Abstract
The TrkC/NT-3 receptor/ligand pair is believed to be part of the classic neurotrophic theory claiming that neuronal death occurs by default when neurotrophic factors become limited, through loss of survival signals. Here, we show that TrkC is a dependence receptor and, as such, induces caspase-dependent apoptotic death in the absence of NT-3 in immortalized cells, a proapoptotic activity inhibited by the presence of NT-3. This proapoptotic activity of TrkC relies on the caspase-mediated cleavage of the intracellular domain of TrkC, which permits the release of a proapoptotic fragment. This fragment induces apoptosis through a caspase-9-dependent mechanism. Finally, we show that the death of dorsal root ganglion (DRG) neurons provoked by NT-3 withdrawal is inhibited when TrkC-proapoptotic activity is antagonized. Thus, the death of neurons upon disappearance of NT-3 is not only due to a loss of survival signals but also to the active proapoptotic activity of the unbound TrkC dependence receptor.
Keywords: neurotrophin-3, sensory neurons, programmed cell death, tyrosine kinase
The classic neurotrophic theory usually proposes that neuronal survival depends on neurotrophic factors, such as neurotrophins (1, 2). This theory also claims that death triggered when these neurotrophic factors become limited is due to a loss of survival signals (3). Neurotrophins include NGF, BDNF, NT-3, and NT-4 (2). These proteins have been shown to be crucial for the development of the nervous system, especially by controlling the massive developmental loss of neurons that are produced in excess and that fail to adequately connect their targets. The current neurotrophic model holds that the main neurotrophin receptors, TrkA, TrkB, and TrkC, generate survival signals via the PI3K/Akt and Ras/MEK/MAPK pathways upon neurotrophin binding (3). This binding is thought to inhibit the naturally occurring apoptotic death of neurons. However, a weakness in this theory is that the molecular nature of the “default apoptotic state” of the developing neurons is not understood. One mechanism could be that a death signal is actively generated in the neurons. When bound by the ligand, death receptors of the tumor necrosis receptor family trigger caspase activation and death of many cell types, but there is little evidence of their involvement in the nervous system. However, recent observations support that, depending on the availability of the ligand, some receptors initiate two completely opposite signaling pathways: in the presence of ligand, these receptors transduce a positive signal of differentiation, guidance, or survival, whereas in the absence of ligand, they induce an active process of apoptotic cell death. These receptors, called dependence receptors, include p75ntr, DCC (deleted in colorectal cancer), UNC5H, Patched, Neogenin, and the tyrosine kinase receptor RET (4–10). The proapoptotic activity of these receptors, observed in the absence of their respective ligand, has been speculated to be important to dictate the adequate territories of neuron migration or localization during the development of the nervous system and to inhibit tumor growth in adult. This activity has been exemplified in vivo with the dependence receptor Patched and the survival of neuroepithelial cells in the developing spinal cord (4) as well as for the netrin-1 receptors DCC and/or UNC5H in colorectal tumorigenesis (5). Here, we provide evidence that the protein tyrosine kinase receptor TrkC, a main cognate receptor for NT-3, is also a dependence receptor.
Results
TrkC Is a Dependence Receptor.
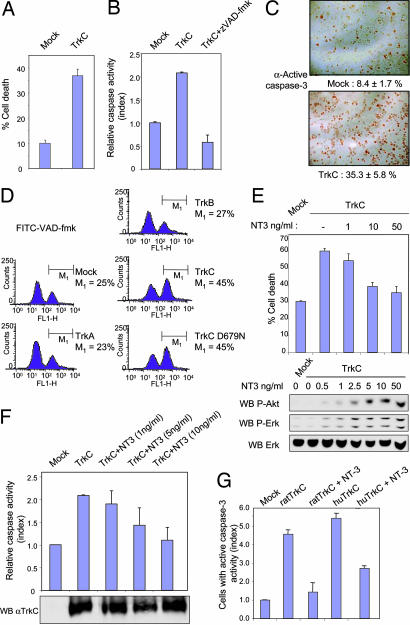
We first transiently expressed full-length rat TrkC in HEK293T cells (in which HEK stands for “human embryonic kidney”) or in immortalized olfactory neuroblast 13.S.24 cells. TrkC expression was detected only when these cells were transfected with a TrkC-encoding construct [supporting information (SI) Fig. 5 and Fig. 1 A and F]. As shown in Fig. 1A, cell death induction was associated with the expression of TrkC. TrkC-induced cell death was defined as apoptosis because TrkC expression induced (i) an increased caspase activity [determined by the measurement of DEVD-AFC cleavage in cell lysate (Fig. 1B), by the quantification of cells stained with anti-active caspase-3 antibody (Fig. 1C), or by measuring the cleavage of a FITC-VAD-fmk caspase substrate in living cells (Fig. 1D)] and (ii) an increased DNA condensation [determined by the percentage of cells stained with an anti-single stranded DNA antibody (SI Fig. 5B)]. This apoptosis is caspase-dependent because addition of the general caspase inhibitors zVAD-fmk or boc-aspartyl(OMe)-fluoromethylketone (BAF) fully inhibit TrkC-induced apoptosis (Fig. 1B and data not shown). Interestingly, such a death-promoting effect is not observed when TrkA or TrkB is expressed instead of TrkC (Fig. 1D), even though TrkA, TrkB, and TrkC are present at the cell membrane at a similar level (SI Fig. 5A and data not shown). To exclude the possibility that apoptosis induction could be caused by abnormal autoactivation of TrkC, a kinase-dead mutant, TrkC D679N, was expressed instead of TrkC wild type. This mutant, which fails to induce Erk or Akt phosphorylation in response to NT-3 (see Fig. 4C), displays a similar proapoptotic activity to TrkC wild type (Fig. 1D). Thus, TrkC expression drives apoptotic cell death that is not caused by TrkC kinase activity.
Fig. 1.
TrkC is a dependence receptor. HEK293T (A–C) or 13.S.24 (D–G) cells were transfected with the mock plasmid (Mock) or expression plasmid TrkA, TrkB, or TrkC. (A) Cell death induction by TrkC measured by trypan blue exclusion. Standard deviations are indicated (n = 3). (B) TrkC induced increased caspase activity, monitored by cleavage of the Ac-DEVD-AFC substrate. zVAD-fmk, a general and potent caspase inhibitor, was also used. (C) TrkC induces caspase-3 activation as measured by immunostaining with antiactive caspase-3 antibody. Representative images are shown. Quantification is indicated together with standard deviations (n = 3). (D) Transfected cells were labeled with FITC-VAD-fmk. As a control, a kinase-inactive (see Fig. 4C) TrkC (TrkC D679N) was also transfected. A representative flow cytometry analysis is shown. Note that TrkC induces caspase activation, whereas TrkA and TrkB behave like mock transfection. (E–G) Mock 13.S.24 cells or rat (E and F)/human (G) TrkC transfected 13.S.24 cells were treated with increasing doses of NT-3 (0.5, 1, 2.5, 5, 10, and 50 ng/ml as indicated for rat TrkC and 10 ng/ml for human TrkC). (E) Cell death induction by TrkC measured by trypan blue exclusion (Upper). Phosphorylation of Akt and Erk is shown by immunoblot with anti-phospho Akt and anti-phospho Erk (Lower). A loading control is indicated by immunoblot on total Erk. (F) NT-3 inhibits TrkC-induced caspase activity, as monitored by using the Ac-DEVD-AFC substrate. Note that the level of TrkC expression is similar in the different tested conditions as shown by Western blot. (G) Human TrkC instead of rat TrkC was expressed in 13.S.24 cells. Cell death was measured by enumerating the cells labeled with active caspase-3. A ratio is presented as active caspase-3-positive cells in each condition to the one detected in the mock transfection. Standard deviations are indicated (n = 3).
Fig. 4.
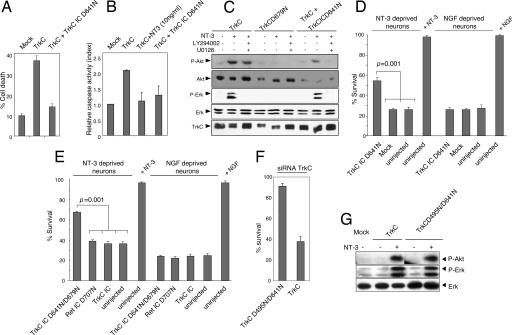
DRG neurons death upon NT-3 loss depends on the proapoptotic activity of TrkC. (A and B) TrkC IC D641N acts as a dominant-negative mutant of TrkC. HEK293T cells were transfected with the mock plasmid (Mock), the TrkC expression plasmid together with the mock plasmid, or the TrkC IC D641N expression plasmid. The dominant-negative effect of TrkC IC D641N was measured by trypan blue exclusion (A) or a caspase activity assay, as in Fig. 1B (B). Standard deviations are indicated (n = 3). (C) 13.S.24 neuroblasts were transfected with TrkC wild type or TrkC kinase-dead (TrkCD679N), or cotransfected with TrkC and the dominant-negative mutant TrkC IC D641N, in the presence or absence of 10 ng/ml NT-3 and with or without the addition of a PI3K inhibitor (LY294002, 10 μM) or a MEK inhibitor (U0126, 10 μM). Akt and Erk phosphorylation was visualized by Western blot with an anti-phosphoAkt and an anti-phosphoErk antibody, respectively. The levels of Akt and Erk kinases were shown by reprobing the membrane with an anti-total Akt antibody or an anti-total Erk antibody, respectively. TrkC immunoblot is also shown. Similar results were obtained by using the FACE-Akt ELISA (Active Motif, Carlsbad, CA). (D and E) Sensory neurons were maintained with NT-3 or NGF, microinjected with a mock plasmid (Mock) or the plasmids encoding TrkC IC D641N (D) or kinase dead TrkC IC D641N/D679N, the dominant-negative mutant of Ret (i.e., Ret IC D707N) or TrkC IC (E), and grown further without NT-3 or NGF. Living neurons were counted 72 h later and expressed as the percentage of initially injected neurons. Experiments shown in D and E were performed separately. (F) Same as D and E: sensory neurons were maintained with NT-3, microinjected with endogenous TrkC siRNA and either TrkC or TrkC D495N/D641N, and grown further in the absence of NT-3. Living neurons were counted 72 h later and expressed as the percentage of initially injected neurons. The standard errors of the means are shown (n = 3). (G) Same as in C, except that Akt/Erk phosphorylation was measured in 13.S.24 cells transfected with TrkC or TrkC D495N/D641N.
We then assessed whether the presence of NT-3 affected TrkC-proapoptotic activity. TrkC-mediated cell death [measured by the trypan blue exclusion assay (Fig. 1E), by caspase activity (Fig. 1F), or by DNA condensation (SI Fig. 5C)] was inhibited, in a dose-dependent manner, by NT-3 used within the range of NT-3 concentration that triggered the classic positive signaling downstream of TrkC [i.e., measured by Akt or Erk phosphorylation (Fig. 1E)]. Hence, NT-3 blocks TrkC-mediated apoptosis. Moreover, the dependence effect is not restricted to rat TrkC; human TrkC also triggers cell death unless NT-3 is present (Fig. 1G). Taken together, these data show that TrkC acts as a dependence receptor.
TrkC Intracellular Domain Is Cleaved by Caspase.
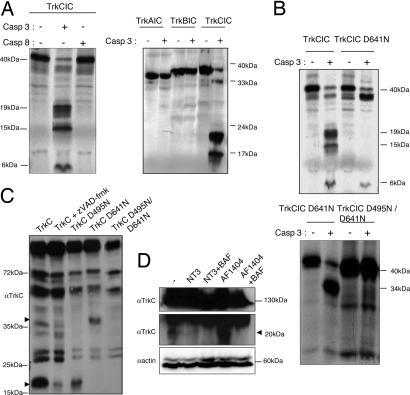
To elucidate the molecular mechanisms of TrkC-induced cell death, we further analyzed the involvement of caspases. The dependence receptors DCC, UNC5H, Patched, and RET were shown to require preliminary caspase cleavage to induce cell death (4, 6–8). We therefore analyzed whether the intracellular domain of TrkC can be cleaved by caspases. The intracellular region of TrkC encompasses the last 372 C-terminal amino acids. This domain was translated in vitro, and the product was incubated with purified active caspase-3 or caspase-8. Fig. 2A shows that the intracellular domain of TrkC is cleaved in vitro by caspase-3 but not by caspase-8. In the same experimental conditions, TrkA and TrkB intracellular domains failed to be significantly cleaved by caspase-3 (Fig. 2A). Hence, TrkC is cleaved in vitro by caspases and particularly by caspase-3-like caspases. Incubation with active caspase-3 leads to the detection of cleavage products that migrate at apparent relative molecular masses of 19, 15, and 6 kDa, suggesting the presence of at least two sites of cleavage. The caspase cleavage sites were mapped by constructing mutants based on preferred P4 and P1′ positions (9) and the apparent relative sizes of the caspase cleavage fragments. Whereas mutation of various aspartic acid (Asp) residues within the intracellular domain of TrkC had no effect on caspase-3 cleavage, the mutation of Asp-641 to Asn completely suppressed the appearance of the 19- and 15-kDa fragments (Fig. 2B). The second caspase site was subsequently located at Asp-495, because the double mutant D641N and D495N was completely resistant to caspase-3 cleavage (Fig. 2B). Thus, TrkC is cleaved by caspases at two sites located at Asp-495 and Asp-641. Interestingly, these aspartic residues appear to be conserved in chick, rat, mouse, and human TrkC, but they were not found at the corresponding positions in TrkA or TrkB. An immunoblot performed on 13.S.24 cells expressing TrkC, by using an antibody raised against a TrkC C-terminal epitope, revealed two bands (around, respectively, 35 and 20 kDa) that failed to be detected when cells were treated with the general caspase inhibitors zVAD-fmk (Fig. 2C) and BAF (SI Fig. 6A). The same two bands were observed when human TrkC was expressed in 13.S.24 cells (SI Fig. 6C). Moreover, the mutation of D495N inhibits the appearance of the 35-kDa fragment (mutation of D641N is associated with the absence of the 20-kDa band), whereas the double mutant expressed in 13.S.24 cells fails to show either of these two fragments (Fig. 2C). Thus, these two bands represent two TrkC fragments resulting from the endogenous caspase cleavage of TrkC at Asp-495 and Asp-641. Interestingly, this caspase cleavage at the two sites is not affected by overexpression of the Trk coreceptor p75ntr (SI Fig. 6D). Yet, the nature of the TrkC-cleaving caspase remains to be shown. Indeed, if in vitro caspase-3 cleaves TrkC, it is probably not only caspase-3 that cleaves TrkC in cells; both caspase-3 inhibitor DEVD-fmk and the use of a dominant-negative mutant for caspase-3 fail to block caspase-dependent cleavage of TrkC in 13.S.24 cells (SI Fig. 6 A and B). To monitor whether the TrkC cleavage by caspases naturally occurs, embryonic mouse dorsal root ganglion (DRG) were semidissociated and maintained overnight in the presence of 10 ng/ml NT-3 or in the presence of a TrkC-blocking antibody together or not with the caspase inhibitor BAF. Whereas a 20-kDa band was detected in normal culture condition, this TrkC fragment disappeared with NT-3 or BAF while it was enhanced by the blocking antibody presence (Fig. 2D). Together, these data support that TrkC is cleaved by caspases in cell-free conditions, in transfected cells, and in DRG.
Fig. 2.
TrkC is a caspase substrate. (A) TrkC is mainly cleaved by caspase-3 in vitro. The in vitro-translated intracellular domain of TrkC, but also of TrkA and TrkB, was incubated in the absence of caspase or with purified caspase-3 or caspase-8 (0.3 μM). An autoradiograph is shown. (B) Aspartic acid residues 641 and 495 are the cleavage sites. As shown in the lower autoradiograph, the TrkC IC D495/D641N mutant is not cleaved by caspase-3. (C) TrkC wild type or mutated at either single (TrkC D495N, TrkC D641N) or both (TrkC D495N/D641N) cleavage sites were expressed in 13.S.24 cells in the presence or absence of z-VAD-fmk. (D) Semidissociated DRG were left untreated (−) or treated overnight with NT-3 or with the TrkC blocking antibody AF1404 together with BAF or not. In C and D, cleavage fragments are observed by Western blot with a TrkC C terminus-directed antibody. D Top shows the full-length TrkC, whereas D Middle shows a 20-kDa fragment. D Bottom is a loading control revealing actin.
Caspase Cleavage of TrkC Releases a TrkC-Proapoptotic Domain.
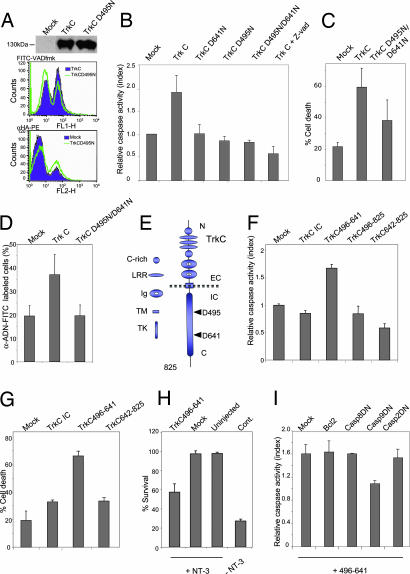
To evaluate the functional importance of the cleavage of the TrkC protein by caspases, we expressed the full-length TrkC D641N mutant, the TrkC D495N mutant, or the TrkC D641N/D495N double mutant in 13.S.24 or HEK293T cells, and cell death was assessed by trypan blue exclusion assay, and by measuring caspase activity or DNA condensation (Fig. 3A–D). Remarkably, although the mutations of one single caspase site and both caspase sites failed to affect expression levels and plasma membrane localization of TrkC (Fig. 3A and data not shown), they were sufficient to fully inhibit TrkC-proapoptotic activity (Fig. 3 B–D). Taken together, these results indicate that the caspase cleavage of TrkC is a prerequisite for TrkC-proapoptotic activity. We next investigated whether this cleavage allows the release or the exposure of a proapoptotic domain (i.e., the dependence domain). The deletion of the region located after Asp-495 was sufficient to abrogate TrkC-proapoptotic activity (data not shown). We then expressed the complete intracellular domain, the region located after the second caspase cleavage site Asp-641, or the fragment encompassed between the two caspase cleavage sites in 13.S.24 cells. As shown in Fig. 3 F and G, expression of the fragment located between Asp-495 and Asp-641 (Fig. 3E) was sufficient to trigger apoptosis, whereas the 642–825 fragment failed to display any proapoptotic activity. Intriguingly, TrkC intracellular domain expression failed to induce apoptosis, whereas the full-length TrkC was proapoptotic, suggesting that the caspase cleavage and the subsequent cell death induction requires transmembrane TrkC (Fig. 3 F and G). Moreover, together with the observation that the mutation of one single caspase cleavage site is sufficient to abrogate TrkC-proapoptotic activity, the fact that the fragment resulting from the two caspase cleavages (i.e., TrkC 496–641) kills cells, whereas the fragment resulting from the single caspase cleavage at Asp-495 (for example, TrkC 496–825) does not, supports the argument that both caspase cleavages are required for TrkC-induced apoptosis (Fig. 3F).
Fig. 3.
TrkC cleavage releases a proapoptotic domain. (A–D) Mutation of one or two caspase cleavage sites of TrkC inhibits the proapoptotic activity of TrkC. (A) Mock plasmid- (Mock), TrkC-, or TrkC D495N-transfected 13.S.24 cells were analyzed by Western blot with anti-HA (HA-TrkC) antibody, by FACS analysis for membrane localization, as in SI Fig. 5A, and by FACS analysis for measurement of caspase activity, as in Fig. 1D. (B) Single or double caspase-site mutants were transfected into HEK293T cells, and caspase activity was measured by DEVD–AFC cleavage in cell lysates, as in Fig. 1B. (C and D) TrkC D495N/D641N mutant is not proapoptotic in HEK293T cells, as measured by trypan blue exclusion (C) or by DNA condensation as in SI Fig. 5B (D). Standard deviations are indicated (n = 3). (E) TrkC is composed of 825 amino acids. Caspase cleavage sites are located at D495 and D641 in the cytoplasmic region of the receptor. EC, extracellular domain; IC, intracellular domain; TK, tyrosine kinase domain; LRR, leucin-rich repeat; Ig, Ig domain; C-rich: cystein-rich domain; TM, transmembrane domain. (F and G) The fragment released by caspase cleavage (496–641) is a potent cell death inducer when expressed in 13.S.24 neuroblasts, as measured by a caspase activity assay based on FITC-VAD-fmk, as in Fig. 1D (F), or by trypan blue exclusion (G). Standard deviations are indicated (n = 3). (H) Primary sensory neurons were maintained with NT-3, microinjected with a mock plasmid (Mock) or with the plasmid encoding TrkC 496–641. Living neurons were counted 72 h later and expressed as the percentage of initially injected neurons. The standard errors of the means are shown (n = 3). (I) 13.S.24 cells were cotransfected with expression plasmids for TrkC 496–641 (or empty vector) and either empty vector (Mock), Bcl-2, or dominant-negative (DN) caspase-2, -8, or -9. Apoptosis was measured by monitoring caspase activity as in Fig. 1D. Caspase activity is presented as the ratio between the TrkC 496–641-transfected population and the control-transfected population for each cotransfection (Mock, Bcl2, Casp2DN, Casp8DN, and Casp9DN). Standard deviations are indicated (n = 3).
To monitor whether this fragment was proapoptotic in a more biological setting, we analyzed whether expression of this dependence domain of TrkC (TrkC 496–641) was proapoptotic in TrkC expressing primary neurons. We analyzed embryonic mouse DRG neurons maintained in culture for 5 days with NT-3 and then as a control deprived of NT-3 (also see Fig. 4). As shown in Fig. 3H, expression of this domain via microinjection was apoptotic in NT-3-maintained DRG neurons, hence surpassing the survival signaling provided by NT-3. Together with the fact that NT-3 inhibits caspase-dependent TrkC cleavage in DRG (Fig. 2D), this observation supports the view of unbound TrkC being cleaved by caspases, resulting in the release of a TrkC-proapoptotic fragment. How the released fragment induces apoptosis remains to be shown. However, it is interesting to note that death induction by this fragment resembles death induction by DCC, another dependence receptor (6). Indeed, TrkC dependence domain-induced 13.S.24 cell death appears independent of the death-receptor pathway because expression of a dominant-negative mutant of caspase-8 that is known to block TNF- or Fas-induced cell death failed to inhibit TrkC-496–641-induced cell death (Fig. 3I). Similarly, TrkC dependence domain-induced cell death was not inhibited by the dominant-negative mutant of another initiator caspase, caspase-2 (Fig. 3I). On the contrary, caspase-9 dominant-negative mutant fully inhibited cell death induced by the TrkC dependence domain (Fig. 3I). The requirement of caspase-9 was rather suggestive of the involvement of the mitochondrial apoptotic pathway. Yet, we failed to observe inhibition of 13.S.24 cell death when Bcl2 was overexpressed (Fig. 3I), hence suggesting that this released domain does not kill through the mitochondria-dependent pathway. This finding is in agreement with the observation that Bcl-XL overexpression failed to block the death of cultured DRG neurons associated with NT-3 withdrawal (L.-Y.Y. and U.A., unpublished data). However, this observation is not supported by the phenotype of NT-3/Bax double knockout mice that show survival of proprioceptive neurons, suggesting a more complex regulation of neuronal death in vivo (10). Even though a more detailed study on the mechanisms used by the TrkC dependence domain to kill cells in vivo and in vitro remains to be done, it is intriguing to relate TrkC-induced cell death with DCC-induced cell death that requires (i) DCC cleavage by caspase, (ii) the release/exposure of a proapoptotic dependence domain, and (iii) interaction of this domain with caspase-9 and activation of caspase-9 (11). Whether the dependence domain of TrkC recruits caspase-9 and activates apoptosis through such a caspase-activating complex remains, however, to be shown.
The Dependence Receptor Activity of TrkC Is a Prerequisite for Sensory Neuron Death.
We then investigated whether the TrkC-proapoptotic activity described here has any implication in the death of primary neurons after withdrawal of NT-3. As also observed with the dependence receptor Patched (Ptc), we first noticed that the expression of a mutant form of TrkC [i.e., the intracellular domain of TrkC bearing a mutation on the caspase site D641 (TrkC IC D641N)] completely inhibits cell death induced by full-length TrkC (Fig. 4 A and B). This dominant-negative effect was specific, because the expression of TrkC IC D641N had no effect on Ptc- or Bax-induced apoptosis (data not shown). Thus, TrkC IC D641N acts as a specific dominant-negative mutant for TrkC-proapoptotic activity. The D641N mutation could theoretically lead to ectopic activation of the TrkC kinase domain when the intracellular region is separated from the whole receptor, and the resulting enhanced survival signaling could prevent apoptosis. To exclude this possibility, we analyzed Erk and Akt phosphorylation in response to NT-3 treatment in TrkC-transfected 13.S.24 cells. As shown in Fig. 4C, the presence of the TrkC IC D641N dominant-negative mutant does not induce activation of Erk/Akt in the absence of NT-3, nor does it interfere with NT-3-dependent TrkC-mediated Erk/Akt activation. Thus, the TrkC IC D641N does not prevent the death via increased survival signaling but, instead, via interfering with death signaling activated by deliganded TrkC.
To check the antiapoptotic effect of TrkC IC D641N on endogenous TrkC, we dissociated DRG from embryonic mice and cultured sensory neurons in the presence of NT-3 for 5 days. Control neurons were maintained with NGF that activates TrkA. Withdrawal of either NGF or NT-3 leads to death of ≈60–70% of the neurons, upon being counted 72 h later. We microinjected the NT-3- or NGF-maintained neurons with either TrkC IC D641N or the mock vector and removed NT-3 or NGF. As shown in Fig. 4D, the dominant-negative mutant dramatically enhanced survival of the NT-3-deprived neurons, although it did not affect the death of NGF-deprived neurons. Interestingly, microinjection of a construct encoding the intracellular domain of TrkC without the D641N mutation had no effect on survival of the NT-3-deprived neurons (Fig. 4E). Thus, the antiapoptotic effect of TrkC IC D641N on NT-3-deprived neurons is not due to overexpression of the ectopic TrkC intracellular domain, which may have forced TrkC kinase catalytic activity. To specifically exclude the role of the tyrosine kinase domain, we microinjected the TrkC IC D641N bearing the additional kinase-inactivating mutation D679N (this mutation abrogates TrkC ability to activate Erk or Akt in response to NT-3; see Fig. 4C). As shown in Fig. 4E, the kinase-inactivating mutation did not abolish the death-suppressing activity of TrkC IC D641N in NT-3-deprived neurons, showing that the tyrosine kinase catalytic activity is not involved here. Moreover, the antiapoptotic effect of TrkC IC D641N on NT-3-deprived neurons is receptor-specific, because the microinjection of a dominant-negative mutant of another dependence receptor, Ret (Ret IC D707N), into NT-3-deprived (and also NGF-deprived) neurons failed to inhibit death (Fig. 4E). To further study whether cell death observed upon NT-3 loss is related to the endogenous proapoptotic activity of unbound TrkC, we performed a replacement study in which endogenous TrkC was inhibited via microinjection of siRNA while ectopic TrkC wild type or TrkC mutated in the two caspase sites TrkC D495N/D641N was expressed. As control experiments, microinjection of control siRNA failed to have significant effect on primary neurons death upon NT-3 loss (data not shown). Moreover, as shown in Fig. 4F, similarly to the control situation, replacement of endogenous TrkC by ectopic TrkC is associated with primary neuron death in response to NT-3 loss. On the other hand, the replacement of endogenous TrkC by the TrkC caspase-dead mutant inhibited cell death induction upon NT-3 withdrawal (Fig. 4F). Moreover, this effect is not due to a possible interference of the caspase site's mutation with TrkC positive signaling, because TrkC wild type and TrkC D495N/D641N show a similar pattern of Akt/Erk phosphorylation in the absence or presence of NT-3 (Fig. 4G). Taken together, these data demonstrate that the cell death observed upon NT-3 loss is not only due to the loss of survival signals but also to an active cell death stimulus triggered by unbound TrkC.
Discussion
Many neurons die physiologically in vivo at different stages of development, a process in which neurotrophins and their receptors play a key role. In developing sensory ganglia, NT-3-dependent neurons are overproduced. Excess neurons are removed through a deficiency in NT-3 during periods of programmed death. Along this line, overexpression of NT-3 in mouse increases the number of neurons in DRG (12–14). The classic view proposes that the death of these neurons is due to loss of the survival signals (i.e., MAPK and/or PI3K pathways) resulting from the loss of kinase activation of neurotrophins receptors. Yet, from our study, it appears tempting to speculate that excess TrkC-expressing NT-3-sensitive neurons die not only because of the loss of these survival signals but also via the unbound TrkC-triggered proapoptotic pathway described here. One interesting hint that fits with this hypothesis is provided by the data obtained from the different knockout mice for neurotrophins and their respective receptors. Indeed, inactivation of TrkA or NGF in mouse results in the same amount of sensory neurons loss at birth (i.e., nociceptive neurons) (15). Similarly, inactivation of either TrkB or BDNF results in an equivalent loss of mechanoceptive neurons (16, 17). On the other hand, neonates invalidated for TrkC present a loss of 30% DRG neurons, whereas NT-3−/− neonates have lost 70% of them (18, 19). The search for an explanation that may fit with the classic view of neurotrophins acting only positively via kinase-dependent signaling has raised a controversy and two hypotheses are currently proposed. Fariñas et al. (20, 21) suggested that the increased loss of DRG neurons in NT-3−/− mice is explained by the ability of NT-3 to signal through TrkA and TrkB (20–22). Alternatively, Ernfors et al. (17) have proposed that the effect is due to the death of neuronal precursors that in their vast majority express TrkC early during gangliogenesis before the different subpopulations are established (24). Yet, an alternative and attractive explanation for this discrepancy between TrkC and NT-3 inactivation could be related to the dependence-receptor facet of TrkC. Indeed, as a common feature of dependence receptors, it has been postulated that inactivation of the ligand of a dependence receptor should be associated with a more profound phenotype than inactivation of the receptor. This discrepency has been further demonstrated for the dependence receptor neogenin (25). In the case of TrkC, neuronal death observed in TrkC mutant mice could then be the result of the loss of the positive/kinase signaling of TrkC, whereas neuronal death observed in NT-3 mutant would be the result of both the loss of the positive pathway and the constitutive proapoptotic activity of TrkC. Such a view would be proven, per se, if the double NT-3/TrkC mutant mice show a less-severe phenotype than NT-3 mutant mice. This possibility needs to be further investigated.
Even though it appears clear that more in vivo data are required to apprehend the relative importance of the loss of survival signals and of the active proapoptotic signal initiated by unbound TrkC, this work brings a twist in the neurotrophic theory that assumes that the loss of neurotrophic factors equals the loss of survival signals, leading to “death by default.” Here, we propose that the mere loss of survival signals is not sufficient to explain physiological neuronal death. An active proapoptotic activity is also necessary to create the “intrinsic apoptotic status” when neurotrophic support is inadequate. In some cases, this active proapoptotic signal is provided by dependence receptor-independent mechanisms, such as the stimulation of p75ntr by unprocessed pro-NGF (26, 27), or the engagement of the Fas receptor by the Fas ligand (28). However, in several cases, as shown here in NT-3-dependent sensory neurons, such proapoptotic activity could be mediated by the unbound dependence receptor TrkC.
However, it can be argued that TrkC, which requires a caspase cleavage to be proapoptotic, is not sufficient to trigger apoptosis by itself but rather acts as an amplifier downstream of a primary apoptotic stimulus. Indeed, how can a receptor initiate apoptosis while it requires, to produce a proapoptotic molecule, a cleavage by caspases that are believed to be the effectors of apoptosis? One possibility is that the process may be initiated by a noncaspase protease, then propagated via caspase cleavage. Only a few cleavage events by a noncaspase protease would then be sufficient to initiate the cell death pathway by locally activating enough caspase to generate a caspase-amplification loop via these receptors. Alternatively, the now-old dogma suggesting that caspases are completely inactive in nonapoptotic cells and are only activated massively upon proapoptotic stimuli might be wrong. Recent findings have shown that caspase zymogens display some protease activity (29), whereas cells express endogenous caspase inhibitors, such as IAP proteins, that prevent the propagation of active caspases. Similarly, local caspase activation without cell death induction is now being documented (30, 31). Cell-death induction could therefore result from caspase amplification rather than from caspase initiation, and this would support the importance of the cellular control of caspase activation/inhibition in cell-fate determination: cell death induction would be the result of a move from low/local caspase activation (i.e., that may have a “positive” input on the cell like cell differentiation) (30) to high/distributed caspase activation. The balance between low/local and high/distributed caspase activation would therefore likely be modulated by endogenous caspase inhibitors such as IAPs and by endogenous caspase amplifiers such as the dependence receptor TrkC.
Interestingly, TrkA- and TrkB-forced expression failed to induce apoptotic death of HEK293T or 13.S.24 cells. Moreover, TrkA and TrkB are not cleaved by caspases in vitro. Thus, whereas TrkC is a prototype dependence receptor, TrkA and TrkB are probably not, suggesting that even closely related receptors like TrkA, TrkB, and TrkC can acquire a completely different activity regarding cell survival/cell death. Beside mono-sided receptors like TrkA and TrkB, which induce only survival when liganded, two-sided receptors like TrkC control both survival and death. It is tempting to speculate that both sides of the TrkC/NT-3 pair play important roles during development of the nervous system. Whereas the positive signaling pathways activated by TrkC upon NT-3 binding are important for cell differentiation, proliferation, or survival, the negative signaling pathway initiated by TrkC in the absence of NT-3 may be part of the normal apoptotic removal of cells during embryogenesis and adult tissue homeostasis.
TrkC is also involved in tumor formation and especially in medulloblastoma. In particular, elevated expression of TrkC by childhood medulloblastomas is associated with favorable clinical outcome, and it has been proposed that this effect may be related to the ability of TrkC to trigger apoptosis. Indeed, overexpression of TrkC inhibits the growth of intracerebral xenografts of a medulloblastoma cell line in nude mice, and TrkC expression by individual tumor cells is highly correlated with apoptosis within primary medulloblastoma biopsy specimens (32, 33). Even though, to date, this implication has been seen under a classic scheme of a receptor activated by its ligand NT-3, it may be worth considering the importance of the dependence receptor side of TrkC in medulloblastoma development. Moreover, the data presented here with the TrkC tyrosine kinase receptor, which will possibly hold for some other tyrosine kinase receptors, also raises questions about the common anticancer strategy, which is based on inhibiting survival pathways by interfering with the kinase activity of receptors. According to our data, inhibiting the kinase activity may not be sufficient to efficiently trigger death of tumor cells. Thus, a cotreatment based on both kinase inhibition and stimulation of the proapoptotic activity of these tyrosine kinase dependence receptors could appear as a more attractive and efficient therapeutic strategy that may bypass some of the currently observed tumor resistance.
Materials and Methods
Detailed materials and methods are provided as SI Materials and Methods.
Cell Cultures, Transfection Procedures, Plasmids, and Products.
Vectors, cell culture, and transfections of HEK 293T and olfactory neuroblasts 13.S.24 cells were performed as previously described (4). Trk plasmids were from S. Meakin (University of Western Ontario, London, ON, Canada), P. Sorensen (University of British Columbia, Vancouver, BC, Canada), and B. Nelkin (The Johns Hopkins University, Baltimore, MD). The other constructs were described in refs. 11 and 34, and TrkC mutants were generated by means of QuikChange strategies (Stratagene, La Jolla, CA). Recombinant proteins and antibodies were purchased as described in SI Materials and Methods.
Cell-Death Analysis.
Cell death was analyzed as described in refs. 6, 8, and 9, or by flow cytometry with FITC-VAD-fmk (Promega, Madison, WI) or anti-ssDNA/APOSTAIN antibody (AbCys) as described in SI Materials and Methods.
Caspase Cleavage in Vitro, in Cell Cultures, and in DRG.
In vitro transcription/translation and incubation with caspase-3 or -8 were performed as described previously (6). Cleavage of TrkC in 13.S.24 cells or in DRG is described in SI Materials and Methods.
MAPK/PI3K Pathways Activation.
Erk and Akt phosphorylation was monitored in cells stimulated or not with NT-3 by using specific phospho-antibodies as described in SI Materials and Methods.
Sensory Neuron Dissociation, Culture, and Injection.
DRGs were prepared essentially as described for trigeminal neurons (35). The neurons were microinjected essentially as described in ref. 34 and SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank P. Dubreuil for helpful comments on this manuscript; G. S. Salvesen, B. Nelkin, P. Sorensen, and S. Meakin for reagents; M. Weiss, A. P. Morel, and I. Durand for FACS analysis; S. Gobert for Erk/Akt experiments; S. Bosch and C. Segura for animal breeding; H. Bilak for text correction; and Transat for siRNA design strategy. This work was supported by the Ligue Contre le Cancer, the National Institutes of Health, the European Specific Targeted Research Projects project HERMIONE, the Institute of Biotechnology of the University of Helsinki, and Academy of Finland program 44896 (Finnish Centre of Excellence, Program 2000–2005). S.T.-D. was supported by an Association Recherche Cancer fellowship and Centre National de la Recherche Scientifique. J.R.C. was supported by Centre National de la Recherche Scientifique and the European STREP project HERMIONE. J.B.-R. was supported by Consejo Nacional de Ciencia y Tecnología Mexico.
Abbreviations
- DRG
dorsal root ganglion
- NGF
nerve growth factor
- BAF
boc-aspartyl(OMe)-fluoromethylketone.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701243104/DC1.
References
- 1.Levi-Montalcini R, Angeletti PU. Dev Biol. 1963;7:653–659. doi: 10.1016/0012-1606(63)90149-0. [DOI] [PubMed] [Google Scholar]
- 2.Huang EJ, Reichardt LF. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaplan DR, Miller FD. Curr Opin Neurobiol. 2000;10:381–391. doi: 10.1016/s0959-4388(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 4.Thibert C, Teillet MA, Lapointe F, Mazelin L, Le Douarin NM, Mehlen P. Science. 2003;301:843–846. doi: 10.1126/science.1085405. [DOI] [PubMed] [Google Scholar]
- 5.Mazelin L, Bernet A, Bonod-Bidaud C, Pays L, Arnaud S, Gespach C, Bredesen DE, Scoazec JY, Mehlen P. Nature. 2004;431:80–84. doi: 10.1038/nature02788. [DOI] [PubMed] [Google Scholar]
- 6.Mehlen P, Rabizadeh S, Snipas SJ, Assa-Munt N, Salvesen GS, Bredesen DE. Nature. 1998;395:801–804. doi: 10.1038/27441. [DOI] [PubMed] [Google Scholar]
- 7.Bordeaux MC, Forcet C, Granger L, Corset V, Bidaud C, Billaud M, Bredesen DE, Edery P, Mehlen P. EMBO J. 2000;19:4056–4063. doi: 10.1093/emboj/19.15.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Llambi F, Causeret F, Bloch-Gallego E, Mehlen P. EMBO J. 2001;20:2715–2722. doi: 10.1093/emboj/20.11.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, et al. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 10.Patel TD, Kramer I, Kucera J, Niederkofler V, Jessell TM, Arber S, Snider WD. Neuron. 2003;38:403–416. doi: 10.1016/s0896-6273(03)00261-7. [DOI] [PubMed] [Google Scholar]
- 11.Forcet C, Ye X, Granger L, Corset V, Shin H, Bredesen DE, Mehlen P. Proc Natl Acad Sci USA. 2001;98:3416–3421. doi: 10.1073/pnas.051378298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ockel M, Lewin GR, Barde YA. Development (Cambridge, UK) 1996;122:301–307. doi: 10.1242/dev.122.1.301. [DOI] [PubMed] [Google Scholar]
- 13.Ringstedt T, Kucera J, Lendahl U, Ernfors P, Ibanez CF. Development (Cambridge, UK) 1997;124:2603–2613. doi: 10.1242/dev.124.13.2603. [DOI] [PubMed] [Google Scholar]
- 14.Wright DE, Zhou L, Kucera J, Snider WD. Neuron. 1997;19:503–517. doi: 10.1016/s0896-6273(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 15.Crowley C, Spencer SD, Nishimura MC, Chen KS, Pitts-Meek S, Armanini MP, Ling LH, McMahon SB, Shelton DL, Levinson AD, et al. Cell. 1994;76:1001–1011. doi: 10.1016/0092-8674(94)90378-6. [DOI] [PubMed] [Google Scholar]
- 16.Minichiello L, Piehl F, Vazquez E, Schimmang T, Hokfelt T, Represa J, Klein R. Development (Cambridge, UK) 1995;121:4067–4075. doi: 10.1242/dev.121.12.4067. [DOI] [PubMed] [Google Scholar]
- 17.Ernfors P, Lee KF, Jaenisch R. Nature. 1994;368:147–150. doi: 10.1038/368147a0. [DOI] [PubMed] [Google Scholar]
- 18.Tessarollo L, Tsoulfas P, Donovan MJ, Palko ME, Blair-Flynn J, Hempstead BL, Parada LF. Proc Natl Acad Sci USA. 1997;94:14776–14781. doi: 10.1073/pnas.94.26.14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tessarollo L, Vogel KS, Palko ME, Reid SW, Parada LF. Proc Natl Acad Sci USA. 1994;91:11844–11848. doi: 10.1073/pnas.91.25.11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farinas I, Wilkinson GA, Backus C, Reichardt LF, Patapoutian A. Neuron. 1998;21:325–334. doi: 10.1016/s0896-6273(00)80542-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fariñas I, Yoshida CK, Backus C, Reichardt LF. Neuron. 1996;17:1065–1078. doi: 10.1016/s0896-6273(00)80240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies AM, Minichiello L, Klein R. EMBO J. 1995;14:4482–4489. doi: 10.1002/j.1460-2075.1995.tb00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ElShamy WM, Ernfors P. Development (Cambridge, UK) 1996;122:2405–2414. doi: 10.1242/dev.122.8.2405. [DOI] [PubMed] [Google Scholar]
- 24.White FA, Silos-Santiago I, Molliver DC, Nishimura M, Phillips H, Barbacid M, Snider WD. J Neurosci. 1996;16:4662–4672. doi: 10.1523/JNEUROSCI.16-15-04662.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsunaga E, Tauszig-Delamasure S, Monnier PP, Mueller BK, Strittmatter SM, Mehlen P, Chedotal A. Nat Cell Biol. 2004;6:749–755. doi: 10.1038/ncb1157. [DOI] [PubMed] [Google Scholar]
- 26.Lee R, Kermani P, Teng KK, Hempstead BL. Science. 2001;294:1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- 27.Nykjaer A, Lee R, Teng KK, Jansen P, Madsen P, Nielsen MS, Jacobsen C, Kliemannel M, Schwarz E, Willnow TE, et al. Nature. 2004;427:843–848. doi: 10.1038/nature02319. [DOI] [PubMed] [Google Scholar]
- 28.Raoul C, Estevez AG, Nishimune H, Cleveland DW, de Lapeyriere O, Henderson CE, Haase G, Pettmann B. Neuron. 2002;35:1067–1083. doi: 10.1016/s0896-6273(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 29.Salvesen GS, Duckett CS. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 30.Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Proc Natl Acad Sci USA. 2002;99:11025–11030. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell DS, Holt CE. Neuron. 2003;37:939–952. doi: 10.1016/s0896-6273(03)00158-2. [DOI] [PubMed] [Google Scholar]
- 32.Grotzer MA, Janss AJ, Phillips PC, Trojanowski JQ. Klin Padiatr. 2000;212:196–199. doi: 10.1055/s-2000-10044. [DOI] [PubMed] [Google Scholar]
- 33.Kim JY, Sutton ME, Lu DJ, Cho TA, Goumnerova LC, Goritchenko L, Kaufman JR, Lam KK, Billet AL, Tarbell NJ, et al. Cancer Res. 1999;59:711–719. [PubMed] [Google Scholar]
- 34.Yu LY, Jokitalo E, Sun YF, Mehlen P, Lindholm D, Saarma M, Arumae U. J Cell Biol. 2003;163:987–997. doi: 10.1083/jcb.200305083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moshnyakov M, Arumae U, Saarma M. Brain Res Mol Brain Res. 1996;43:141–148. doi: 10.1016/s0169-328x(96)00168-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.