DNA damage induces a number of cellular responses in human cells, including DNA repair, transcriptional reprogramming, delay of cell cycle progression, and apoptosis (1). The most common DNA lesions fall into two broad groups: base lesions and single- and double-strand breaks. Both types of lesions are detected by damage sensors that initiate various response reactions. A central question in understanding these responses is the identity of the damage sensor. The study by Derheimer et al. (2) in a recent issue of PNAS, together with previous studies, suggests that RNA polymerase II (RNAP II) stalled at a damaged DNA base may constitute the most specific signal for DNA repair, DNA damage checkpoints, and apoptosis (Fig. 1). We suggest that RNAP II is an ideal damage sensor because it has the highest selectivity of all known DNA damage recognition proteins.
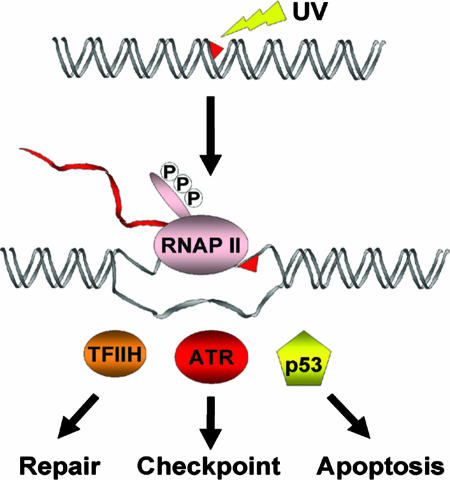
Fig. 1.
RNAP II as the universal high-specificity damage sensor for three major cellular responses to bulky DNA lesions, such as the cyclobutane pyrimidine dimers induced by UV light. RNAP II arrests at a dimer site in the transcribed strand. The resulting structure recruits proteins that initiate repair, cell cycle checkpoints, or apoptosis.
In general, the specificity of a sequence or a structure-specific DNA damage recognition protein is determined by the dissociation off-rate for the target relative to the dissociation off-rate for the nontarget DNA. The off-rates for the vast majority of specific DNA-binding proteins, such as repressors, activators, repair enzymes, and damage recognition proteins, bound at their target sites is in the range of 10−1 to 10−3 s−1. Although these slow off-rates provide considerable specificity, the specificity gained is compromised by the significant competition from the vast excess of undamaged or nontarget DNA. In contrast to the limitations of these direct or matchmaker recognition mechanisms (1), RNAP II, which is not a damage sensor as such, provides damage recognition specificity by a mechanism referred to as “recognition by proxy” (1). During transcription, RNAP II stops when it encounters bulky lesions, such as the UV-induced thymine dimer, and the resulting ternary complex of RNAP II–RNA–DNA has a half-life of ≈20 h or an off-rate of koff ≈ 1 × 10−5 s−1 (3). In contrast, even damage recognition proteins with the highest selectivity, such as UV-DDB, have a half-life of only ≈15 min (koff ≈ 1 × 10−3 s−1) for specific complexes at high affinity sites, such as the (6-4) photoproduct and a half-life in the range of 1–2 s (koff ≈ 2 × 10−1 s−1) for the thymine dimers that constitute ≈90% of the UV-induced photoproducts (4). These values mean that RNAP II can provide damage detection specificity 100- to 10,000-fold higher for UV-damaged DNA, in particular cyclobutane pyrimidine dimers, than any damage recognition protein known to date. Despite this impressive capacity of RNAP II to detect damage, it is generally not considered to be a damage recognition protein, in part because not all of DNA is transcribed, and, even in the transcribed sequences, damage in the coding strand does not block RNAP II. Hence, it was assumed that such stalled complexes would be relatively infrequent. However, whereas RNAP II was once thought to only transcribe the ≈2% of the genome containing protein-coding DNA, recent analysis of the human genome shows that the vast majority of bases can be found in primary transcripts (5). Therefore, RNAP II is an ideal damage sensor in cellular responses to DNA damage because it continuously “scans” the genome by engaging in transcription during all phases of the cell cycle except mitosis. This singularity of RNAP II is in contrast to repair proteins with the sole purpose of damage recognition with much lower specificity and with DNA polymerases that stall at damage sites and provide damage sensor function during S phase but are rapidly replaced by error-prone DNA polymerases carrying out translesion synthesis (6).
The role of RNAP II in repair is well established (7). The stalled RNAP II is recognized by a class of translocases called transcription–repair coupling factors that bind to both the stalled polymerase and nucleotide excision repair factors to recruit the repair factors to the site of damage and accelerate the rate of repair by ≈10-fold (8). The mechanistic details of the transcription–repair coupling are relatively well understood in Escherichia coli cells but not in human cells (8).
The role of stalled RNAP II as a damage sensor in other cellular response reactions, namely apoptosis and DNA damage checkpoints, is not as commonly appreciated as its role in transcription-coupled repair, although these response reactions are perhaps of equal significance in the genomic stability of the cell and survival of multicellular organisms. Previously, it was shown that, in human cell lines with defective transcription-coupled repair, stalled RNAP II causes an increase in p53 levels and eventual induction of apoptosis (9). The report by Derheimer et al. (2) that stalled RNAP II leads to p53 induction in a manner that depends on ATR [ataxia telangiectasia mutated (ATM)- and Rad3-related] and RPA (replication protein A) provides fresh evidence that RNAP II does, in fact, function as a damage sensor for the DNA damage checkpoint response. This study shows that the inhibition of elongation by RNAP II with three DNA damaging agents (UV light, actinomycin D, and psoralen), which cause pyrimidine dimers, base intercalation, and interstrand DNA cross-links, respectively, induces p53 phosphorylation in nonproliferating cells. Thus, the p53 activation is not specific to the type of DNA damage or cell cycle phase, and it is fair to assume that any base lesion that blocks transcription elongation would lead to p53 accumulation. This conclusion was considerably strengthened by experiments in which anti-RNAP II antibodies were microinjected into the nuclei of cells not subjected to any DNA-damaging treatment. Antibodies against the elongating form of RNAP II, which has a phosphorylated C-terminal domain (CTD), led to phosphorylation of p53, whereas antibodies against the preelongating form of RNAP II, which has an unphosphorylated CTD, did not.
The Ser-15 residue of p53 that was phosphorylated as a consequence of inhibition of transcription elongation is known to be the target of the phosphoinositide kinase-like kinase (PIKK) family members, ATM and ATR (1). In general, ATM phosphorylates its target proteins in response to double-strand breaks, and ATR does the same in response to base lesions that block replication or transcription. To ascertain whether the blockage of transcription elongation leads to the phosphorylation of p53 Ser-15 by ATR, Derheimer et al. (2) microinjected anti-ATR antibodies known to block ATR kinase activity together with anti-RNAP II phospho-CTD antibodies. Under this condition, p53 Ser-15 was no longer phosphorylated, indicating that p53 phosphorylation after inhibition of transcription elongation is mediated by ATR. Because several reports have indicated that RPA plays a crucial role in the recruitment of ATR to single-stranded DNA, which is known to be a strong signal for checkpoint activation (10), Derheimer et al. tested the role of RPA in the activation of ATR by stalled RNAP II. They found that the microinjection of anti-RPA antibodies, along with anti-RNAP II antibodies that inhibit transcription elongation, abolished the p53 Ser-15 phosphorylation elicited by stalled elongation complexes.
These findings by Derheimer et al. (2) led them to propose the following model. Stalled RNAP II is associated with single-stranded DNA at the transcription bubble that is bound by RPA, which then recruits ATR to DNA, activating the DNA damage checkpoint. Indeed, a previous study (11) using a ChIP assay showed that RPA, ATR, and other “damage sensor” checkpoint proteins are recruited to transcribed sequences after DNA-damaging treatments that produce bulky base lesions, and the authors concluded that stalled RNAP II elongation complexes can activate the checkpoint response in the absence of replication or repair.
It must be noted, however, that, although the paper by Derheimer et al. (2) mechanistically and teleologically makes sense, it is in apparent disagreement with a number of reports on the nature of the signal for recruitment of ATR to the site of damage and activation of the checkpoints, of which p53 Ser-15 phosphorylation is just one manifestation (Fig. 2). First, several studies have claimed that DNA-damaging agents that produce bulky base adducts activate the ATR-mediated checkpoint response in the G1 (or G0) phase of the cell cycle only when the damage is excised by nucleotide excision repair, which produces ≈30-nt single-stranded gaps (12–15). Second, although it has been suggested that RPA binds to single-stranded DNA in the transcription bubble (2), another study failed to detect preferential binding of RPA to transcribed DNA (16). Finally, a recent report (11) suggests that the primary DNA base damage itself can be recognized by the checkpoint sensors to activate the DNA damage checkpoints. A follow-up in vitro study (17) supports these in vivo findings by demonstrating that, in the presence of TopBP1, ATR is recruited to the primary base damage and is activated as a kinase. It also is quite likely that the discrepancies between this and some of the other studies regarding the nature of the damage-sensing mechanisms are in large part caused by the different experimental approaches for analyzing the checkpoint response, all of which, including the one used by Derheimer et al., have certain limitations. Undoubtedly, further studies will reconcile these apparent discrepancies among the various reports and will result in a unified model that encompasses more than one way of sensing the DNA damage (Fig. 2).
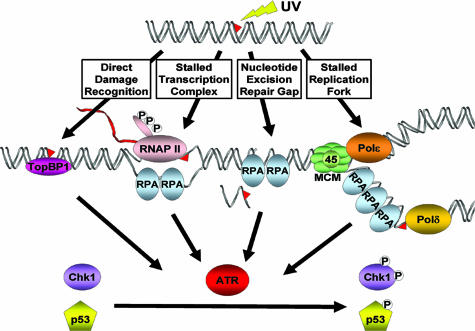
Fig. 2.
Potential DNA damage detection mechanisms for initiating checkpoint responses. The primary damage itself may be recognized by damage-specific proteins that may activate the checkpoint directly. Alternatively, the stalled RNAP II constitutes the checkpoint signal and recruits checkpoint kinase ATR. RPA-coated single-stranded DNA gaps generated by nucleotide excision repair and the extensive RPA-coated single-stranded DNA generated by replication forks stalled at a damage site are known to be strong signals for checkpoint activation.
In conclusion, the paper by Derheimer et al. (2) serves two important purposes. First, it presents strong evidence that stalled RNAP II can activate p53 and initiate apoptosis and perhaps cell cycle checkpoints. Second, it reemphasizes the role of stalled RNAP II, and RNA polymerases in general, as a major sensor for all DNA damage response reactions and hopefully will draw more attention to this important function.
Footnotes
The authors declare no conflict of interest.
See companion article on page 12778 in issue 31 of volume 104.
References
- 1.Sancar A, Lindsey-Boltz LA, Ünsal-Kaçmaz K, Linn S. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 2.Derheimer FA, O'Hagan HM, Krueger HM, Hanasoge S, Paulsen MT, Ljungman M. Proc Natl Acad Sci USA. 2007;104:12778–12783. doi: 10.1073/pnas.0705317104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selby CP, Drapkin R, Reinberg D, Sancar A. Nucleic Acids Res. 1997;25:787–793. doi: 10.1093/nar/25.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reardon JT, Nichols AF, Keeney S, Smith CA, Taylor JS, Linn S, Sancar A. J Biol Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 5.The ENCODE Project Consortium. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prakash S, Johnson RE, Prakash L. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 7.Hanawalt PC. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 8.Selby CP, Sancar A. Methods Enzymol. 2003;371:300–324. doi: 10.1016/S0076-6879(03)71023-4. [DOI] [PubMed] [Google Scholar]
- 9.Ljungman M, Zhang F. Oncogene. 1996;13:823–831. [PubMed] [Google Scholar]
- 10.Zou L. Genes Dev. 2007;21:879–885. doi: 10.1101/gad.1550307. [DOI] [PubMed] [Google Scholar]
- 11.Jiang G, Sancar A. Mol Cell Biol. 2006;26:39–49. doi: 10.1128/MCB.26.1.39-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 13.Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. Proc Natl Acad Sci USA. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto M, Yaginuma K, Igarashi A, Imura M, Hasegawa M, Iwabuchi K, Date T, Mori T, Ishizaki K, Yamashita K, et al. J Cell Sci. 2007;120:1104–1112. doi: 10.1242/jcs.03391. [DOI] [PubMed] [Google Scholar]
- 15.Marini F, Nardo T, Giannattasio M, Minuzzo M, Stefanini M, Plevani P, Falconi MM. Proc Natl Acad Sci USA. 2006;103:17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, Li G, Schrum JP, Manis JP, Alt FW. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 17.Choi J-H, Lindsey-Boltz LA, Sancar A. Proc Natl Acad Sci USA. 2007;104:13301–13306. doi: 10.1073/pnas.0706013104. [DOI] [PMC free article] [PubMed] [Google Scholar]