Abstract
Recent studies indicate that T cell cross-priming preferentially occurs against long-lived, stable proteins. We have studied cross-priming by using the glycoprotein (GP) of lymphocytic choriomeningitis virus (LCMV), a protein that normally is not MHC class I cross-presented. This study shows that a C-terminally truncated, noncleavable variant of LCMV-GP led to the accumulation of stable, soluble GP trimers in the endoplasmic reticulum (ER) of the antigen donor cell, and thereby converted LCMV-GP into a potent immunogen for cytotoxic T lymphocyte cross-priming. Immunization of mice with tumor cells expressing an ER-retained LCMV-GP variant cross-primed protective antiviral cytotoxic T lymphocyte responses in vivo at least 10,000-fold better than immunization with cells expressing the cross-presentation-“resistant” wild-type LCMV-GP. Thus the ER is a cellular compartment that can provide antigen for cross-presentation, and modifications affecting stability and subcellular localization of the antigen significantly increase its availability for MHC class I cross-presentation. These findings impinge on vaccine strategies.
Keywords: cross-presentation, cytotoxic T lymphocytes, vaccination, antigen presentation
CD8+ T cells specifically recognize and eliminate infected or transformed cells that display antigenic peptides on their surface MHC class I molecules. Induction of strong CD8+ T cell responses is, therefore, a major goal in the development of preventive and therapeutic vaccines against persistent viruses and tumors. Most MHC class I positive cells exclusively present peptides generated during the degradation of endogenously synthesized proteins. In contrast, professional antigen-presenting cells (APCs), the key initiators of CD8+ T cell responses (1), can additionally present peptides derived from exogenous antigens taken up from the extracellular milieu via a pathway termed cross-presentation (2–4). Cross-presentation is most efficient for particulate or cell-associated antigens that target phagocytic uptake by APCs. Antigenic peptides may be directly generated by endosomal proteases; however, the main cross-presentation pathway in vivo appears to involve antigen translocation from the phagosome into the cytosol and degradation by the proteasome (2, 3). Although the mechanisms governing the uptake and processing of exogenous antigen are now being elucidated at the molecular level (5–14), much less is known about whether the nature or subcellular localization of the antigen influence its ability to enter the cross-presentation pathway.
One critical parameter limiting the ability of a given antigen to be cross-presented very likely is its half-life. Indeed, several recent studies indicate that mature, stable protein represents a major source of antigen for cross-presentation in vivo. For instance, cytotoxic T lymphocyte (CTL) responses were found to be preferentially cross-primed against peptide determinants of long-lived proteins but not of rapidly degraded signal sequences (15). Conversely, cross-priming is enhanced by inhibition of proteasomal degradation of proteins (16). Cross-presentation was also found not to require antigen neosynthesis by the antigen donor cell (17). Last, it was shown that the cross-priming antigen can be removed from cell lysates by depletion of the intact protein, further suggesting that cross-presented peptides were derived from stable protein (18). Taken together, these studies imply that effective vaccines for cross-priming CTL responses should aim at maximal metabolic stability of the antigen.
We have tested whether this concept could be exploited to augment cross-presentation of antigens that normally do not enter the cross-presentation pathway. Previous studies investigating CTL priming against lymphocytic choriomeningitis virus glycoprotein (LCMV-GP) as a cell-associated antigen (19–21) have consistently failed to demonstrate a role for cross-presentation in these responses. Therefore, we attempted to increase the stability of LCMV-GP and generated a soluble, noncleavable, and trimer-stabilized GP variant. Our findings indicate that these modifications not only led to an unexpected retention and accumulation of the variant protein within the endoplasmic reticulum (ER) of the antigen donor cell but also drastically enhanced its potential to enter the cross-presentation pathway and to induce protective CTL responses in vivo.
Results
A Soluble, Noncleavable Variant of LCMV-GP Is Retained in the ER.
Wild-type (WT) LCMV-GP (Fig. 1A) is translated as a membrane-bound single precursor protein (GPc) into the ER, where it undergoes folding, glycosylation, and trimerization before ER export (22, 23). Within the Golgi compartment, the GPc trimers are proteolytically processed by S1P/SKI-1 (24) into a peripheral receptor-binding subunit GP-1 and a transmembrane subunit GP-2 (Fig. 1A). Both subunits stay noncovalently associated and form metastable heterodimeric trimers that are exported to the cell surface, where they become incorporated into budding virions. Similar to other class I viral fusion glycoproteins, proteolytically activated GP-1/GP-2 trimers serve two major functions in the virus life cycle: (i) receptor-binding and virus uptake is mediated by GP-1, and (ii) viral membrane fusion at low endosomal pH by GP-2 (25). By following a recently published approach (22), we have expressed LCMV-GP as a soluble trimeric complex (GPER, Fig. 1B) by replacing the transmembrane region and the C-terminal half of GP-2 with a small trimerization-promoting domain derived from phage T4 fibritin. In addition, we mutated the S1P/SKI-1 protease recognition site to prevent proteolytic activation, and thus conversion of the trimeric complex into the metastable low-pH-sensitive conformation. When expressing GPER in murine MC57 fibroblast cells we unexpectedly found that the variant protein was not secreted into the cell culture supernatant, but was retained intracellularly as stable trimeric complex. Western blot analysis of cell lysate but not of culture supernatant detected GPER as a single band migrating between 150 and 250 kDa (Fig. 1C). Thus, GPER had undergone complete trimerization, and the GP-1 subunit was correctly folded to allow recognition by the conformational antibody KL25 (Fig. 1C).
Fig. 1.
C-terminally truncated LCMV-GP is retained within the ER. (A) Cartoon of full-length WT LCMV-GP. LCMV-GP is translated as a single precursor protein (GPc) into the lumen of the ER. The leader sequence (L) at the N terminus is indicated. After transport to the Golgi complex, GPc is cleaved by the protease S1P/SKI-1 into the peripheral subunit GP-1 and the transmembrane subunit GP-2. Several features within GP-2 are highlighted: an N-terminal fusion peptide (green), a central coiled-coil core (dark blue), and a helical region (light blue) before the transmembrane region (TM). (B) Truncated LCMV-GP fibritin fusion protein (GPER). The S1P/SKI-1 cleavage site is mutated to avoid proteolytic processing, and the C-terminal half of GP-2 is replaced by a trimerization-promoting fibritin sequence (F). (C) Native Western blot analysis of total cell lysate (CL) and of concentrated cell supernatant (SN) of MC57 cells stably expressing GPER. (D–O) Subcellular localization of WT LCMV-GP in stably transfected MC57 cells (MC-GP) and of GPER in stably transfected MC57 cells (MC-GPER). GP expression is detected with the GP-1-specific mAb KL25 (red). (D and E) MC-GP transfected with GFP-KDEL (green) being either permeabilized (D) or nonpermeabilized (E) before KL25 staining. (F and G) KL25 stained, permeabilized MC-GPER, either costained with the ER marker calnexin (green) (F) or transfected with GFP-KDEL (green) (G). (H–O) Triple staining of MC-GP (H–K) and MC-GPER (L–O) with KL25 (red), GFP-KDEL (green), and calnexin (blue). (Scale bars: 2 μm.) (P) HeLa cells transiently transfected with WT LCMV-GP (GP) or with truncated fibritin fusion protein (GPER) were radioactively labeled and chased for 3 h before lysis and immunoprecipitation with the GP-1-specific Ab KL25. Immunoprecipitates were treated with peptide-N-glycosidase F, treated with Endo H, or left untreated as indicated. Bands representing the glycosylated or deglycosylated precursor protein (gGPc or GPc), the glycosylated or deglycosylated GP-1 subunit (gGP-1 or GP-1), the glycosylated or deglycosylated GP-2 subunit (gGP-2 or GP-2), and the glycosylated or deglycosylated truncated GPER fusion protein (gGPER or GPER) are indicated by arrows. (Q) MC-GPER (black line), MC-GP (gray line), and parental MC57 cells (gray solid line) as assessed by FACS after intracellular KL25 staining.
To determine the subcellular location of the retained GPER, we generated stable MC57 cell lines expressing GPER (MC-GPER) or WT GP (MC-GP). Subcloned MC-GPER and control MC-GP were transfected with GFP-KDEL or stained for calnexin before immunofluorescence microscopy analysis (Fig. 1 D–O). In contrast to WT GP, which was detectable throughout the complete secretory pathway (Fig. 1 D and H) and showed marked cell surface expression (Fig. 1E), GPER completely colocalized with GFP-KDEL and did thus not reach the medial Golgi (Fig. 1 G and L–N). Moreover, as we observed GFP-KDEL-positive sites that were GPER-negative, and because GPER did not colocalize with COPII (data not shown), we concluded that GPER was fully retained within the ER. GPER was predominantly retained in calnexin-positive ER regions, indicating that it could be associated with chaperones (Fig. 1 F and L). To further demonstrate that GPER was completely retained within the ER, we performed a pulse–chase experiment and tested for endoglycosidase H (Endo H) sensitivity (Fig. 1P). Because the majority of newly translated WT GP has left the ER after 3 h as judged by S1P/SKI-1 cleavage (26), we analyzed the Endo H sensitivity of GPER 3 h after translation. As expected, the majority of the control WT GPc was proteolytically processed into GP-1 and GP-2, and thus had passed the Golgi, whereas GPER was uncleaved. GPER and remaining uncleaved WT GPc were found to be Endo H-sensitive, in contrast to GP-1 and GP-2, which were Endo H-resistant, but peptide-N-glycosidase F-sensitive. Thus, GPER was completely retained within the ER and did not reach the Golgi compartment. Finally, we tested whether ER retention caused increased GPER levels in the MC-GPER cells compared with WT GP in the MC-GP cells. Indeed, intracellular retention of GPER resulted in ≈10-fold higher total protein levels (Fig. 1Q).
GPER Is a Potent Immunogen for Cross-Priming.
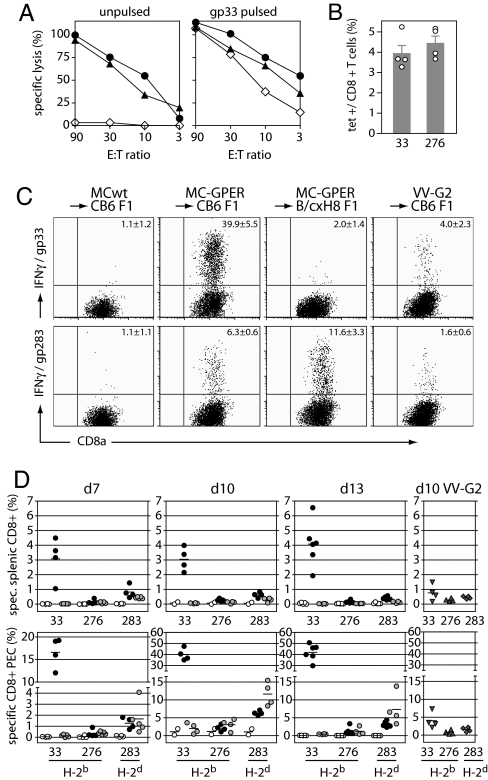
MC-GPER did not differ from MC-GP in the ability to present endogenously derived GP epitopes via MHC class I, and they were readily lysed by gp33-specific CD8+ T cell lines or primary LCMV-WE-specific effector CTLs obtained from day 8 infected mice (Fig. 2A). We next tested the ability of MC-GPER to induce CTL responses in vivo by immunizing C57BL/6 mice with MC-GPER, and we found a strong priming of GP-specific CTL responses as determined by enumeration of gp33- and gp276-specific effector CD8+ T cells (Fig. 2B). These effector cells also exhibited a highly activated phenotype as demonstrated by down-regulation of CD62L and their ability to cause beta-islet destruction in the RIP-GP mouse model (19) of T cell-mediated type I diabetes [see supporting information].
Fig. 2.
ER-retained GPER is cross-presented in vivo. (A) Lysis of unpulsed (Left) and gp33 peptide-pulsed (Right) MC-GPER (circles) and MC-GP (triangles) cells by GP-specific CTLs. Open diamonds, parental MC57 cells. E:T ratio, ratio of effector to target cells. (B) LCMV-GP-specific tetramer-positive CD8+ T cells in blood of C57BL/6 mice on day 9 after priming with MC-GPER. (C and D) GP-specific CTL responses in CB6 F1 and B/c × H8 F1 mice after priming i.p. with 5 × 106 MC-GPER. The frequency of specific IFNγ-producing CD8+ T cells in peritoneal exudate (C and D Lower) and spleen (D Upper) on days 7, 10, and 13 (d7, d10, and d13) after immunization was measured after in vitro restimulation for 6 h with the indicated peptides. (C) FACS plots of peritoneal exudate cells from individual mice on day 10. (D) Spleen cells (Upper) and peritoneal exudate cells (PEC) (Lower). Open circles, MC57-immunized CB6 F1 mice; filled circles, MC-GPER-immunized CB6 F1 mice; shaded circles, MC-GPER-immunized B/c × H8 F1 mice. Mice infected with GP-recombinant vaccinia virus (VV-G2, grey triangles and diamonds) were used as positive controls. Symbols represent individual mice.
Cells expressing GPER induced CTL via cross-priming as follows. For this purpose H-2b × H-2d F1 mice were immunized with H-2b expressing MC-GPER and induction of H-2d-restricted CTL responses was monitored. GP-specific CTL responses were not detected in mice immunized with the parental MC cell line; however, priming with 5 × 106 MC-GPER induced H-2b-restricted gp33-specific as well as H-2d-restricted gp283-specific CTL responses (Fig. 2 C and D). The cross-primed CTL responses were more easily detected in cells from peritoneal exudates, where gp283-specific CTLs accumulated to represent 6.3 ± 0.6% of total CD8+ T cells at day 10 after immunization (Fig. 2 C and D). Although these data indicated that GPER can be cross-presented in vivo, it might not be valid to compare an immunodominant CTL epitope with a subdominant CTL epitope to evaluate the efficiency in CTL priming against epitopes that are either directly presented by tumors (such as the dominant H-2b epitope gp33) or depend on cross-presentation by host APCs (such as the subdominant H-2d epitope gp283). To analyze cross-priming of gp283-specific CTL in absence of the gp33-specific CTL response, responses were studied in H-2bxd F1 mice, which are selectively tolerant to gp33. BALB/c mice (H-2d) were intercrossed with the H8 mouse strain (H-2b), which ubiquitously expresses the gp33 CTL epitope as a transgene. As expected, B/c × H8 F1 mice immunized with MC-GPER lacked gp33-specific CTL responses and exhibited slightly increased responses to another H-2Db-restricted epitope (i.e., gp276). In addition, B/c × H8 F1 mice mounted enhanced CD8+ T cell responses against the cross-presented H-2Kd-restricted gp283 epitope, such that these cells represented 11.6 ± 3.3% of the peritoneal CD8+ T cells at the peak of the response (Fig. 2 C and D).
Immunization of H-2bxd F1 mice with either MC-GPER or MC-GP induced the expansion of H-2b-restricted gp33-specific CTL (Fig. 3) either by direct priming or by cross-priming. Nevertheless, compared with MC-GPER, a 10-fold higher priming dose of MC-GP was required, likely reflecting the known differences in antigen expression by these cell lines (Fig. 1P). Although the weaker CTL priming with MC-GP could be compensated with application of higher doses for the induction of gp33-specific CTL responses, this result was not achieved for gp283-specific H-2d-restricted CTL responses that depend entirely on cross-presentation. Strikingly, no cross-presentation was observed even if the number of injected MC-GP was increased to 5 × 107 cells (Fig. 3). Thus, in contrast to WT LCMV-GP, GPER was efficiently cross-presented in vivo.
Fig. 3.
Cross-presentation of GPER does not result from increased antigen dose. CB6 F1 and B/c × H8 F1 mice were immunized with titrated numbers of MC-GPER or MC-GP, and IFNγ-producing CD8+ T cells in spleen (data not shown) and peritoneal exudate cells (data of individual mice are shown) on day 10 after immunization were analyzed after in vitro restimulation using the indicated peptides.
Cross-Primed CTL Responses Confer Protection Against LCMV Infection in Mice.
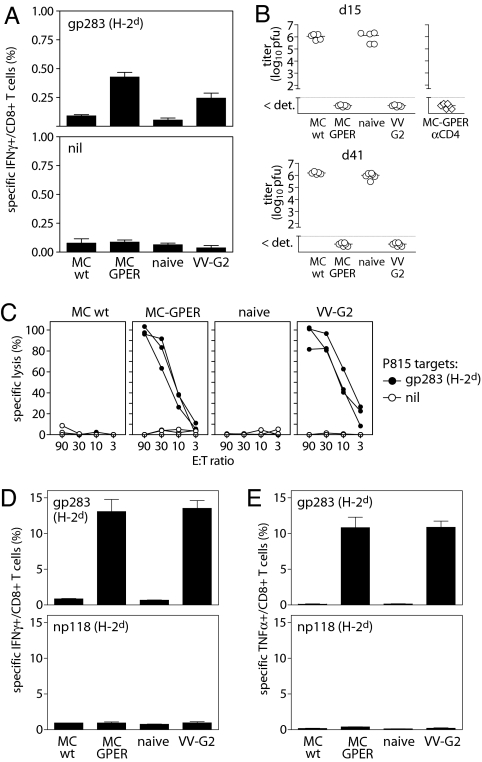
We next sought to test the functionality of the cross-primed CTL responses in a more physiological situation, and to evaluate their protective capacity against a challenge infection with LCMV-WE. To exclude any contribution of direct priming by the tumor cell itself, we injected the H-2b+ MC-GPER into H-2d+ BALB/c recipient mice. Immunization of BALB/c mice with MC-GPER primed gp283-specific CTL responses, to a comparable extent as that observed after infection with a recombinant vaccinia virus expressing LCMV-GP (VV-G2; Fig. 4A). Although low in frequency at 15 days after immunization, the gp283-restricted CTLs expanded drastically after challenge infection with LCMV-WE and were able to confer full protection against this virus (Fig. 4 B–E). We could exclude that the gp283-specific CTL response reflected a primary response induced by the LCMV challenge infection rather than a secondary response of cross-primed CTL induced by tumor cells. Immunization of the mice showed no CTL responses against the LCMV nucleoprotein-derived np118 epitope, which is the immunodominant H-2d epitope. In addition, specific CTL were not detected at this time point in mice receiving virus challenge alone (Fig. 4 D and E). Four days after infection, spleens of MC-GPER primed mice were free of virus and contained high numbers of cytokine producing gp283-specific CD8+ T cells that lysed gp283-labeled target cells directly ex vivo. Furthermore, cross-primed gp283-specific CTL induced by MC-GPER immunization protected against LCMV infection not only during the effector phase (day 15 after immunization) but also during the memory phase (day 41 after immunization) of the immune response (Fig. 4B). A contribution of CD4+ T cell responses to antiviral protection could be excluded because tumor cells and virus replication were controlled in CD4+ T cell-depleted animals (Fig. 4B). Thus, immunization with cells expressing ER-retained GPER cross-primed strong CTL responses that protected against infection with LCMV-WE.
Fig. 4.
Protective CTL responses induced by GPER-expressing tumor cells. BALB/c mice were i.p. immunized with parental MC57 (MCwt), MC-GPER, GP-recombinant vaccinia virus (VV-G2), or were left naïve as controls, and challenged 15 or 41 days later with 200 pfu of LCMV-WE. Virus titers (days 15 and 41 are shown) and T cell responses (day 15 is shown) were assessed in spleens on day 4 after LCMV challenge. (A) gp283-specific CD8+ T cell responses on day 15 after immunization with MC-GPER. (B) Virus titers in mice that received the LCMV challenge either 15 days (Upper) or 41 days (Lower) after being immunized as indicated (open circles). CD4-depleted (open diamonds) animals were immunized with MC-GPER and challenged 15 days later with LCMV-WE. (C–E) Ex vivo cytotoxicity (C) and frequency of IFNγ-producing (D) or TNFα-producing (E) gp-283 and np118-specific CD8+ T cells in spleens on day 4 after virus challenge.
Modifications of GPER Determine Cross-Priming Potential.
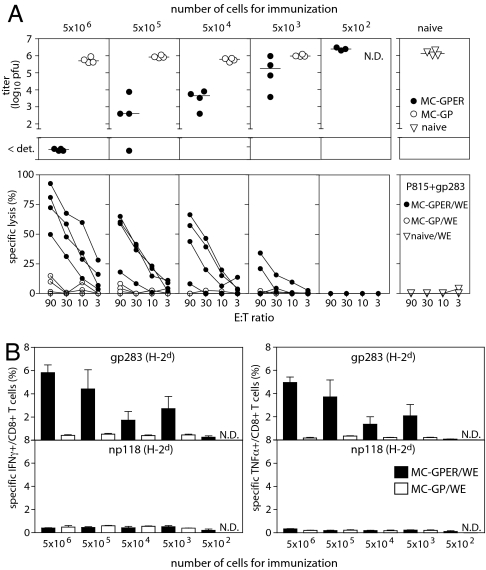
Given that other studies failed to demonstrate cross-priming after immunization with tumor cells expressing full-length WT LCMV-GP (20, 21), we set out to determine whether it was the altered stability and subcellular localization, or the total quantity of GPER, that enabled cross-presentation. To address this question, BALB/c mice were immunized with titrated numbers of MC-GP or MC-GPER. CTL responses were analyzed by their ability to produce cytokines, to lyse target cells directly ex vivo, and to protect against in vivo challenge with a low-dose infection of LCMV-WE (Fig. 5). Consistent with the observations of others (20, 21), we found that MC-GP cells expressing WT LCMV-GP did not cross-prime CTL responses, and consequently failed to provide protection against challenge infection. In contrast, antiviral protection after immunization with MC-GPER was consistently observed at a priming dose of as few as 50,000 cells, and could be detected even in a subset of the animals immunized with 5,000 cells (Fig. 5A). Spleens of these mice also contained LCMV-GP-specific cross-primed CTLs capable of cytokine production and of lysing gp283-labeled targets directly ex vivo (Fig. 5). These data demonstrate that GPER-expressing cells were at least 1,000- to 10,000-fold more potent at inducing CTL cross-priming than WT GP-expressing cells. Our earlier data showed that GPER is expressed at ≈10-fold greater levels than WT LCMV-GP in murine fibrosarcoma cells (Fig. 1P). Therefore, the enhanced cross-presentation potential of GPER not only was determined by the increased antigen dose but very likely resulted from the modifications affecting its solubility, stability, and subcellular localization.
Fig. 5.
ER-retained GP, but not WT GP, is a potent immunogen for cross-priming of protective antiviral CTLs. BALB/c (H-2d) mice were primed with titrated numbers of MC-GPER (filled circles) or MC-GP (open circles) and challenged with LCMV-WE on day 15 after immunization. Four days later, virus titers (A Upper), ex vivo cytotoxicity (A Lower), and IFNγ-producing (B Left) or TNFα-producing (B Right) virus-specific CD8+ T cells were assessed using spleen cells. Filled bars, MC-GPER; open bars, MC-GP. N.D., not determined. Symbols represent individual mice in A, the mean ± SEM of groups of 3–4 mice in B.
Discussion
We have previously performed extensive studies of CTL priming against cell-associated LCMV-GP without detecting a contribution of cross-presentation to the induction of GP-specific CD8+ T cells in response to GP-expressing fibroblasts (20, 21) or splenocytes derived from GP-transgenic mice (21). In addition, and in contrast to similar studies employing other model antigens, such as ovalbumin (27, 28), LCMV-GP is also not cross-presented when expressed in pancreatic islet cells (19, 29). In the present study we show that the normally cross-presentation-“resistant” LCMV-GP can be converted into a potent antigen for CTL cross-priming by several modifications that affect the solubility and stability of the trimeric GP complex, its subcellular localization, and the quantity expressed on a per-cell basis.
The variant GPER differed from the WT GP by its lack of a transmembrane domain, the addition of a phage-derived trimerization-promoting domain, and by its resistance to processing into metastable GP-1/GP-2 complexes because of a mutated protease cleavage site. Although all of these modifications very likely have increased the stability of the GP trimeric complex, they also led to the retention and accumulation of the variant protein within the ER. Because the capacity of both proteins to cross-prime CTL represented a difference of >3–4 orders of magnitude, the increased CTL cross-priming efficiency of GPER cannot be attributed to the accumulation of ≈10-fold higher protein levels alone. Instead, both the presence of soluble, noncleavable, and trimer-stabilized GP complexes and the protein's retention in the ER are likely to be responsible for its acquired ability to be cross-presented.
Cross-presented antigens are preferentially long-lived proteins (15–18). Presumably GPER formed stable, soluble trimeric complexes that might survive longer in the endosomal pathway of the APC, which in turn could increase the chance of translocation of sufficient amounts into the cytosol for antigen processing and cross-presentation. That degradation in the APC phagosome critically limits the availability of protein for cross-presentation is illustrated by the observation that blocking phagosome acidification enhances CTL cross-priming in vivo (30). Importantly, the proteolytically activated GP-1/GP-2 complexes of WT GP dissociate on endosomal acidification (25) and expose the N-terminal sequences of GP-2 containing the gp283 epitope. Premature degradation of this CTL epitope may therefore be limited or prevented in noncleavable GPER where gp283 remains buried in the interior of the GP-1/GP-2 complex. In addition to its increased stability, the ER retention of GPER may have also promoted cross-presentation. Although both cell membrane-bound WT GP and ER-retained GPER would be phagocytosed as cell-associated antigens, accumulation of GPER within the ER could enhance cross-presentation by providing the antigen in a more concentrated manner. Furthermore, it is conceivable that, on cell death, the GPER might be enclosed within ER-derived membranes or vesicles that could further protect the protein from rapid degradation within the endocytic pathway. Yet another aspect of ER retention should be considered. Although GPER had trimerized and folded overall correctly to generate the conformational GP-1 epitope detected by the antibody KL25 (see Fig. 1C), the observed ER-retention phenotype was likely caused by some minor folding and/or export defect. This phenotype, in turn, would lead to reassociation with chaperones, which has been reported to enhance delivery of proteins and peptides to APCs for cross-presentation (12, 17). In addition, misfolded proteins are retrotranslocated into the cytosol and destroyed via the ER-associated degradation pathway (ERAD) (31). Thus, GPER-expressing donor cells may contain higher numbers of antigenic peptides derived from continuous degradation of misfolded and accumulated protein.
Finally, our observations may also signify a more general phenomenon. Although CTL cross-priming was not observed for LCMV under WT conditions, other virus infections may well induce overexpression and accumulation of viral proteins in the ER, and thus trigger the ER-stress response (32). While this cellular response to unfolded proteins is known to assist protein folding by up-regulating molecular chaperones, it may also promote apoptosis of infected cells after prolonged exposure to ER stress (33). In view of our data, it is tempting to speculate that viral proteins accumulating in the ER of infected cells undergoing ER stress-triggered apoptosis could be used by the immune system as a source of antigen for cross-presentation. Thus, the ER quality control might be linked to antigen cross-presentation via the ER stress response, in a manner similar to that already recognized for direct MHC I presentation via the ERAD.
In summary, our findings identify the ER as a cellular compartment that efficiently provides antigen for CTL cross-presentation, and they show that modifications affecting the stability and subcellular localization of an antigen can drastically enhance its delivery into the cross-presentation pathway.
Materials and Methods
Cell Lines and Constructs.
All cell lines were originally obtained from the American Type Culture Collection (ATCC) (Manassas, VA). LCMV-GP constructs were prepared by using a full-length human codon-optimized GP sequence provided by D. von Laer (34). GPER is composed of amino acids 1–353 of LCMV-GP with a mutated SKI-1/S1P cleavage site (R262A; ref. 24) fused to the phage T4-derived fibritin sequence GYIPEAPRDGQAYVRKDGEWVLLSTFL (35).
Immunofluorescence Microscopy.
The monoclonal antibody KL25 (36), the polyclonal α-calnexin rabbit serum (37), and the GFP-KDEL expression plasmid (38) have been described. Cells were fixed with 2% formaldehyde, blocked and permeabilized for 20 min with 10% goat serum/5% FCS/10 mM glycine/0.05% saponin in PBS, followed by staining with primary antibodies (KL25 at 10 μg/ml and anti-calnexin serum at 1:1,000) for 2 h. Cells were washed, and stained with secondary antibodies (anti-rabbit IgG AlexaFluor 647, anti-rabbit IgG AlexaFluor 488, and anti-mouse IgG AlexaFluor 594; all from Molecular Probes, Invitrogen, Basel, Switzerland; at 1:400) for 45 min. Coverslips were rinsed with water and mounted with ImmuMount (Thermo Shandon, Pittsburgh, PA). Images were recorded with a confocal microscope (model LSM510 m; Zeiss, Feldbach, Switzerland) equipped with a ×100/1.40 Plan-Apochromat objective.
Pulse–Chase.
Transfected HeLa cells were starved for 2 h with DMEM lacking methionine and cysteine, labeled with 72 μCi/ml [35S]methionine/[35S]cysteine (1 Ci = 37 GBq) for 30 min, washed, and then chased for 3 h with normal medium. Cells were lysed (1% Triton X-100), nonsoluble material was removed by centrifugation, and supernatants were incubated overnight with protein A-Sepharose and KL25 at 4°C. Peptide-N-glycosidase F and Endo H were used as described by the supplier (New England Biolabs, Frankfurt am Main, Germany). Samples were reduced and analyzed by SDS/PAGE on 10–16% polyacrylamide gels, followed by phosphorimaging using a BAS 1500 (Fuji, Tokyo, Japan) for 35S-labeled proteins.
Mice and Viruses.
C57BL/6 mice, 318 TCR-transgenic mice (39), RIP-GP mice (19), and H8 mice (40) were bred at the Institut für Labortierkunde, University of Zurich. BALB/c (H-2d) mice and CB6F1 (H-2bxd) mice were from Harlan (Horst, The Netherlands). BALB/c and H8 mice were crossed to obtain B/c × H8 F1 (H-2bxd) mice. All animal experiments were performed according to institutional guidelines and Swiss federal regulations, and were approved by the veterinary office of the Kanton of Zurich. T cell responses of tumor-immunized mice were assessed in peritoneal exudate cells and spleens. For challenge experiments, tumor-primed mice were infected intravenously with 200 pfu of LCMV-WE as indicated. After 4 days, spleens were collected for analysis of T cell responses and virus titers (41). RIP-GP mice (19) were injected with tumor cells or LCMV, and blood glucose concentration was monitored by using a Glucometer Elite (Bayer, Zurich, Switzerland). LCMV-WE was propagated on L929 cells. Vaccinia virus expressing the full-length LCMV-GP (VV-G2) was propagated on BSC40 cells.
Native Western Blotting.
GPER complexes were assessed in MC-GPER cell lysates and in 40-fold concentrated cell supernatant. Cells were disrupted in lysis buffer (1% octyl glucopyranoside/0.15 M NaCl in PBS) by Dounce homogenization. Samples were not heated and were separated by SDS/PAGE under nonreducing conditions, blotted onto nitrocellulose membranes, and detected by using KL25 (5 μg/ml) followed by secondary HRP-coupled goat anti-mouse IgG1 (Zymed, San Francisco, CA).
Flow Cytometry Staining.
Lymphocytes were stained with H-2Db/gp33 or H-2Db/gp276 tetramers in FACS buffer (FB = PBS/2% FCS/20 mM EDTA) for 10 min at 37°C. Then anti-CD62L-FITC and anti-CD8a-APC (both BD Pharmingen, Basel, Switzerland) were added, and staining continued at 4°C for 30 min. Cells were washed, fixed (FACS Lysis Solution, BD Pharmingen), and analyzed on a FACSCalibur (BD Biosciences, Basel, Switzerland) by using the CellQuest software. To assess GP expression, 106 fixed tumor cells were permeabilized in permeabilization buffer (PB = FB containing 0.1% wt/vol saponin) for 5 min at 4°C, washed, and stained with KL25 in PB at 4°C for 60 min. Cells were then washed, and bound antibody was detected by FITC-labeled goat anti-mouse IgG (BD Pharmingen).
Intracellular Cytokine Staining.
Spleen cells or peritoneal exudate cells were cultured in IMDM containing 5 μg/ml brefeldin A, 50 units/ml IL-2, 10% FCS, and antibiotics for 6 h at 37°C, either with or without specific peptide (10−6 M). Cells were first stained with anti-CD8a-PE (BD Pharmingen) for 20 min at 4°C, then fixed with 4% paraformaldehyde for 5 min at 4°C, permeabilized with PB for 5 min at 4°C, and stained with anti-mouse IFNγ-APC and anti-mouse TNFα-FITC (both BD Pharmingen) in PB for 60 min at 4°C. After three washes, cells were resuspended in FB and analyzed by flow cytometry as stated above.
CTL Assay.
Cytotoxic T cell responses in spleens of tumor- and virus-primed mice were tested against peptide-pulsed and nonpulsed 51Cr-labeled P815 (H-2d) cells in a standard chromium-release assay directly ex vivo. Direct MHC class I presentation of GP-derived CTL epitopes by GP-expressing cells was tested by using LCMV-specific CTLs as effectors and 51Cr-labeled tumor cells as targets either with or without pulse with gp33 peptide.
Supplementary Material
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation and the Deutsche Forschungsgemeinschaft (to S.F.).
Abbreviations
- APC
antigen-presenting cell
- CTL
cytotoxic T lymphocyte
- Endo H
endoglycosidase H
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- GP
glycoprotein
- GPc
GP precursor
- GPER
ER-retained LCMV-GP
- LCMV
lymphocytic choriomeningitis virus
- MC-GP
murine MC57 fibrosarcoma cells expressing LCMV-GP
- MC-GPER
MC57 cells expressing LCMV-GPER
- RIP-GP
LCMV-GP expressed under control of the rat insulin promoter
- VV-G2
recombinant vaccinia virus expressing LCMV-GP.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704423104/DC1.
References
- 1.Jung S, Unutmaz D, Wong P, Sano G, De los Santos K, Sparwasser T, Wu S, Vuthoori S, Ko K, Zavala F, et al. Immunity. 2002;17:211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rock KL, Shen L. Immunol Rev. 2005;207:166–183. doi: 10.1111/j.0105-2896.2005.00301.x. [DOI] [PubMed] [Google Scholar]
- 3.Heath WR, Belz GT, Behrens GM, Smith CM, Forehan SP, Parish IA, Davey GM, Wilson NS, Carbone FR, Villadangos JA. Immunol Rev. 2004;199:9–26. doi: 10.1111/j.0105-2896.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- 4.den Haan JM, Bevan MJ. Curr Opin Immunol. 2001;13:437–441. doi: 10.1016/s0952-7915(00)00238-7. [DOI] [PubMed] [Google Scholar]
- 5.Norbury CC, Hewlett LJ, Prescott AR, Shastri N, Watts C. Immunity. 1995;3:783–791. doi: 10.1016/1074-7613(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 6.Regnault A, Lankar D, Lacabanne V, Rodriguez A, Thery C, Rescigno M, Saito T, Verbeek S, Bonnerot C, Ricciardi-Castagnoli P, et al. J Exp Med. 1999;189:371–380. doi: 10.1084/jem.189.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, et al. Nat Med. 2001;7:297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 8.Guermonprez P, Saveanu L, Kleijmeer M, Davoust J, Van Endert P, Amigorena S. Nature. 2003;425:397–402. doi: 10.1038/nature01911. [DOI] [PubMed] [Google Scholar]
- 9.Houde M, Bertholet S, Gagnon E, Brunet S, Goyette G, Laplante A, Princiotta MF, Thibault P, Sacks D, Desjardins M. Nature. 2003;425:402–406. doi: 10.1038/nature01912. [DOI] [PubMed] [Google Scholar]
- 10.Shen L, Sigal LJ, Boes M, Rock KL. Immunity. 2004;21:155–165. doi: 10.1016/j.immuni.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Ackerman AL, Kyritsis C, Tampe R, Cresswell P. Nat Immunol. 2005;6:107–113. doi: 10.1038/ni1147. [DOI] [PubMed] [Google Scholar]
- 12.Binder RJ, Srivastava PK. Nat Immunol. 2005;6:593–599. doi: 10.1038/ni1201. [DOI] [PubMed] [Google Scholar]
- 13.Neijssen J, Herberts C, Drijfhout JW, Reits E, Janssen L, Neefjes J. Nature. 2005;434:83–88. doi: 10.1038/nature03290. [DOI] [PubMed] [Google Scholar]
- 14.Touret N, Paroutis P, Terebiznik M, Harrison RE, Trombetta S, Pypaert M, Chow A, Jiang A, Shaw J, Yip C, et al. Cell. 2005;123:157–170. doi: 10.1016/j.cell.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 15.Wolkers MC, Brouwenstijn N, Bakker AH, Toebes M, Schumacher TN. Science. 2004;304:1314–1317. doi: 10.1126/science.1096268. [DOI] [PubMed] [Google Scholar]
- 16.Norbury CC, Basta S, Donohue KB, Tscharke DC, Princiotta MF, Berglund P, Gibbs J, Bennink JR, Yewdell JW. Science. 2004;304:1318–1321. doi: 10.1126/science.1096378. [DOI] [PubMed] [Google Scholar]
- 17.Basta S, Stoessel R, Basler M, van den Broek M, Groettrup M. J Immunol. 2005;175:796–805. doi: 10.4049/jimmunol.175.2.796. [DOI] [PubMed] [Google Scholar]
- 18.Shen L, Rock KL. Proc Natl Acad Sci USA. 2004;101:3035–3040. doi: 10.1073/pnas.0308345101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohashi PS, Oehen S, Buerki K, Pircher H, Ohashi CT, Odermatt B, Malissen B, Zinkernagel RM, Hengartner H. Cell. 1991;65:305–317. doi: 10.1016/0092-8674(91)90164-t. [DOI] [PubMed] [Google Scholar]
- 20.Kundig TM, Bachmann MF, DiPaolo C, Simard JJ, Battegay M, Lother H, Gessner A, Kuhlcke K, Ohashi PS, Hengartner H, et al. Science. 1995;268:1343–1347. doi: 10.1126/science.7761853. [DOI] [PubMed] [Google Scholar]
- 21.Ochsenbein AF, Sierro S, Odermatt B, Pericin M, Karrer U, Hermans J, Hemmi S, Hengartner H, Zinkernagel RM. Nature. 2001;411:1058–1064. doi: 10.1038/35082583. [DOI] [PubMed] [Google Scholar]
- 22.Eschli B, Quirin K, Wepf A, Weber J, Zinkernagel R, Hengartner H. J Virol. 2006;80:5897–5907. doi: 10.1128/JVI.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchmeier MJ. Arenaviruses: Protein Structure and Function. 1st Ed. Vol 1. Berlin: Springer; 2002. [DOI] [PubMed] [Google Scholar]
- 24.Beyer WR, Popplau D, Garten W, von Laer D, Lenz O. J Virol. 2003;77:2866–2872. doi: 10.1128/JVI.77.5.2866-2872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Simone C, Zandonatti MA, Buchmeier MJ. Virology. 1994;198:455–465. doi: 10.1006/viro.1994.1057. [DOI] [PubMed] [Google Scholar]
- 26.Wright KE, Spiro RC, Burns JW, Buchmeier MJ. J Virol. 1992;177:175–183. doi: 10.1016/0042-6822(90)90471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo D, Freedman J, Hesse S, Palmiter RD, Brinster RL, Sherman LA. Eur J Immunol. 1992;22:1013–1022. doi: 10.1002/eji.1830220421. [DOI] [PubMed] [Google Scholar]
- 28.Kurts C, Kosaka H, Carbone FR, Miller JF, Heath WR. J Exp Med. 1997;186:239–245. doi: 10.1084/jem.186.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oldstone MB, Nerenberg M, Southern P, Price J, Lewicki H. Cell. 1991;65:319–331. doi: 10.1016/0092-8674(91)90165-u. [DOI] [PubMed] [Google Scholar]
- 30.Accapezzato D, Visco V, Francavilla V, Molette C, Donato T, Paroli M, Mondelli MU, Doria M, Torrisi MR, Barnaba V. J Exp Med. 2005;202:817–828. doi: 10.1084/jem.20051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai B, Ye Y, Rapoport TA. Nat Rev Mol Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- 32.He B. Cell Death Differ. 2006;13:393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 33.Schroder M, Kaufman RJ. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 34.Beyer WR, Miletic H, Ostertag W, von Laer D. J Virol. 2001;75:1061–1064. doi: 10.1128/JVI.75.2.1061-1064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letarov AV, Londer YY, Boudko SP, Mesyanzhinov VV. Biochemistry. 1999;64:817–823. [PubMed] [Google Scholar]
- 36.Bruns M, Cihak J, Muller G, Lehmann-Grube F. Virology. 1983;130:247–251. doi: 10.1016/0042-6822(83)90135-6. [DOI] [PubMed] [Google Scholar]
- 37.Hammond C, Helenius A. J Cell Biol. 1994;126:41–52. doi: 10.1083/jcb.126.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sbalzarini IF, Mezzacasa A, Helenius A, Koumoutsakos P. Biophys J. 2005;89:1482–1492. doi: 10.1529/biophysj.104.057885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pircher H, Burki K, Lang R, Hengartner H, Zinkernagel RM. Nature. 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 40.Ehl S, Hombach J, Aichele P, Hengartner H, Zinkernagel RM. J Exp Med. 1997;185:1241–1251. doi: 10.1084/jem.185.7.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battegay M, Cooper S, Althage A, Banziger J, Hengartner H, Zinkernagel RM. J Virol Methods. 1991;33:191–198. doi: 10.1016/0166-0934(91)90018-u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.