Abstract
HIV protease inhibitors (HIV-PIs) target the HIV aspartyl protease, which cleaves the HIV gag-pol polyprotein into shorter proteins required for the production of new virions. HIV-PIs are a cornerstone of treatment for HIV but have been associated with lipodystrophy and other side effects. In both human and mouse fibroblasts, we show that HIV-PIs caused an accumulation of prelamin A. The prelamin A in HIV-PI-treated fibroblasts migrated more rapidly than nonfarnesylated prelamin A, comigrating with the farnesylated form of prelamin A that accumulates in ZMPSTE24-deficient fibroblasts. The accumulation of farnesyl-prelamin A in response to HIV-PI treatment was exaggerated in fibroblasts heterozygous for Zmpste24 deficiency. HIV-PIs inhibited the endoproteolytic processing of a GFP-prelamin A fusion protein. The HIV-PIs did not affect the farnesylation of HDJ-2, nor did they inhibit protein farnesyltransferase in vitro. HIV-PIs also did not inhibit the activities of the isoprenyl-cysteine carboxyl methyltransferase ICMT or the prenylprotein endoprotease RCE1 in vitro, but they did inhibit ZMPSTE24 (IC50: lopinavir, 18.4 ± 4.6 μM; tipranavir, 1.2 ± 0.4 μM). We conclude that the HIV-PIs inhibit ZMPSTE24, leading to an accumulation of farnesyl-prelamin A. The inhibition of ZMPSTE24 by HIV-PIs could play a role in the side effects of these drugs.
Keywords: lamins, lipodystrophy, Zmpste24, lopinavir
HIV protease inhibitors (HIV-PIs) block the HIV aspartyl protease, a viral enzyme that cleaves the gag-pol polyprotein into smaller proteins with essential roles in viral replication (1). HIV-PIs are a cornerstone of multidrug HIV treatment regimens referred to as “highly active antiretroviral therapy” (HAART) (2). HAART regimens including HIV-PIs dramatically reduce HIV titers in blood and improve survival (2, 3). However, significant side effects have been encountered, including a metabolic syndrome associated with partial lipodystrophy, hyperlipidemia, insulin resistance, and atherosclerotic disease (4, 5).
Both HIV-PIs and other antiretroviral drugs (e.g., reverse transcriptase inhibitors) have been implicated in the metabolic/lipodystrophy syndrome (6). This syndrome is poorly understood, although multiple underlying mechanisms have been proposed, including effects on the glucose transporter Glut4, intracellular trafficking of apolipoprotein B, and adipocyte differentiation (7).
In 2003, Caron et al. (8) proposed that the HIV-PIs interfere with the processing of lamin A/C, a component of the nuclear lamina. A role for lamin A/C metabolism in the HIV-PI-associated metabolic/lipodystrophy syndrome is attractive, because missense mutations in LMNA, the gene for lamin A/C, cause a similar syndrome (9). Also, genetic defects in the conversion of prelamin A to mature lamin A (e.g., ZMPSTE24 deficiency) cause progeroid syndromes that include lipodystrophy (10–12).
Caron et al. (8) incubated a mouse preadipocyte cell line with several HIV-PIs and found prelamin A in treated but not untreated cells. The prelamin A was detected with sensitive Western blots using a prelamin A-specific antibody. However, Western blots with a lamin A/C-specific antibody revealed only mature lamin A and no prelamin A, suggesting that the amount of prelamin A accumulation and the level of inhibition of prelamin A processing were negligible. The biochemical basis for the prelamin A accumulation was not determined.
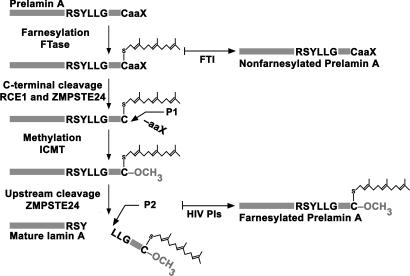
In the current study, we pursued a possible “HIV-PI/prelamin A connection,” with three goals in mind. First, we wanted to determine whether HIV-PIs, at physiologically relevant concentrations, cause significant accumulation of prelamin A relative to mature lamin A. Second, if we observed significant amounts of prelamin A, we wanted to determine whether it had the electrophoretic mobility of farneylsated or nonfarnesylated prelamin A. This is an important issue, because farnesylated prelamin A adversely affects mammalian tissues (13). Third, if the HIV-PIs caused significant prelamin A accumulation in cells, we wanted to determine the mechanism. Lamin A biogenesis is complex (Fig. 1), and a drug that interfered with any one of three different enzymes [protein farnesyltransferase (FTase), isoprenyl-cysteine carboxyl methyltransferase (ICMT), or ZMPSTE24] could potentially cause prelamin A accumulation (14–16). Thus, identifying the enzymatic step affected by HIV-PIs is important.
Fig. 1.
Biogenesis of lamin A from prelamin A. Prelamin A undergoes four posttranslational processing steps (13). First, the cysteine of the C-terminal CaaX motif is farnesylated by protein FTase. Second, the last three amino acids (-aaX) are clipped off (cleavage P1), a redundant activity of RCE1 and ZMPSTE24 (13). Third, the farnesylcysteine is carboxyl-methylated by ICMT. Fourth, the last 15 amino acids of prelamin A (including the farnesylcysteine methyl ester) are clipped off by ZMPSTE24 (cleavage P2), releasing mature lamin A. Lonafarnib, an FTI, blocks all of the processing steps and leads to the accumulation of nonfarnesylated prelamin A. In this study, we tested the hypothesis that HIV-PIs inhibit ZMPSTE24, causing an accumulation of farnesyl-prelamin A in cells.
Results
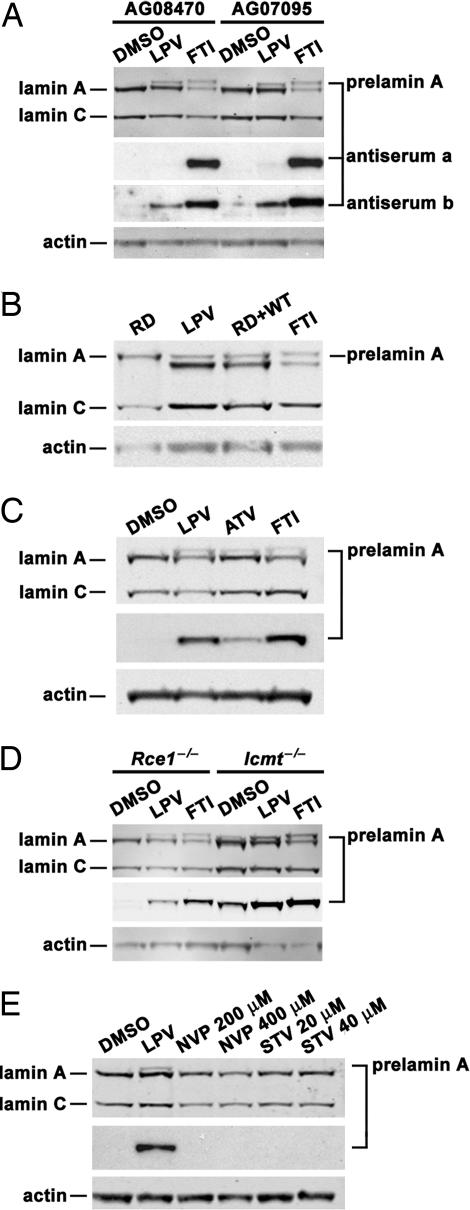
Treatment of human fibroblasts with HIV-PIs resulted in a significant accumulation of prelamin A, easily detectable on Western blots with a lamin A/C antibody (averaging 31% of lamin C, as judged by Li-Cor image quantification) (Figs. 2 A and B). The electrophoretic mobility of the prelamin A in the lopinavir (LPV)-treated cells was more rapid than the nonfarnesylated prelamin A that accumulates in cells treated with a FTase inhibitor (FTI), suggesting that it was probably farnesylated (Fig. 2 A and B). Indeed, the prelamin A protein that was induced by LPV was detectable with a prelamin A antiserum that binds farnesylated and nonfarnesylated human prelamin A but not by a prelamin A antiserum that is specific for nonfarnesylated prelamin A (Fig. 2A). Also, the prelamin A in LPV-treated cells comigrated with the farnesyl-prelamin A that accumulates in human restrictive dermopathy (RD) fibroblasts (ZMPSTE24-deficient fibroblasts) (11) (Fig. 2B). HIV-PI treatment of mouse fibroblasts yielded a similar increase in prelamin A (Fig. 2 C and D). Again, the prelamin A in the HIV-PI-treated cells migrated slightly more rapidly than the prelamin A in FTI-treated cells. The accumulation of prelamin A with LPV treatment was also observed in Rce1−/− fibroblasts (Fig. 2D). In Icmt−/− fibroblasts, there is some prelamin A accumulation at baseline, reflecting inefficient conversion of prelamin A to lamin A (15); the amount of prelamin A increased further with LPV treatment (Fig. 2D). Prelamin A accumulation was not observed in cells treated with nevirapine (NVP) or stavudine (STV), two reverse transcriptase inhibitors (Fig. 2E).
Fig. 2.
Accumulation of prelamin A in fibroblasts treated with an FTI and HIV-PIs. (A) Western blots of extracts from WT human fibroblast cell lines (AG08470 and AG07095) that had been treated for 10 days with LPV (20 μM), an FTI (5 μM), or vehicle alone (DMSO). The prelamin A to lamin C ratio, as judged by quantitative image analysis, averaged 0.31 in the LPV-treated cells and 0.39 in the FTI-treated cells; the prelamin A to mature lamin A ratio averaged 0.23 in the LPV-treated cells and 0.96 in the FTI-treated cells. We used an antibody against lamin A/C, two different rabbit antisera against the C terminus of mouse prelamin A (“a” and “b”), and an antibody against actin. Note the greater distance between the prelamin A and lamin A bands in the FTI-treated cells compared with the LPV-treated cells. One of the anti-prelamin A antisera, “a,” bound only nonfarnesylated human prelamin A, whereas the other, “b,” bound to both farnesylated and nonfarnesylated human prelamin A (see Materials and Methods). (B) Western blots of human fibroblasts treated with LPV or an FTI. Again, the prelamin A in LPV-treated cells migrates further than the prelamin A in FTI-treated cells. The lane labeled “RD + WT” represents a mixture of RD (ZMPSTE24-deficient) and WT cell extracts, each treated with DMSO. Note that the prelamin A from the RD extracts migrates at the same position as the prelamin A from the LPV-treated fibroblasts. (C) Western blots of extracts from mouse fibroblasts treated for 48 h with LPV (20 μM), atazanavir (ATV) (20 μM), an FTI (5 μM), or vehicle (DMSO). (D) Western blot analysis of Rce1−/− and Icmt−/− mouse fibroblasts that had been treated for 24 h with LPV (20 μM), an FTI (5 μM), or vehicle (DMSO). Icmt deficiency is associated with some prelamin A accumulation (15), which is increased further with LPV. (E) Western blots of extracts from WT mouse fibroblasts treated for 10 days with LPV (20 μM), NVP (200 μM and 400 μM), STV (20 μM and 40 μM), or vehicle (DMSO).
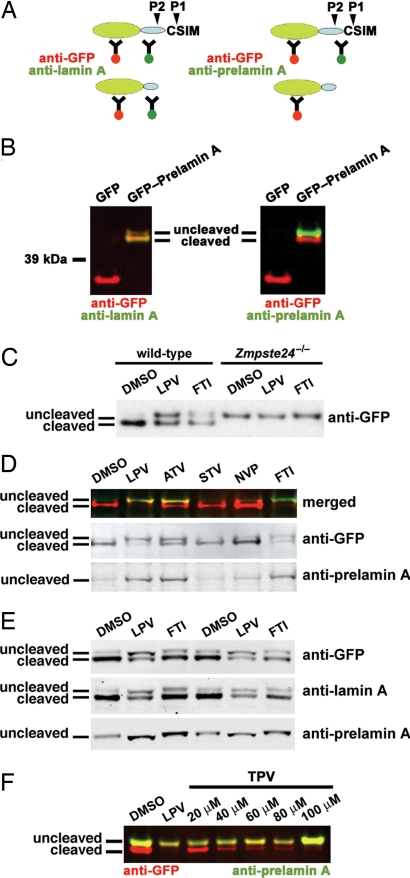
To further explore this issue, we examined the effects of HIV-PIs on the processing of a GFP-prelamin A fusion protein containing the C-terminal 118 aa of prelamin A. In HEK293 cells, the uncleaved fusion protein could be visualized with antibodies against GFP, mature lamin A, and prelamin A, whereas the cleaved fusion protein was visualized only with antibodies against GFP and mature lamin A (Fig. 3 A and B). Most of the fusion protein was processed in wild-type mouse fibroblasts, but none was processed in Zmpste24−/− fibroblasts (Fig. 3C). Both HIV-PIs and an FTI interfered with the processing of the fusion protein, resulting in an accumulation of the uncleaved protein (Fig. 3 C–E); NVP and STV had no effect (Fig. 3D). The uncleaved fusion protein in the HIV-PI-treated cells migrated more rapidly than the nonfarnesylated uncleaved fusion protein in FTI-treated cells (Fig. 3E), just as the prelamin A in HIV-PI-treated fibroblasts migrated more rapidly than the prelamin A in FTI-treated cells (Fig. 2). Tipranavir (TPV), like LPV, inhibited the processing of the fusion protein (Fig. 3F).
Fig. 3.
HIV-PIs block the processing of an enhanced GFP-prelamin A fusion protein. (A) Schematic representations of the GFP-prelamin A fusion protein and its cleavage by ZMPSTE24. The cDNA sequences encoding amino acids 548–665 from mouse prelamin A (represented by the blue oval) were ligated in-frame to GFP (represented as a green oval). The location of the prelamin A cleavage reaction carried out exclusively by ZMPSTE24 (cleavage P2) is indicated, as are the locations of epitopes for antibodies against GFP, mature lamin A, and prelamin A. (B) Western blot analysis of HEK293 cells transiently transfected with a GFP expression vector (pEGFP-C1) or the GFP-prelamin A fusion construct. Antibody binding was detected with an Odyssey system (Li-Cor Biosciences). (Left) Red signal indicates binding of the anti-GFP antibody, and green signal indicates binding of the anti-mature lamin A antibody. The merged signals result in a yellow/green color. (Right) Red signal indicates binding of the anti-GFP antibody, and green signal indicates binding of the anti-prelamin A antibody. (C) Western blot, using an antibody against GFP, of WT and Zmpste24−/− fibroblasts transiently transfected with the GFP-prelamin A fusion construct and treated overnight with the vehicle (DMSO), LPV (20 μM), or the FTI (5 μM). (D) Western blot analysis of HeLa cells transiently transfected with the GFP-prelamin A fusion construct and treated overnight with vehicle (DMSO), LPV (20 μM), atazanavir (ATV) (20 μM), STV (20 μM), NVP (20 μM), or an FTI (5 μM). Western blot was performed with antibodies against GFP and prelamin A (Middle and Bottom). Top shows the merged image; the anti-GFP signal is red, and the anti-prelamin A signal is green. (E) Western blots of HEK293 cells transiently transfected with the GFP-prelamin A construct and treated overnight with LPV (20 μM), the FTI (5 μM), or vehicle (DMSO). (F) Western blots of HEK293 cells transiently transfected with the GFP-prelamin A construct and treated overnight with LPV (20 μM), TPV (20–100 μM), or DMSO.
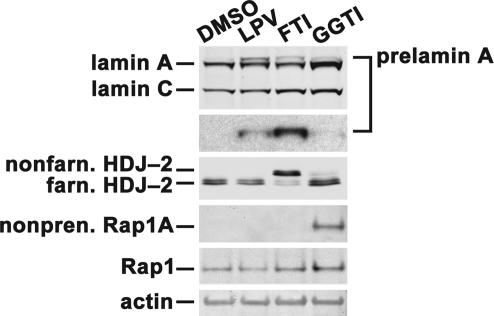
The distinct electrophoretic mobilities of prelamin A and the GFP-prelamin A fusion in FTI- and HIV-PI-treated cells made it unlikely that HIV-PIs inhibit protein FTase. Indeed, treatment of cells with LPV had no effect on the farnesylation of HDJ-2 (Fig. 4). Similarly, LPV also had no effect on the isoprenylation of Rap1A (Fig. 4). Also, in direct biochemical assays, HIV-PIs had no significant effect on the ability of FTase to farnesylate a recombinant H-Ras substrate, whereas an FTI blocked farnesylation at concentrations as low as 0.1 μM (Fig. 5A).
Fig. 4.
Western blots showing that LPV has no effect on protein farnesylation or geranylgeranylation. Wild-type fibroblasts were treated for 24 h with LPV (20 μM), an FTI (5 μM), a geranylgeranyltransferase type I inhibitor (GGTI-298) (15 μM), or vehicle (DMSO). LPV had no effect on the isoprenylation of HDJ-2 or Rap1A.
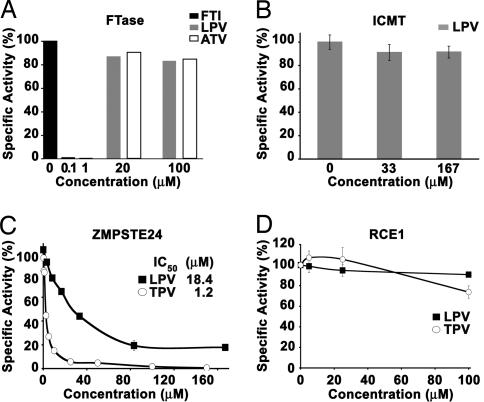
Fig. 5.
HIV-PIs inhibit ZMPSTE24 but not FTase, ICMT, or RCE1 in vitro. (A) Effect of LPV and ATV on the farnesylation of H-Ras by FTase (mean of triplicate determinations). (B) Effect of LPV on the methylation of N-acetyl-farnesylcysteine by ICMT (mean of three experiments, each point in quadruplicate ± SD). (C) Abilities of LPV and TPV to inhibit the enzymatic activity of mouse ZMPSTE24. Shown are results from a coupled endoproteolysis/methylation assay (17) that tested the ability of membranes from Δste24Δrce1 yeast overexpressing mouse ZMPSTE24 to cleave a yeast a-factor substrate, rendering it susceptible to methylation by Ste14p. Each assay was repeated four to seven times, with each point in duplicate, ± SD. (D) Effect of the HIV-PIs LPV and TPV on the activity of mouse RCE1. This assay tested the ability of membranes from Δste24Δrce1 yeast overexpressing RCE1 to cleave an a-factor substrate and then be methylated by Ste14p. Each assay was performed three times, each point in duplicate, ± SD.
A complete deficiency of ICMT partially inhibits the conversion of prelamin A to mature lamin A (15), so it was conceivable that HIV-PIs inhibited ICMT. However, this was not the case. Even at high concentrations, LPV did not block the enzymatic activity of human ICMT (Fig. 5B).
We suspected that the HIV-PIs inhibited ZMPSTE24, which is absolutely required for the conversion of farnesyl-prelamin A to mature lamin A (13, 16). To explore this possibility, we took advantage of the fact that mouse ZMPSTE24 cleaves the last three amino acids (cleavage P1; see Fig. 1) from a farnesylated a-factor substrate (17). The cleavage of the a-factor peptide by ZMPSTE24 can be quantified in a coupled endoproteolysis/methylation assay (17). Using this assay, we found that LPV and TPV inhibited ZMPSTE24 activity (Fig. 5C). The IC50s for LPV and TPV were 18.4 ± 4.6 and 1.2 ± 0.4 μM, respectively. LPV did not change Zmpste24 mRNA levels in fibroblasts, as judged by quantitative PCR (not shown), nor did it change ZMPSTE24 protein levels, as judged by Western blotting [supporting information (SI) Fig. 7]. HIV-PIs had only a marginal effect on the activity of the prenylprotein endoprotease RCE1 (Fig. 5D).
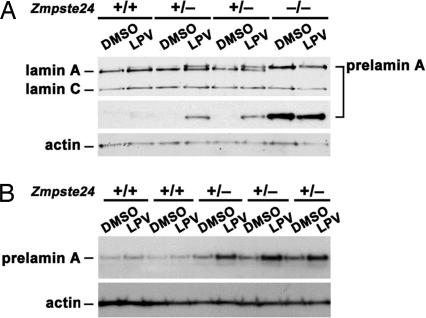
That HIV-PIs inhibit ZMPSTE24 activity (but not FTase, RCE1, or ICMT) explains the accumulation of prelamin A in HIV-PI-treated cells and explains why the prelamin A migrated more rapidly than nonfarnesylated prelamin A. We reasoned that fibroblasts with lower amounts of ZMPSTE24 expression (e.g., cells heterozygous for Zmpste24 deficiency) might be particularly sensitive to the HIV-PIs. Indeed, this was the case; primary fibroblasts from Zmpste24+/− embryos accumulated significantly more prelamin A in response to HIV-PIs than fibroblasts from littermate wild-type embryos (Fig. 6).
Fig. 6.
Mouse fibroblasts heterozygous for Zmpste24 deficiency (Zmpste24+/−) exhibit higher levels of prelamin A accumulation with LPV treatment. Primary cultures of Zmpste24+/+, Zmpste24+/−, and Zmpste24−/− fibroblasts, prepared from littermate embryos, were treated for 10 days with LPV (20 μM) or vehicle (DMSO). A and B show two independent experiments with different cell lines. Quantitative PCR studies showed that Zmpste24 mRNA levels in Zmpste24+/− cells were half-normal (not shown).
Discussion
We demonstrated that HIV-PIs caused a significant accumulation of prelamin A in cells, easily detectable in Western blots with a lamin A/C antibody with ≈31% of the intensity of lamin C. Our data differ from the report by Caron et al. (8); they identified prelamin A in HIV-PI-treated preadipocytes in Western blots with a prelamin A-specific antibody, but the amount of prelamin A accumulation appeared to be miniscule, because no prelamin A could be seen in their lamin A/C Western blots. Importantly, we found that the electrophoretic mobility of the prelamin A in HIV-PI-treated cells was more rapid than the nonfarnesylated prelamin A in FTI-treated cells, comigrating with the farnesyl-prelamin A that accumulates in human RD (ZMPSTE24-deficient) fibroblasts (13). We also found that HIV-PIs interfered with the processing of a GFP-prelamin A fusion in transfected cells; again, the electrophoretic mobility of the uncleaved fusion protein was more rapid in HIV-PI- than in FTI-treated cells. HIV-PIs had no effect on FTase or on ICMT, a methyltransferase that is required for highly efficient conversion of prelamin A to mature lamin A. However, the HIV-PIs clearly inhibited ZMPSTE24, a metalloproteinase that converts farnesyl-prelamin A to mature lamin A. Thus, the accumulation of farnesyl-prelamin A in HIV-PI-treated cells is caused by inhibition of ZMPSTE24.
That the HIV-PIs inhibit ZMPSTE24 is intriguing, given that the HIV protease is a soluble aspartyl protease (1), and ZMPSTE24 is a zinc metalloproteinase of the endoplasmic reticulum with seven predicted transmembrane helices (18, 19). In the case of the HIV protease, the crystal structure has been solved and the binding of HIV-PIs is understood at the molecular level (20). In contrast, there is almost no information regarding the structure of ZMPSTE24, the domains of the enzyme involved in catalysis, or how HIV-PIs might interfere with enzyme activity. ZMPSTE24 contains a zinc-binding HEXXH motif (residues 335–339) that is required for catalysis (18, 19), and missense mutations at residues 265 and 340 dramatically reduce enzymatic activity (10, 21). However, whether HIV-PIs bind in the vicinity of those residues is unknown.
This study did not attempt to determine whether the HIV-PI-induced accumulation of prelamin A in fibroblasts is relevant to the metabolic/lipodystrophy syndrome that occurs in some patients on HAART regimens. The mechanisms for HAART-related side effects are likely heterogeneous and involve multiple drugs (7), and whether the prelamin A accumulation underlies some of the side effects is unknown. However, genetic studies suggest that a prelamin A connection is plausible. Patients with Hutchinson–Gilford progeria syndrome (HGPS) accumulate a truncated farnesyl-prelamin A and develop multiple disease phenotypes, including lipodystrophy (22). In HGPS fibroblasts, the farnesyl-prelamin A band is less intense than the lamin A band, similar to the situation in HIV-PI-treated fibroblasts (22). Also, ZMPSTE24 mutations that partially block the conversion of farnesyl-prelamin A to mature lamin A cause severe progeroid syndromes (10, 21).
Our studies involved cultured fibroblasts and in vitro biochemical assays, but the levels of HIV-PIs that were tested in our studies are similar to those achieved in human patients (1). In the future, it will be important to define the extent of ZMPSTE24 inhibition and prelamin A accumulation in the tissues of HIV-PI–treated patients.
Our studies showed that different HIV-PIs differ in their capacity to inhibit ZMPSTE24. With this knowledge in hand, it should be possible to determine whether the ZMPSTE24-blocking properties of different HIV-PIs correlate with drug side effects in HIV-treated patients. If so, the biochemical approaches described here provide a springboard for identifying new HIV-PIs that lack the capacity to block ZMPSTE24.
The accumulation of prelamin A in response to the HIV-PIs is exaggerated in fibroblasts with half-normal levels of ZMPSTE24. This finding is intriguing for two reasons. First, this finding reinforces biochemical data showing that HIV-PIs cause prelamin A accumulation by inhibiting ZMPSTE24. Second, these data raise the possibility that individuals with relatively low ZMPSTE24 expression levels could be at the greatest risk for developing side effects from HIV-PIs.
In summary, our studies show that HIV-PIs inhibit ZMPSTE24, resulting in a significant accumulation of prelamin A in cells. Future studies are required to define the importance of this finding to the HIV-PI side effects.
Materials and Methods
Cell Culture, Drug Treatment, and Transfection.
Human fibroblasts AG07095 and AG08470 were purchased from Coriell Cell Repository (Camden, NJ). Human ZMPSTE24-deficient (RD) fibroblasts (11) were provided by Casey Moulson and Jeffrey Miner (Washington University School of Medicine, St. Louis, MO). Mouse embryonic fibroblasts were prepared from Zmpste24-, Rce1-, and Icmt-deficient mice (16, 23, 24). All fibroblasts were cultured in 5% CO2 and at 37°C in MEM (Cellgro; Mediatech, Herndon, VA) or DMEM (Gibco/Invitrogen, Carlsbad, CA) containing 10% FBS (HyClone, Logan, UT), 2 mM l-glutamine (Gibco/Invitrogen), and vitamin and amino acid supplements (Cellgro). For transfection studies, HeLa cells, HEK293 cells, and immortalized mouse fibroblasts were grown to 80% confluency and transfected with Fugene HD Transfection Reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions with a 5:2 ratio of transfection reagent (microliters) to DNA (micrograms) in Opti-MEM I Reduced Serum Medium (Gibco). Four hours after transfection, the medium was changed to MEM with 10% FBS, and the cells were incubated with drugs for 18 h.
Reagents.
All antiretroviral drugs were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (www.aidsreagent.org/Index.cfm). The HIV-PIs were prepared as 20 mM stock solutions in DMSO; NVP was prepared as a stock solution at 200 mM in DMSO, and STV was prepared at 20 mM in water. The FTI lonafarnib (Schering–Plough, Kenilworth, NJ) and the geranylgeranyltransferase inhibitor GGTI-298 (Sigma, St. Louis, MO) were prepared as 10 mM solutions in DMSO.
Antibodies and Western Blots.
Cell extracts were separated on NuPAGE 4–12% Bis-Tris gels (Invitrogen) and then transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA). For the HDJ-2 Western blots, samples were run on 15% acrylamide and 0.08% bisacrylamide SDS/PAGE gels (25). Antibodies from Santa Cruz Biotechnology (Santa Cruz, CA) were diluted as follows: anti-lamin A/C goat polyclonal antibody SC-6215 (1:400), anti-lamin A rabbit polyclonal SC-20680 (1:400), anti-GFP mouse monoclonal antibody SC-9996 (1:1,000), anti-Rap1A goat polyclonal SC-1482 (1:200), anti-Rap1 rabbit polyclonal SC-65 (1:200), anti-actin goat polyclonal SC-1616 (1:1,000). An anti-HDJ-2 mouse monoclonal antibody (clone KA2A5.6; Lab Vision, Fremont, CA) was diluted 1:400; an anti-ZMPSTE24 antibody from Novus (Littleton, CO) was used at 1:100. Two prelamin A-specific antibodies were raised against a nonfarnesylated mouse prelamin A peptide, LLGNSSPRSQSSQN. One of the two prelamin A-specific antisera, “a,” bound to human nonfarnesylated prelamin A (in FTI-treated human fibroblasts) but not to human farnesyl-prelamin A (which accumulates in human ZMPSTE24-deficient cells), whereas the other antiserum, “b,” detected both nonfarnesylated prelamin A and farnesyl-prelamin A. Antibody “b” was used in all experiments except as indicated; both antisera were used at 1:4,000. The following antibody detection systems were used: anti-rabbit and anti-goat IgG horseradish peroxidase linked antibodies (GE Healthcare, Piscataway, NJ) diluted 1:4,000 in combination with Amersham ECL Plus Western Blotting Detection system (GE Healthcare); IR-Dye 800CW and IR-Dye 700DX conjugated anti-goat, anti-mouse, and anti-rabbit polyclonal antibodies (Rockland, Gilbertsville, PA) were used at 1:8,000 and detected on an Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE).
GFP-Prelamin A Fusion Construct.
A GFP-prelamin A fusion protein was generated by ligating a prelamin A cDNA fragment encoding amino acids 548–665 downstream of the coding sequences of enhanced GFP (pEGFP-C1; Invitrogen). This fragment was amplified from a full-length prelamin A clone (IMAGE:4240057) with oligonucleotides 5′-GGTAGATCTACCATGGTTGAGGAC-3′ and 5′-TAGAATTCTATTCATTTACATGATGCTGC-3′ and cloned into the BglII and EcoRI sites of pEGFP-C1.
Endoprotease-Coupled Methylation Assays and Other Enzymatic Assays.
Membrane fractions (26) were prepared from the following yeast strains: Δste24Δrce1 (17); Δste24Δrce1 overexpressing mouse ZMPSTE24 (pMB4) (17) or mouse RCE1 (pCH10HA-N-Δ1–21-mRCE1) (17); CH2733, Δste24Δrce1 overexpressing Ste14p (pCHH10m3N-Ste14) (27); and CH2766, Δste14 yeast overexpressing human ICMT (pCHH10m3N-hICMT) (27). The substrate for the endoprotease-coupled methylation reaction was a farnesylated 15-mer a-factor peptide [YIIKGVFWDPA(farnesyl)CVIA] (synthesized by California Peptide Research, Napa, CA). Endoprotease-coupled methylation reactions (17) were assembled on ice by mixing 5 μg of Δste24Δrce1 overexpressing mouse ZMPSTE24 or RCE1, 8 μg of CH2733 membranes, the farnesylated peptide (25 μM for mouse RCE1 reactions and 5 μM for mouse ZMPSTE24 reactions), and 20 μM S-adenosyl-l-[methyl-14C]methionine (55 Ci/mol, Amersham; GE Healthcare) in 100 mM Tris·HCl, pH 7.5, for a final volume of 60 μl. HIV-PIs do not inhibit yeast Ste14p (not shown). Where noted, the reactions also contained HIV-PIs at the indicated concentration. After incubating the reactions at 30°C for 30 min, the reactions were stopped with 50 μl of 1 M NaOH/1% SDS; the reactions were then spotted on a pleated filter paper, and base-releasable [14C]methanol was quantified by a vapor diffusion assay (28). IC50 values were calculated by using GraphPad 4.0 (San Diego, CA). Human ICMT activity was measured by incubating 5 μg of CH2766 membranes with 25 μM N-acetyl-S-farnesyl-l-cysteine and 20 μM S-adenosyl-l-[methyl-14C]methionine in 100 mM Tris·HCl, pH 7.5, for 30 min at 30°C (27). FTase activity measurements were performed as described (29). Reaction mixtures containing 50 ng of FTase, 500 nM [3H]FPP, and 1 μM H-Ras (an FTase substrate) were incubated at 30°C for 10 min. After protein precipitation, the samples were glass-fiber-filtered, and the radiolabeled isoprenoid was detected by scintillation counting (29).
Acknowledgments
We thank Drs. Casey Moulson and Jeffrey Miner for the RD fibroblasts; Carolyn A. Weinbaum and Dr. Patrick J. Casey (Duke University, Durham, NC) for measuring FTase activities; and Heather B. Hodges-Loaiza (Purdue University) for providing human ICMT-expressing yeast membranes. This research was supported by National Institutes of Health Grants AR050200, HL76839, HL86683, and CA099506; by grants from the Progeria Research Foundation; and by a grant-in-aid from the American Heart Association, Western States Affiliate (0665016Y).
Abbreviations
- HIV-PI
HIV protease inhibitor
- HAART
highly active antiretroviral therapy
- LPV
lopinavir
- FTase
farnesyltransferase
- FTI
FTase inhibitor
- RD
restrictive dermopathy
- TPV
tipranavir
- NVP
nevirapine
- STV
stavudine
- ICMT
isoprenyl-cysteine carboxyl methyltransferase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.F.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704212104/DC1.
References
- 1.Flexner C. N Engl J Med. 1998;338:1281–1292. doi: 10.1056/NEJM199804303381808. [DOI] [PubMed] [Google Scholar]
- 2.Mondy K, Tebas P. Annu Rev Med. 2007;58:141–155. doi: 10.1146/annurev.med.58.072905.180040. [DOI] [PubMed] [Google Scholar]
- 3.Grinspoon S, Carr A. N Engl J Med. 2005;352:48–62. doi: 10.1056/NEJMra041811. [DOI] [PubMed] [Google Scholar]
- 4.Vigouroux C, Gharakhanian S, Salhi Y, Nguyên TH, Adda N, Rozenbaum W, Capeau J. Diabetes Metab. 1999;25:383–392. [PubMed] [Google Scholar]
- 5.Green ML. J Gen Intern Med. 2002;17:797–810. doi: 10.1046/j.1525-1497.2002.20201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr A. Aids. 2003;17(Suppl 1):S141–S148. [PubMed] [Google Scholar]
- 7.Nolan D, John M, Mallal S. Antivir Ther. 2001;6:145–160. [PubMed] [Google Scholar]
- 8.Caron M, Auclair M, Sterlingot H, Kornprobst M, Capeau J. AIDS. 2003;17:2437–2444. doi: 10.1097/00002030-200311210-00005. [DOI] [PubMed] [Google Scholar]
- 9.Hegele RA. Mol Genet Metab. 2000;71:539–544. doi: 10.1006/mgme.2000.3092. [DOI] [PubMed] [Google Scholar]
- 10.Agarwal AK, Fryns J-P, Auchus RJ, Garg A. Hum Mol Genet. 2003;12:1995–2001. doi: 10.1093/hmg/ddg213. [DOI] [PubMed] [Google Scholar]
- 11.Moulson CL, Go G, Gardner JM, van der Wal AC, Smitt JH, van Hagen JM, Miner JH. J Invest Dermatol. 2005;125:913–919. doi: 10.1111/j.0022-202X.2005.23846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navarro CL, Cadinanos J, De Sandre-Giovannoli A, Bernard R, Courrier S, Boccaccio I, Boyer A, Kleijer WJ, Wagner A, Giuliano F, et al. Hum Mol Genet. 2005;14:1503–1513. doi: 10.1093/hmg/ddi159. [DOI] [PubMed] [Google Scholar]
- 13.Young SG, Fong LG, Michaelis S. J Lipid Res. 2005;46:2531–2558. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Beck LA, Hosick TJ, Sinensky M. J Cell Biol. 1990;110:1489–1499. doi: 10.1083/jcb.110.5.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young SG, Clarke S, Bergo M, Philips M, Fong LG. In: The Enzymes. Clarke S, Tamanoi F, editors. Vol 24. San Diego, CA: Academic; 2005. pp. 273–301. [Google Scholar]
- 16.Bergo MO, Gavino B, Ross J, Schmidt WK, Hong C, Kendall LV, Mohr A, Meta M, Genant H, Jiang Y, et al. Proc Natl Acad Sci USA. 2002;99:13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung GK, Schmidt WK, Bergo MO, Gavino B, Wong DH, Tam A, Ashby MN, Michaelis S, Young SG. J Biol Chem. 2001;276:29051–29058. doi: 10.1074/jbc.M102908200. [DOI] [PubMed] [Google Scholar]
- 18.Boyartchuk VL, Ashby MN, Rine J. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 19.Fujimura-Kamada K, Nouvet FJ, Michaelis S. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eder J, Hommel U, Cumin F, Martoglio B, Gerhartz B. Curr Pharmacol Des. 2007;13:271–285. doi: 10.2174/138161207779313560. [DOI] [PubMed] [Google Scholar]
- 21.Shackleton S, Smallwood DT, Clayton P, Wilson LC, Agarwal AK, Garg A, Trembath RC. J Med Genet. 2005;42:e36. doi: 10.1136/jmg.2004.029751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, et al. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim E, Ambroziak P, Otto JC, Taylor B, Ashby M, Shannon K, Casey PJ, Young SG. J Biol Chem. 1999;274:8383–8390. doi: 10.1074/jbc.274.13.8383. [DOI] [PubMed] [Google Scholar]
- 24.Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Gomes AQ, Seabra MC, Young SG. J Biol Chem. 2001;276:5841–5845. doi: 10.1074/jbc.C000831200. [DOI] [PubMed] [Google Scholar]
- 25.Cox AD, Solski PA, Jordan JD, Der CJ. Methods Enzymol. 1995;255:195–220. doi: 10.1016/s0076-6879(95)55023-2. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt WK, Tam A, Michaelis S. J Biol Chem. 2000;275:6227–6233. doi: 10.1074/jbc.275.9.6227. [DOI] [PubMed] [Google Scholar]
- 27.Anderson JL, Frase H, Michaelis S, Hrycyna CA. J Biol Chem. 2005;280:7336–7345. doi: 10.1074/jbc.M410292200. [DOI] [PubMed] [Google Scholar]
- 28.Clarke S, Vogel JP, Deschenes RJ, Stock J. Proc Natl Acad Sci USA. 1988;85:4643–4647. doi: 10.1073/pnas.85.13.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor JS, Reid TS, Terry KL, Casey PJ, Beese LS. EMBO J. 2003;22:5963–5974. doi: 10.1093/emboj/cdg571. [DOI] [PMC free article] [PubMed] [Google Scholar]