiaE2H protein with buffered sodium dithionite in the presence of phenazine methosulfate as a mediator resulted in a slight increase in the intensity of the gav = 1.83 signal. At maximal conversion, 50% of dinuclear iron clusters were in the mixed-valent state. When methyl viologen was used as a mediator, a new EPR signal appeared at g ≈ 15 (data not shown), arising from an integer spin (S = 4) system and indicating the formation of a fully reduced diferrous center (19). This redox state is under investigation.
Fig. 2.
UV-visible absorption spectrum of purified MiaE2H. The buffer was 100 mM Tris·HCl (pH 8), containing 30 mM NaCl and 5% glycerol. The sample concentration was 0.6 mg/ml. Arrows indicate the 320-nm and 370-nm iron charge transfer bands.
Fig. 3.
Characterization of MiaE2H protein by EPR spectroscopy. (A) X-band EPR spectrum of the MiaE2H protein (1.9 mM) in 100 mM Tris·HCl (pH 8) containing 30 mM NaCl and 5% glycerol. Experimental conditions: temperature 4 K, microwave power 1 mW, modulation amplitude 10 mT. The weak signal indicated by a star at g ≈ 2.00 is contaminating Cu2+. (B) Low-frequency region of the X-band HYSCORE spectrum of the mixed-valent state [FeII–FeIII] of MiaE2H protein (1.9 mM) in 100 mM Tris·HCl (pH 8) containing 30 mM NaCl and 5% glycerol. Experimental conditions: magnetic field 3,900 G (g = 1.778), frequency 9.7 GHz, and temperature 4 K.
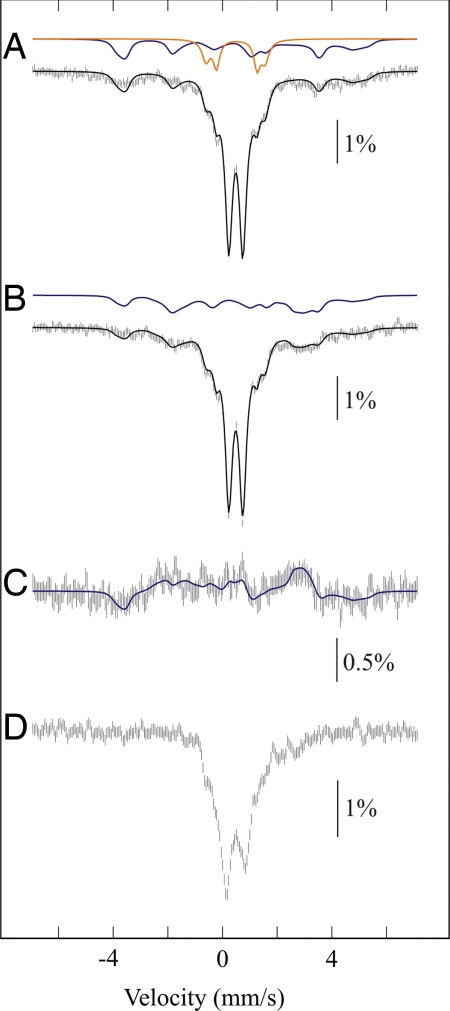
The presence of a diiron cluster was further supported by Mössbauer spectroscopy. Fig. 4 displays the Mössbauer spectra of the MiaE2H enzyme isolated from bacteria grown in the presence of 57Fe (as described in Materials and Methods), recorded at 4.2 K under a small magnetic field applied both parallel (Fig. 4A) or perpendicular (Fig. 4B) to the γ-rays. Spectra were also recorded at 77 (Fig. 4D) and 120 K (not shown) without applied field. These spectra clearly indicate the presence of several iron entities that can be accounted for in the context of dinuclear centers as suggested by the EPR experiments. The spectra are dominated by a quadrupole doublet, the parameters of which (δ ≈ 0.50 (1) mm·s−1, ΔEQ = 0.51 (1) mm·s−1) clearly correspond to high-spin ferric ions. This doublet may be associated with two indistinguishable antiferromagnetically coupled ferric ions of a single diamagnetic S = 0 species. It accounts for ≈54 (3)% of total Fe. Interestingly at 77 K (Fig. 4D), this doublet experienced a significant broadening and this effect was further increased at 120 K. This behavior can be explained by the contribution of thermally accessible excited spin states of diferric species. This is indicative of a system which is moderately antiferromagnetically coupled as has been observed for hydroxo-bridged entities (20, 21). The presence of a hydroxo-bridged diferric unit would be consistent with the low ΔEQ value (22, 23). This major doublet is flanked by a pair of nearly symmetric shoulders weighing altogether ≈16 (3)% of total Fe. They are associated with two equally intense quadrupole doublets: δ = 0.54 (2) mm·s−1 with ΔEQ = 1.49 (2) mm·s−1 and δ = 0.49 (2) mm·s−1 with ΔEQ = 2.16 mm·s−1 (2). The large quadrupole splittings of theses ions are the signature of a μ-oxo-diferric system (24). Finally, a broad magnetic contribution, reminiscent of those observed for enzymes with a localized mixed-valent [FeII–FeIII] center (MMOH, uteroferrin and myo-inositol oxygenase), was observed (20, 21, 25), this contribution could be fitted as well. However, because the limited contribution of this species [30 (5)%] and the spread of its spectrum over a wide velocity range, the Mössbauer parameters cannot be reliably determined with a high enough accuracy. The simulated curves shown in Fig. 4 have been obtained from the spin Hamiltonian parameters reported for the mixed-valent center of myo-inositol oxygenase complexed with myo-inositol (MIOX-MI, see Table 2) (25). The excellent agreement between the experimental and simulated spectra is further shown in the difference spectrum obtained by subtracting the parallel from the perpendicular spectrum (Fig. 4C), in which the contributions of the diamagnetic components are cancelled out. A precise quantitation of the various dinuclear entities is difficult owing to the fact the adventitious high spin ferric center detected in EPR was not distinguished by Mössbauer spectroscopy. It could contribute a tenth of the major quadrupole doublet if it were a fast relaxing high-spin ferric center. Alternatively it would overlap the magnetic spectrum of the mixed-valent species. To summarize, these Mössbauer studies point to the coexistence of three forms of a diiron center in the as-isolated MiaE2H enzyme: a major probably hydroxo-bridged diferric species, a minor μ-oxo-bridged diferric species and a mixed-valent [FeII–FeIII] species.
Fig. 4.
Mössbauer spectra of MiaE2H (1.9 mM) in 100 mM Tris·HCl (pH 8) containing 30 mM NaCl and 5% glycerol. Experimental conditions: spectra were recorded at 4.2 K in a magnetic field of 0.60 mT applied parallel to the γ beam (A), or 0.22 mT applied perpendicular to the γ beam (B), or at 77K and zero applied field (D). Spectrum C is obtained by subtraction (A − B). The solid black lines are spin-Hamiltonian simulations, generated by using the parameters described below. The solid colored lines are contributions from the oxodiferric clusters (orange) and the mixed-valence [FeII–FeIII] clusters (blue). Contribution from the majority diferric species (central doublet) is not shown.
Table 2.
Comparison of spectroscopic properties of the diiron center in MiaE2H, methane monooxygenase, myo-inositol oxygenase (MIOX-MI), R2 subunit of aerobic ribonucleotide reductase, stearoyl carrier Δ9-desaturase protein, and uteroferrin
| Diiron center | g values | δ (ΔEQ), mm/s FeIII | δ (ΔEQ), mm/s FeII/FeIII | Ref. |
|---|---|---|---|---|
| Mixed-valent form [FeII–FeIII] | ||||
| MiaE2H | 1.91, 1.78, 1.68 | This work | ||
| MIOX-MI | 1.95, 1.81, 1.81 | 0.49 (−1.11) | 1.12 (+2.68) | 25 |
| Ufr | 0.54 (−1.85) | 1.24 (+2.68) | 20 | |
| MMOH | 21 | |||
| Oxidized forms [FeIII–FeIII] | ||||
| MiaE2H (minor) | 0.52 (1.49) | 0.48 (2.16) | This work | |
| RNR | 0.55 (−1.62) | 0.45 (−2.44) | 52 | |
| Ufo | 0.55 (1.69) | 0.48 (2.17) | 20 | |
| MiaE (major) | 0.49 (0.51) | |||
| Δ9D (minor) | 0.49 (0.72) | 23 | ||
| MMOH | 0.50 (0.87) | 0.51 (1.16) | 21 | |
As shown by EPR and Mössbauer spectroscopies, the as-isolated MiaE2H protein contains up to 25% to ≈30% of the diiron cluster in the mixed-valent [FeII–FeIII] state. The latter was analyzed by HYSCORE spectroscopy to further characterize the coordination sphere of the diiron center. One of the main advantages of this two-dimensional pulsed EPR technique resides in its ability to distinguish three types of nuclei: the strongly (|aN|/2 > νN) and weakly (|aN|/2 < νN) coupled ones and the “distant” nuclei, which are characterized by very low hyperfine couplings (26, 27). In the latter case, the corresponding peaks lie on the diagonal of the (+, +) quadrant, whereas the strongly and weakly coupled nuclei appear in the (−,+) and the (+, +) quadrants, respectively (28). In Fig. 3B, the HYSCORE spectrum of the mixed-valent state shows a symmetrical set of features in the (−, +) quadrant. These peak patterns and their positions are characteristic of at least one strongly coupled nitrogen atom (an I = 1 nucleus with quadrupolar coupling) (29). From the position of the so-called double quanta-double quanta correlation peaks (asterisk in Fig. 3B) at (9.2; −4.8) and (4.8; −9.2) MHz, it is possible to obtain a good estimation of the isotopic hyperfine coupling |aN| by using the relationship νdq± = 2[((νN ± aN/2)2 + K2(3 + η2)]1/2, where K is the quadrupole coupling constant, and η is the asymmetry parameter (30). The value obtained (|aN| = 6.4 MHz) is characteristic of a nitrogen atom directly bound to the metal center. Upon close examination, a pair of peaks at (6.9; −2.8) and (2.8; −6.9) MHz (# in Fig. 3B) could be attributed to a double quanta–double quanta correlation from another strongly coupled nitrogen with |aN| equal to 4.1 MHz, indicative of the presence of a second nitrogen ligand. Observation of hyperfine coupling of two nitrogens from histidine ligands in similar systems was reported previously (31). Hyperfine values obtained here are compatible with those obtained for the [FeII–FeIII] clusters of the cryoreduced R2 protein (3.16 and 7.31 MHz) and of the cryoreduced stearoyl carrier Δ9-desaturase protein (3.34 and 9.1 MHz) (31). Similar values were obtained for both the His-tagged and untagged forms of MiaE2.
Discussion
The results presented here provide the first and complete characterization of the recombinant MiaE2H protein from S. typhimurium, a monooxygenase involved in specific tRNA modification. MiaE catalyzes an allylic hydroxylation converting ms2i6A to ms2io6A at position 37 within the tRNA substrate (Scheme 1). In this article, we have clearly established that MiaE2H belongs to the class of carboxylate-bridged nonheme diiron enzymes, the most prominent members of which are MMO hydroxylase and the R2 RNR subunit (32, 33).
This type of diiron center can exist in different forms. Indeed, each iron atom can accommodate either the +2 or the +3 state and the two ions may be bridged by either an oxo ligand or corresponding protonated forms, OH or OH2. Moreover, it is not unusual that such diiron enzymes exist as a mixture of such forms (34). Detailed spectroscopic investigation of pure preparations of the as-isolated MiaE2H protein revealed the presence of three different forms of the dinuclear site: (i) an EPR-silent hydroxo-bridged FeIII–OH–FeIII center, with characteristic Mössbauer spectroscopic properties; (ii) an EPR-silent oxo-bridged FeIII–O–FeIII center, characterized by oxo-to-iron charge transfer bands in the UV-visible spectrum and by large quadrupole splittings of the corresponding doublets in the Mössbauer spectrum; (iii) a hydroxo-bridged FeII–OH–FeIII mixed-valent center, remarkably stable and displaying a characteristic rhombic signal at g values <2 in the EPR spectrum as well as a broad magnetic signal in the Mössbauer spectrum, in agreement with a S = ½ ground state. Using this EPR signal, we showed by HYSCORE spectroscopy that each iron is bound by a nitrogen atom (from histidine), as is generally observed in this class of enzyme (33, 34). By analogy, we suspect these histidines to belong to the EXXH (His-139 or His-140 and His-222) conserved sequence motifs (see below) and make the hypothesis that the other ligands are carboxylate groups from glutamate and aspartate.
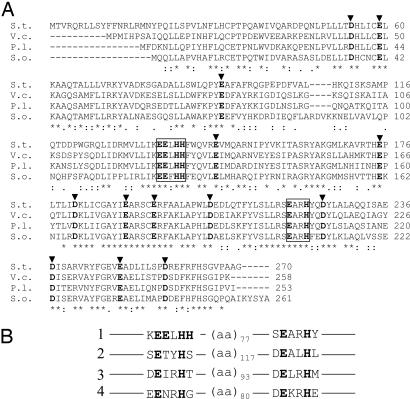
A BLAST search revealed limited sequence homologies of MiaE with the known diiron enzymes. Nevertheless, it showed that MiaE contains two conserved copies of the primary sequence motif EXXH separated by ≈77 aa residues at least (Scheme 2B), which is a hallmark of this large group of enzymes (33–38) in which one can find not only the so-called diiron monooxygenases, such as methane monooxygenase, phenol hydroxylase, toluene monooxygenase, and alkane ω-hydroxylase, but also desaturases such as stearoyl-ACP (acyl carrier protein) desaturase, which inserts a double bond into a protein-bound fatty acid (39), and the R2 subunit of ribonucleotide reductase, which catalyzes an intramolecular one-electron oxidation of tyrosine (40). The amino acid residues Glu (E) and His (H) provide the ligands to the diiron center in all these enzymes (33, 38, 41). Moreover, using the BLAST search algorithm, we were able to identify many putative MiaE proteins from different organisms, showing strict conservation of the EXXH motif for distantly related species (Scheme 2A). It is therefore likely that the glutamate and histidine residues of these motifs are the actual ligands of the iron pair in MiaE.
Scheme 2.
Multiple sequence alignment of MiaE proteins. (A) Amino acid alignments of MiaE proteins from S. typhimurium (S.t, Q08015) and putative tRNA ms2io6A hydroxylases from Vibrio cholerae (V.c, Q9KQT8); Photorhabdus luminescens subsp. Laumondii (P.l, Q7MB84) and Shewanella oneidensis (S.o, Q8CX43). The alignments were performed with the ClustalW program. Totally conserved amino acid residues are indicated by asterisks. The conserved EXXH motifs are framed. Black arrowheads indicate amino acid residues that could be potential ligands for diiron center. The numbers refer to amino acid residues for S. typhimurium protein. (B) Primary sequence homologies of diiron carboxylate proteins. 1, MiaE from S. typhimurium (Q08015); 2, R2 subunit of ribonucleotide reductase from E. coli (P69924); 3, hydroxylase component of methane mono oxygenase from Methylosinus trichosporium (P27353); 4, stearoyl carrier Δ9-desaturase protein from Ricinus communis (P22337).
The similarity of MiaE with methane monooxygenase (MMO), a prototype of this class of enzymes, is likely to extend to its mechanism of action. MMO catalyzes the insertion of one oxygen atom from dioxygen into its substrate (methane), whereas the second atom ends up as a molecule of water (42). This reaction was extensively analyzed and shown to involve a two-electron reductive activation of the dioxygen molecule and to require a source of electrons. In the case of MMO, the electrons are provided by NADH and are transferred to the active site through a specific reductase (32, 43). Expression of MiaE2H in E. coli provides it with the ability to produce ms2io6A, indicating that E. coli contains reductase activities that can be used by MiaE2H for oxygen activation. This is further confirmed by in vitro assays showing that ms2i6A-to-ms2io6A conversion can be achieved by incubating pure MiaE2H and the tRNA substrate with E. coli cell-free extracts under air. More recently, a different mechanism was discovered in the case of myo-inositol oxygenase (44), another diiron enzyme with a slightly different active site, which activates dioxygen through its mixed-valent FeII–FeIII state (25, 45). The sequence homology of MiaE with the hydroxylase component of MMO and the fact that it can function in vitro with hydrogen peroxide as the oxidant, in the absence of a source of electrons, strongly suggests that MiaE operates in a similar way to MMOH through a two-electron O2 reductive activation mechanism. However, as MiaE2H preparations contain a mixture of diiron sites, as discussed above, it is still unknown whether the active form is hydroxo- or oxo-bridged. This issue, as well as the functionality of the mixed-valent form of the center will be addressed in future studies.
Diiron monooxygenases are involved in the oxidation of a wide variety of substrates. Intriguingly, there are no examples of such enzymes for the oxidation of biological macromolecules. In all cases reported so far, the substrates are low-molecular-weight compounds: methane, alkanes, aromatics, alkenes, and myo-inositol. On the other hand, a number of macromolecules, such as proteins or nucleic acids, are known to be extensively chemically modified through controlled and selective hydroxylation reactions catalyzed by important monooxygenases. For example, prolyl- and lysyl hydroxylases are involved in the posttranslational oxidation of collagen. Similarly, prolyl- and asparaginyl hydroxylases are involved in the posttranslational oxidation of the hypoxia-inducible transcription factor (HIF). Protein demethylation, occurring in histones, is initiated by the hydroxylation of the methyl group to be removed (46). DNA is also subject to hydroxylation, for example during the conversion of methylated adenine to adenine, a reaction catalyzed by the protein AlkB and involved in DNA repair (47–49). It is remarkable that all these reactions are catalyzed by Fe(II)- and 2-oxoglutarate-dependent monoiron dioxygenases. Thus, we conclude that MiaE is a unique diiron monooxygenase as it is the first member of this family to achieve the direct and selective hydroxylation of a biological macromolecule, here a tRNA, and thus expands the range of natural substrates for this family. This raises many interesting questions regarding how the active site of MiaE controls the transfer of an activated oxygen atom to the dimethylallyl group of adenine-37 of a tRNA substrate. This, in particular, will require the determination of the three-dimensional structure of MiaE2H, both alone and in complex with the tRNA substrate. This is currently under investigation. Finally, this study provides an additional illustration of the richness of the redox chemistry used for tRNA modification. The first reported metalloenzyme involved in tRNA modification was MiaB, an iron–sulfur protein (50, 51). MiaE, which uses a nonheme diiron center, is the second one.
Considering that inactivation of the miaE gene gives rise to important phenotypes associated with iron metabolism and aerobic growth (3), further studies should address the question of the biological significance of the hydroxylation reaction catalyzed by MiaE.
Materials and Methods
Strains.
E. coli DH5α was used for routine DNA manipulations and as a naturally occurring hydroxylase-deficient strain. E. coli BL21(DE3) was used to produce the recombinant protein MiaE.
Cloning of the miaE Gene.
The ORF encoding the MiaE protein was PCR amplified by using S. typhimurium genomic DNA, Pwo polymerase (Roche, Indianapolis, IN). Full experimental details are provided in SI Materials and Methods.
Miscellaneous Methods.
Protocols and references for expression, purification, in vivo and in vitro MiaE activity, protein determination assays, SDS/PAGE, and iron determination in protein preparations are provided in SI Materials and Methods.
Preparation of MiaE2H Samples.
Parallel Mössbauer and EPR samples of native MiaE2H were prepared aerobically. The MiaE2H used for these studies was purified from bacteria grown in minimal medium (M9) containing 0.1 mM57Fe as the only source of iron. Chemical reduction of the as-isolated MiaE2H was carried out under anaerobic conditions.
Spectroscopic Measurements.
Full experimental details for EPR, Mössbauer, and HYSCORE spectrocopies are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Glenn R. Bjork for helpful discussions and Gunhild Layer for help with preparing the figures.
Abbreviation
- MIOX-MI
myo-inositol oxygenase complexed with myo-inositol.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704338104/DC1.
References
- 1.Agris PF. Prog Nucleic Acid Res Mol Biol. 1996;53:79–129. doi: 10.1016/s0079-6603(08)60143-9. [DOI] [PubMed] [Google Scholar]
- 2.Curran JF. Modified Nucleosides in Translation. Washington, DC: Am Soc Microbiol; 1998. [Google Scholar]
- 3.Bjork GR, Durand JM, Hagervall TG, Leipuviene R, Lundgren HK, Nilsson K, Chen P, Qian Q, Urbonavicius J. FEBS Lett. 1999;452:47–51. doi: 10.1016/s0014-5793(99)00528-1. [DOI] [PubMed] [Google Scholar]
- 4.Farabaugh PJ, Bjork GR. EMBO J. 1999;18:1427–1434. doi: 10.1093/emboj/18.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjork GR, Rasmusen T. Links Between tRNA Modification and Modified Nucleosides as Tumor Markers. Washington, DC: Am Soc Microbiol; 1998. [Google Scholar]
- 6.Winkler ME. Genetic and Regulation of Base Modification in the tRNA and rRNA of Prokaryotes and Eukaryotes. Washington, DC: Am Soc Microbiol; 1998. [Google Scholar]
- 7.Pierrel F, Bjork GR, Fontecave M, Atta M. J Biol Chem. 2002;277:13367–13370. doi: 10.1074/jbc.C100609200. [DOI] [PubMed] [Google Scholar]
- 8.Pierrel F, Douki T, Fontecave M, Atta M. J Biol Chem. 2004;279:47555–47563. doi: 10.1074/jbc.M408562200. [DOI] [PubMed] [Google Scholar]
- 9.Pierrel F, Hernandez HL, Johnson MK, Fontecave M, Atta M. J Biol Chem. 2003;278:29515–29524. doi: 10.1074/jbc.M301518200. [DOI] [PubMed] [Google Scholar]
- 10.Grosjean H, Nicoghosian K, Haumont E, Soll D, Cedergren R. Nucleic Acids Res. 1985;13:5697–5706. doi: 10.1093/nar/13.15.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck M, Ames BN. Cell. 1984;36:523–531. doi: 10.1016/0092-8674(84)90245-9. [DOI] [PubMed] [Google Scholar]
- 12.Persson BC, Bjork GR. J Bacteriol. 1993;175:7776–7785. doi: 10.1128/jb.175.24.7776-7785.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Persson BC, Olafsson O, Lundgren HK, Hederstedt L, Bjork GR. J Bacteriol. 1998;180:3144–3151. doi: 10.1128/jb.180.12.3144-3151.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopp DA, Lippard SJ. Curr Opin Chem Biol. 2002;6:568–576. doi: 10.1016/s1367-5931(02)00366-6. [DOI] [PubMed] [Google Scholar]
- 15.Gehrke CW, Kuo KC. J Chromatogr. 1989;471:3–36. doi: 10.1016/s0021-9673(00)94152-9. [DOI] [PubMed] [Google Scholar]
- 16.Petersson L, Graslund A, Ehrenberg A, Sjoberg BM, Reichard P. J Biol Chem. 1980;255:6706–6712. [PubMed] [Google Scholar]
- 17.Cadieux E, Vrajmasu V, Achim C, Powlowski J, Munck E. Biochemistry. 2002;41:10680–10691. doi: 10.1021/bi025901u. [DOI] [PubMed] [Google Scholar]
- 18.Newman LM, Wackett LP. Biochemistry. 1995;34:14066–14076. doi: 10.1021/bi00043a012. [DOI] [PubMed] [Google Scholar]
- 19.Pikus JD, Studts JM, Achim C, Kauffmann KE, Munck E, Steffan RJ, McClay K, Fox BG. Biochemistry. 1996;35:9106–9119. doi: 10.1021/bi960456m. [DOI] [PubMed] [Google Scholar]
- 20.Sage JT, Xia YM, Debrunner PG, Keough DT, Dejersey J, Zerner B. J Am Chem Soc. 1989;111:7239–7247. [Google Scholar]
- 21.Fox BG, Hendrich MP, Surerus KK, Andersson KK, Froland WA, Lipscomb JD, Munck E. J Am Chem Soc. 1993;115:3688–3701. [Google Scholar]
- 22.Dewitt JG, Bentsen JG, Rosenzweig AC, Hedman B, Green J, Pilkington S, Papaefthymiou GC, Dalton H, Hodgson KO, Lippard SJ. J Am Chem Soc. 1991;113:9219–9235. [Google Scholar]
- 23.Shu LJ, Broadwater JA, Achim C, Fox BG, Munck E, Que L. J Biol Inorg Chem. 1998;3:392–400. [Google Scholar]
- 24.Kurtz DM. Chem Rev. 1990;90:585–606. [Google Scholar]
- 25.Xing G, Hoffart LM, Diao Y, Prabhu KS, Arner RJ, Reddy CC, Krebs C, Bollinger JM., Jr Biochemistry. 2006;45:5393–5401. doi: 10.1021/bi0519607. [DOI] [PubMed] [Google Scholar]
- 26.Samoilova RI, Kolling D, Uzawa T, Iwasaki T, Crofts AR, Dikanov SA. J Biol Chem. 2002;277:4605–4608. doi: 10.1074/jbc.C100664200. [DOI] [PubMed] [Google Scholar]
- 27.Gambarelli S, Luttringer F, Padovani D, Mulliez E, Fontecave M. Chembiochem. 2005;6:1960–1962. doi: 10.1002/cbic.200500182. [DOI] [PubMed] [Google Scholar]
- 28.Schweiger A, Jeschke G. Principles of Pulse Electron Paramagnetic Resonance. Oxford: Oxford Univ Press; 2001. [Google Scholar]
- 29.Maryasov AG, Bowman MK. J Phys Chem B. 2004;108:9412–9420. [Google Scholar]
- 30.Tyryshkin AM, Dikanov SA, Reijerse EJ, Burgard C, Huttermann J. J Am Chem Soc. 1999;121:3396–3406. [Google Scholar]
- 31.Davydov R, Behrouzian B, Smoukov S, Stubbe J, Hoffman BM, Shanklin J. Biochemistry. 2005;44:1309–1315. doi: 10.1021/bi048599t. [DOI] [PubMed] [Google Scholar]
- 32.Baik MH, Newcomb M, Friesner RA, Lippard SJ. Chem Rev. 2003;103:2385–2419. doi: 10.1021/cr950244f. [DOI] [PubMed] [Google Scholar]
- 33.Nordlund P, Eklund H. Curr Opin Struct Biol. 1995;5:758–766. doi: 10.1016/0959-440x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 34.Kurtz DM. J Biol Inorg Chem. 1997;2:159–167. [Google Scholar]
- 35.Nordlund P, Sjoberg BM, Eklund H. Nature. 1990;345:593–598. doi: 10.1038/345593a0. [DOI] [PubMed] [Google Scholar]
- 36.Nordlund P, Eklund H. J Mol Biol. 1993;232:123–164. doi: 10.1006/jmbi.1993.1374. [DOI] [PubMed] [Google Scholar]
- 37.Rosenzweig AC, Frederick CA, Lippard SJ, Nordlund P. Nature. 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 38.Rosenzweig AC, Nordlund P, Takahara PM, Frederick CA, Lippard SJ. Chem Biol. 1995;2:409–418. [PubMed] [Google Scholar]
- 39.Fox BG, Lyle KS, Rogge CE. Acc Chem Res. 2004;37:421–429. doi: 10.1021/ar030186h. [DOI] [PubMed] [Google Scholar]
- 40.Fontecave M, Nordlund P, Eklund H, Reichard P. Adv Enzymol Relat Areas Mol Biol. 1992;65:147–183. doi: 10.1002/9780470123119.ch4. [DOI] [PubMed] [Google Scholar]
- 41.Lindqvist Y, Huang W, Schneider G, Shanklin J. EMBO J. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- 42.Merkx M, Kopp DA, Sazinsky MH, Blazyk JL, Muller J, Lippard SJ. Angew Chem Int Ed. 2001;40:2782–2807. doi: 10.1002/1521-3773(20010803)40:15<2782::AID-ANIE2782>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 43.Wallar BJ, Lipscomb JD. Chem Rev. 1996;96:2625–2657. doi: 10.1021/cr9500489. [DOI] [PubMed] [Google Scholar]
- 44.Xing G, Diao Y, Hoffart LM, Barr EW, Prabhu KS, Arner RJ, Reddy CC, Krebs C, Bollinger JM., Jr Proc Natl Acad Sci USA. 2006;103:6130–6135. doi: 10.1073/pnas.0508473103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown PM, Caradoc-Davies TT, Dickson JM, Cooper GJ, Loomes KM, Baker EN. Proc Natl Acad Sci USA. 2006;103:15032–15037. doi: 10.1073/pnas.0605143103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider J, Shilatifard A. ACS Chem Biol. 2006;1:75–81. doi: 10.1021/cb600030b. [DOI] [PubMed] [Google Scholar]
- 47.Mishina Y, Chen LX, He C. J Am Chem Soc. 2004;126:16930–16936. doi: 10.1021/ja045066z. [DOI] [PubMed] [Google Scholar]
- 48.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 49.Yu B, Edstrom WC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF. Nature. 2006;439:879–884. doi: 10.1038/nature04561. [DOI] [PubMed] [Google Scholar]
- 50.Fontecave M, Ollagnier-de-Choudens S, Mulliez E. Chem Rev. 2003;103:2149–2166. doi: 10.1021/cr020427j. [DOI] [PubMed] [Google Scholar]
- 51.Hernandez HL, Pierrel F, Elleingand E, Garcia-Serres R, Huynh BH, Johnson MK, Fontecave M, Atta M. Biochemistry. 2007;46:5140–5147. doi: 10.1021/bi7000449. [DOI] [PubMed] [Google Scholar]
- 52.Lynch JB, Juarez-Garcia C, Munck E, Que L., Jr J Biol Chem. 1989;264:8091–8096. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.