Abstract
MicroRNAs are predicted to regulate ≈30% of all human genes by targeting sequences in their 3′ UTR. Polymorphisms in 3′ UTR of several genes have been reported to affect gene expression, but the mechanism is not fully understood. Here, we demonstrate that 829C→T, a naturally occurring SNP, near the miR-24 binding site in the 3′ UTR of human dihydrofolate reductase (DHFR) affects DHFR expression by interfering with miR-24 function, resulting in DHFR overexpression and methotrexate resistance. miR-24 has a conserved binding site in DHFR 3′ UTR. DHFR with WT and 3′ UTR containing the 829C→T mutation were expressed in DG44 cells that lack DHFR. Overexpression of miR-24 in cells with WT DHFR resulted in down-regulation of DHFR protein, whereas no effect on DHFR protein expression was observed in the mutant 3′ UTR-expressing cells. Inhibition of endogenous miR-24 with a specific inhibitor led to up-regulation of DHFR in WT and not in mutant cells. Cells with the mutant 3′ UTR had a 2-fold increase in DHFR mRNA half-life, expressed higher DHFR mRNA and DHFR protein, and were 4-fold more resistant to methotrexate as compared with WT cells. SNP-829C→T, therefore, leads to a decrease in microRNA binding leading to overexpression of its target and results in resistance to methotrexate. We demonstrate that a naturally occurring miRSNP (a SNP located at or near a microRNA binding site in 3′ UTR of the target gene or in a microRNA) is associated with enzyme overproduction and drug resistance.
Keywords: 3′ UTR, drug resistance, single nucleotide polymorphism, translational regulation, mRNA stability
MicroRNAs (miRNAs) are generally 22 nt in length, evolutionarily conserved, and a highly abundant class of small noncoding RNAs found in diverse organisms. miRNAs play key roles in regulating the translation and degradation of mRNAs through base pairing to partially complementary sites in the 3′ UTR of the message (1–4). Degradation of the miRNA-bound mRNA can result in reduced steady-state levels of the corresponding transcript (5, 6). The processed miRNA binds to the highly conserved Argonaute (Ago) protein family, in an RNA-induced silencing complex (RISC) that drives gene-silencing events by forming an imperfect hybrid with the target mRNA. Imperfections in the central portions of miRNA–mRNA duplex preclude RNAi-like cleavage and leads to translational repression, frequently accompanied by a considerable degradation of the mRNA by a non-RNAi mechanism (7, 8).
The biological functions of most miRNAs are not yet fully understood. It has been suggested that miRNAs are involved in various biological processes, including cell proliferation, cell death, stress resistance, and fat metabolism through the regulation of gene expression (9). miRNAs may also be differentially expressed in human cancers; changes in miRNA levels have been reported in chronic lymphocytic leukemia cells (10, 11), lymphoma, and solid tumors (12). Recently, it has been shown that miRNAs act as oncogenes and tumor suppressors by targeting key regulators of cell growth (13, 14).
miRNAs are predicted to regulate 30% of all genes and to target sequences in the 3′ UTR of genes. Many 3′ UTR polymorphisms have been reported to be associated with differential gene expression, but the underlying mechanism is not fully understood (reviewed in ref. 15). We propose that some of the 3′ UTR polymorphisms may be in the vicinity of an miRNA binding site and may interfere with miRNA function leading to differential gene expression. Dihydrofolate reductase (DHFR) is a key enzyme in intracellular folate metabolism and target of methotrexate (MTX), an important chemotherapeutic agent widely used in the treatment of several malignancies, including acute lymphocytic leukemia, non-Hodgkin's lymphoma, osteosarcoma, and choriocarcinoma (16, 17).
In the clinic, some patients respond to MTX treatment, whereas some are intrinsically resistant to MTX treatment (18). Several mechanisms contributing to MTX resistance have been identified in cell-culture model systems and in clinical samples from MTX resistant patients (19–22). Because of its central role in DNA synthesis, DHFR would be expected to be highly regulated, but there are only few reports addressing the 3′ UTR-mediated regulation of DHFR (23, 24). Recently, an SNP- 829C→Τ was reported to occur in the 3′ UTR of DHFR gene at a 14.2% allelic frequency in the Japanese population, associated with an increase in the expression of DHFR message (24).
Here, we demonstrate that SNP-829C→T is located near the miR-24 binding site in DHFR 3′ UTR and acts as a loss-of-function mutation. The SNP affects DHFR expression by interfering with miR-24 function, resulting in DHFR overexpression and MTX resistance. Our data allow the suggestion that DHFR mRNA and protein levels are at least in part regulated by miR-24.
Results
DHFR Protein Level Is Regulated by miRNA miR-24 (hsa-miR-24).
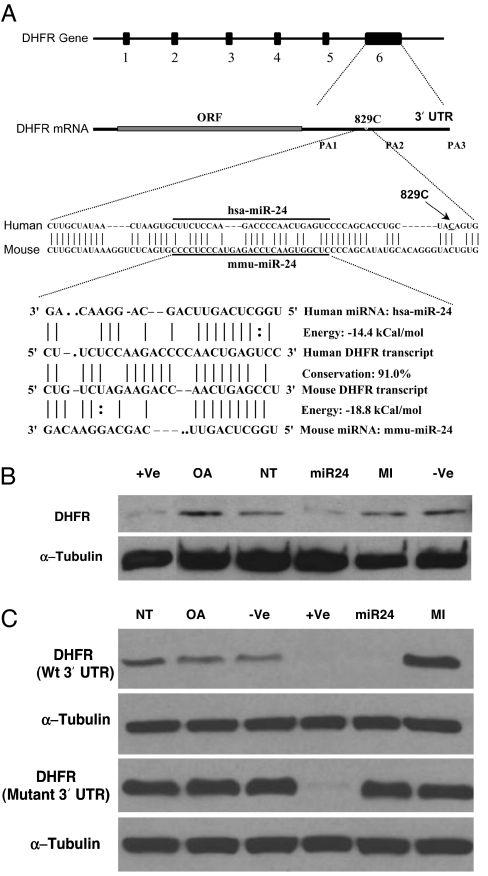
Using miRanda web server (25), we determined that the DHFR 3′ UTR harbors a putative mir-24 microRNA binding site (Fig. 1A). miR-24 mimics (double-stranded processed miRNA) and inhibitors (complementary single-stranded miRNA inhibitors) were transfected in HT1080 human fibrosarcoma cells. DHFR and α-tubulin (control) protein levels were determined using Western blot analysis. Overexpression of the miR-24 mimic led to down-regulation of DHFR protein (Fig. 1B). siRNA specific to DHFR 3′ UTR, transfected as positive control, also down-regulated DHFR protein as expected. Transfection of a mixture of nonspecific duplex miRNA and Oligofectamine alone as negative controls had no effect on DHFR, indicating that DHFR protein was specifically down-regulated by miR-24 miRNA mimics.
Fig. 1.
SNP-829C→T affects DHFR expression by interfering with miR-24 function. (A) The DHFR gene has six exons and is located on chromosome 5. The 3′ UTR of DHFR mRNA harbors a putative miRNA miR-24 binding site, which is 91% conserved in mice and human. An SNP-829C site is located 14 bp downstream of the miR-24 binding site in the DHFR 3′ UTR and is conserved in mice and humans. The sequences of human and mouse miR-24, its binding sites, and energies are shown. (B) Overexpression of miR-24 mimic (processed miRNA) leads to down-regulation of the DHFR protein in HT1080 human fibrosarcoma cells (miR-24) as determined by Western blot analysis. An siRNA specific to DHFR 3′ UTR was transfected as a positive control (+Ve). A nonspecific miRNA cel-miR-67 (−Ve) and Oligofectamine alone (OA), nontransfected cells (NT), were used as negative controls. (C) DG44 cells stably expressing WT and the 829C→T mutation in the 3′ UTR were transfected with a miR-24 mimic and inhibitor (complementary single stranded RNA) (see Materials and Methods). An siRNA specific to DHFR 3′ UTR was transfected as a positive control (+Ve). A nonspecific miRNA cel-miR-67 (−Ve) and Oligofectamine alone (OA), nontransfected cells (NT), were used as negative controls. The experiments were repeated twice in two different cell lines, the human fibrosarcoma cell line HT1080 and the CHO-DG44 cell line.
SNP-829C→T Is Located Near a miR-24 Binding Site in DHFR 3′ UTR.
The miR-24 binding site is 91% conserved in mouse, rat, and human DHFR 3′ UTRs (Fig. 1A). The cytosine at position 829, located 14 bp downstream of the miRNA binding site, is conserved in mice and humans. This is likely because miRNA-mRNA binding is mediated by the RISC complex, and upstream and downstream regions of miRNA binding site may interact with RISC, which mediates miRNA–mRNA binding (26). A polymorphism in the 829C site (SNP-829C→T) is located near the miRNA binding site.
SNP-829C→T Affects DHFR Expression by Interfering with miR-24 Function.
To determine the effect of SNP-829C→T on miR-24-mediated regulation of DHFR, WT DHFR with a WT 3′ UTR and DHFR cDNA with the mutation 829C→T were cloned in a pCDNA3.1 vector. WT and mutant DHFR constructs were transfected into DG44 CHO cells that lack endogenous DHFR (27), to compare DHFR levels in the absence of an endogenous DHFR background. Individual G418 resistant clones were picked and expanded to cell lines. miR-24 mimics and inhibitors were then transfected in WT and mutant DHFR 3′ UTR-expressing DG44 cells. Overexpression of the miR-24 mimics in WT cells led to down-regulation of DHFR protein (Fig. 1C). In cells containing the 829C→T SNP, overexpression of the miR-24 mimic had no effect on the expression level of DHFR protein, suggesting that miR-24-mediated regulation of DHFR was lost in mutant DHFR-expressing cells. Notably, inhibition of endogenous miRNA by overexpressing the miR-24 inhibitor led to up-regulation of DHFR protein in WT but not in mutant cells (Fig. 1C). siRNA specific to DHFR 3′ UTR, transfected as a positive control, down-regulated DHFR in both WT and mutant cells as expected. Transfection of a nonspecific miRNA cel-miR-67 and Oligofectamine alone as negative controls showed no effect on DHFR protein levels. Also, cells expressing mutant DHFR 3′ UTR had higher steady-state levels of DHFR protein expression as compared with WT DHFR 3′ UTR-expressing cells (Fig. 1C). We conclude that DHFR protein levels are regulated by endogenous miR-24 and the miRNA-mediated regulation is lost when SNP-829C→T is introduced near the miR-24 binding site in DHFR 3′ UTR.
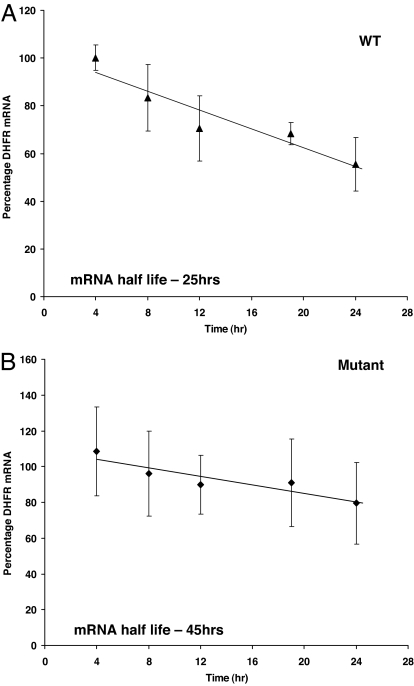
SNP-829C→T Leads to Increased mRNA Stability.
miRNA-bound mRNA may be degraded, resulting in reduced steady-state levels of the corresponding transcript (28). In cells with 829C→T SNP, miR-24 mediated regulation may be lost, a result of impaired binding of miRNA to the region, leading to enhanced mRNA stability in mutant clones. To test this hypothesis, DHFR mRNA half-life was measured in DG44 cells with and without the 829C→T SNP. The presence of SNP-829C→T in DHFR 3′ UTR was associated with a two-fold increase in half-life (WT, 25 h; mutant, 45 h) of the DHFR message (Fig. 2). The half-life of DHFR with the WT 3′ UTR was similar to that reported in refs. 29–31.
Fig. 2.
SNP-829C→T in the miR-24 binding site in DHFR 3′ UTR of DHFR gene leads to enhanced DHFR mRNA stability. DHFR mRNA decay in five WT (A) and five mutant clones (B) were determined by real-time quantitative RT-PCR.
When putative mRNA folding patterns of WT and mutant DHFR 3′ UTRs were determined by using mfold program (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html), the mutant DHFR 3′ UTR mRNA had a more stable folding pattern (ΔG-120.64) compared to that of WT (ΔG-128.19).
Cells Expressing SNP-829C→T Adjacent to the miR-24 Binding Site Overexpress DHFR mRNA Without an Increase in DHFR Gene Copy Number.
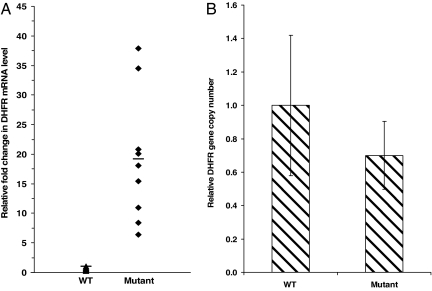
Quantitative real-time RT-PCR was used to determine the ratio of DHFR mRNA levels to β-actin in five WT and nine mutant DHFR 3′ UTR-expressing stable clones. An average 19-fold increase in mRNA level was observed in clones expressing the 829C→T mutation as compared with WT clones (Fig. 3A).
Fig. 3.
SNP-829C→T leads to overexpression of DHFR mRNA without a change in gene copy number. (A) Clones expressing the mutant 829C→T showed average 19-fold higher levels of DHFR mRNA as compared with WT (P < 0.003). Real-time quantitative RT-PCR was used to compare mRNA levels of the clones (see Materials and Methods). (B) Comparison of WT and mutant-expressing clone gene copy numbers (P < 0.92) as detected by real-time quantitative PCR. The gene copy number of five WT DHFR clones was averaged to one and relative fold differences in gene copy number of clones expressing the mutant DHFR were determined.
To rule out gene amplification as a cause of the increase in mRNA, DHFR gene copy numbers were compared by using quantitative PCR using primers and probes designed to amplify DHFR cDNA and genomic β-actin as the control. The average DHFR genomic DNA level in the WT DHFR-expressing clones (n = 5) was found to be comparable to mutant clones (Fig. 3B). Thus, higher levels of DHFR mRNA in mutants could not be attributed to increased DHFR gene copy number.
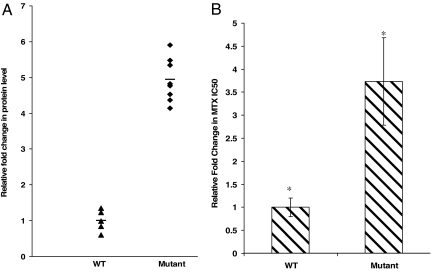
Clones Expressing the Mutant 829C→T Express Increased Levels of DHFR, and as a Result, Are More Resistant to MTX.
Corresponding to higher message levels, DHFR protein was, on average, 5-fold higher in clones expressing the 829C→T SNP as compared with WT clones (Fig. 4A). We tested whether the SNP near the miRNA binding site leads to drug resistance because increased DHFR protein levels are associated with drug resistance (16). 829C→T cell lines were significantly more resistant to MTX: average IC50 for MTX in WT cells was 0.29 μM (±0.2), and in mutants, IC50 was 1.03 μM (±0.27). Thus, SNP-829C→T near the miR-24 binding site in the 3′ UTR of DHFR stabilizes the DHFR message and leads to high mRNA levels, high DHFR protein levels, and increased MTX resistance (Fig. 4 A and B).
Fig. 4.
SNP-829C→T in DHFR 3′ UTR leads to overexpression of DHFR protein levels and confers MTX resistance. (A) A 5-fold increase in DHFR protein level was observed in the mutant clones as compared with WT (*, P < 0.0001) as assayed by Western blotting. The bands were quantitated with Kodak Image Station 2000MM. The band intensities of five WT DHFR clones were averaged to one, and relative fold difference in band intensity of clones expressing the mutant DHFR was determined. (B) Comparison of SNP-829C→T-expressing cells with WT cells to MTX cytotoxicity (*, P < 0.005). IC50 values were determined by a cytotoxicity assay (see Materials and Methods).
Discussion
miRNAs are known to inhibit gene expression by binding to the 3′ UTR of the target transcript. 3′ UTR polymorphisms are reported to be associated with various diseases that include hereditary thrombophilia (32), α-thalassemia (33), insulin sensitivity (34), urolithiasis (35), human papilloma virus infection (36, 15), and increased sensitivity to 5-fluorouracil chemotherapy (37, 38). Recently, it has been shown that a G-to-A transition in the 3′ UTR of the myostatin gene in Texel sheep creates a potential illegitimate miRNA target site, which recruits miR1 and miR206 binding and leads to translational inhibition of the gene, resulting in muscular hypertrophy (39). There are no reports of polymorphisms at or in close proximity to miRNA binding sites leading to loss of miRNA-mediated regulation of a gene.
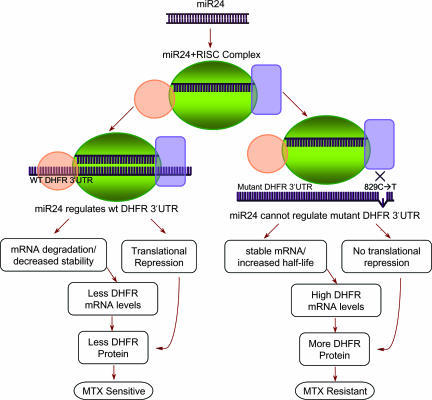
In this report, we present evidence that SNP-829C→T adjacent to the miR-24 binding site in DHFR 3′ UTR affects DHFR expression by interfering with miR-24 function, resulting in DHFR overexpression and MTX resistance. Furthermore, we propose that miRSNPs located in an miRNA or at or near the miRNA binding site in 3′ UTRs of many important genes that are drug targets may affect the drug response in patients and may lead to drug resistance or drug sensitivity (Fig. 5). The miR-24 miRNA-mRNA binding site is 91% conserved in mice, rat, and human DHFR 3′ UTR. By overexpressing miR-24 mimics and inhibiting endogenous miR-24, we demonstrate that SNP-829C→T acts as a loss-of-function mutation and results in the enhanced expression of DHFR mRNA and protein levels in mutant-expressing cells by affecting the binding of miR-24 to 3′ UTR of DHFR (Fig. 5).
Fig. 5.
A model for miR-24-mediated regulation of DHFR. The processed miR-24 miRNA binds to the RISC. The miR-24–RISC complex then binds to WT DHFR 3′ UTR and regulates DHFR mRNA and protein expression, resulting in less net DHFR protein levels in the cell. The miRSNP-829C→T leads to loss of miR-24-mediated regulation of DHFR, which may be a result of weak or no binding between the miR-24–RISC complex and DHFR 3′ UTR. This occurrence leads to comparatively stable DHFR message, a high net DHFR mRNA and protein levels in mutant cells, leading to MTX resistance.
In animals, unlike plants, miRNAs were believed to act by causing translational repression rather than mRNA degradation. However, a recent report provided evidence that miRNA-bound mRNA not only inhibits translation, but also is degraded more rapidly, resulting in reduced levels of the corresponding transcript (40). Here, we also show that the loss of miRNA miR-24 regulation of DHFR leads to slower degradation of mutant transcript and leads to enhanced message stability and higher steady-state DHFR mRNA levels in SNP-829C→T-expressing cells. There are other examples in the literature describing a 2- to 4-fold difference in mRNA half-life resulting in a significant increase in mRNA and protein abundance (as reviewed in ref. 41). Hence, an average 20-h increase in the half-life of mutant DHFR mRNA likely explains the 10- to 20-fold difference in steady-state levels of DHFR mRNA in mutant-expressing clones.
The miRNAs use the RNAi machinery for their action by forming an imperfect hybrid with the target mRNA. The translational repression and gene-silencing events are mediated by RISC, which comprises Ago and P-element-induced wimpy testis (PIWI) protein subfamilies. Ago proteins are characterized by two domains: an ≈20-kDa N-terminal PIWI-Ago-Zwille (PAZ) domain and an ≈40-kDa C-terminal PIWI domain. PAZ is the RNA-binding domain of Ago complex. Crystal structures demonstrated that PAZ interacts with RNA and serves as the anchor for the 3′ end of small RNAs. The PIWI domain also interacts with small RNA and forms few contacts with the target mRNA strand (42, 43). A mutation of the G1-binding pocket in human Ago ablates slicer activity of the complex (44). The 829C site is conserved in mice and humans and located 14 bp from the 3′ end of the miR-24 complementary site. The C→T polymorphism may affect the binding of the miR-24–Ago protein complex to DHFR mRNA.
The finding that an miRNA, miR-24, regulates expression of the DHFR gene by binding to DHFR 3′ UTR provides further insights into the regulation of this important therapeutic target. Further inquiry in various ethnic groups as to its presence and its effect on treatment outcome and or toxicity will be of importance. These findings also raise the possibility that miR-24 mimics may be of value alone or in combination with MTX in the treatment of human disease.
Materials and Methods
Cell Transfections with miRNA Mimics and Inhibitors.
miRanda web server (25) and miRBase (Sanger Institute, Cambridge, U.K.) were used to predict the possible miRNA binding sites in DHFR 3′ UTR. DHFR 3′ UTR was found to have a binding site for miRNA miR-24. miR-24 mimics and inhibitors were obtained from Dharmacon (Chicago, IL) and transfected in HT1080 human fibrosarcoma cells by using Oligofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Duplex siRNA specific to DHFR 3′ UTR was synthesized (Integrated DNA Technologies, Coralville, IA) and transfected as a positive control (sense 5′-rUrUrCrUrCrCrArArGrArCrCrCrCrArArCrUrGrArGrUrCrC-3′, antisense 5′-rGrGrArCrUrCrArGrUrUrGrGrGrGrUrCrUrUrGrGrArGrArA-3′). A nonspecific duplex miRNA cel-mir-67 (sense 5′-rUrCrUrArCrUrCrUrUrUrCrUrArGrGrArGrGrUrUrGrUrGrA-3′, antisense 5′-rUrCrArCrArArCrCrUrCrCrUrArGrArArArGrArGrUrArGrA-3′) and Oligofectamine alone were transfected as negative controls. The transfected HT1080 cells were grown in RPMI medium 1640 with 10% FBS and 1% penicillin-streptomycin. Thirty-six hours after transfection, cell pellets were collected and cell lysates were prepared. DHFR protein levels were determined by using Western blot analysis using monoclonal anti-DHFR antibody (BD Biosciences, San Jose, CA). α-Tubulin protein levels were determined by using anti-α-tubulin antibody (Sigma–Aldrich, St. Louis, MO).
Cloning and Site-Directed Mutagenesis.
WT DHFR was cloned in a pCR2.1 cloning vector. Total RNA was isolated from HCT-8 cells by standard techniques. First-strand cDNA was generated by using total RNA. By using the gene bank sequence of DHFR mRNA, the following oligonucleotide primers were designed to amplify the WT DHFR gene with 3′ UTR by PCR: forward primer 5′-CTGTCATGGTTGGTTCGC-3′, and reverse primer 5′-AAGCTTTTGGTATTTCCA-3′. The PCR product was cloned in the pCR2.1 vector per the manufacturer's protocol. Positive clones were selected by restriction digestion using BamHI and XhoI and confirmed by DNA sequencing.
After WT cloning, QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to generate the mutation 829C→T in the 3′ UTR of DHFR gene by PCR using the WT DHFR construct as the template. The following primers containing the mutation 829C→T were designed and used for site-directed mutagenesis according to the manufacturer's protocol: forward primer 5′-CCCCAGCACCTGCTATAGTGAGCTGCCATT-3′, and reverse primer 5′-GAATGGCAGCTCACTATAGCAGGTGCTGGGG-3′ (the mutation is underlined).
Because pCR2.1 vector was a cloning vector, the WT and mutant DHFR was subcloned in a mammalian expression vector pCDNA3.1 (+) (Invitrogen) between BamHI and XhoI restriction sites by using a rapid DNA-ligation kit (Roche Applied Science, Indianapolis, IN).
Cell Transfections and Generation of Stable Clones.
The mutant construct (pCDNA3.1–829C→T) was transfected in to the CHO cell line DG44, deleted for both alleles of DHFR, to compare DHFR levels with no endogenous background. Also, WT DHFR (pCDNA3.1-WT DHFR) and vector alone (pCDNA3.1) constructs are transfected in DG44 cells as controls in culture medium RL with 10% FBS, 1% penicillin/streptomycin and 1% l-glutamine. All transfections were performed by using N-[1-(2, 3-dioleoyloxy)propyl]-N, N, N-trimethyl-ammonium methylsulfate (DOTAP) transfection reagent (Roche Applied Science) according to the manufacturer's protocol and selected in media containing 750 μg/ml G418. Individual clones were isolated by ring cylinders and expanded into stable resistant cell lines ≈14 d after transfection. Cell pellets of monolayer cultures (70–80% confluent) were harvested, total RNA and genomic DNA were isolated from the pellets, and cell lysate was prepared to detect DHFR protein levels according to standard protocols.
Transfections of WT and Mutant Cells with miR-24 Mimics and Inhibitors.
miR-24 mimics and inhibitors were obtained from Dharmacon and transfected in WT and mutant-expressing DG44 cells by using Oligofectamine (Invitrogen) according to the manufacturer's protocol. Duplex siRNA specific to DHFR 3′ UTR (through supra) was transfected as a positive control and a nonspecific duplex miRNA cel-mir-67 and Oligofectamine alone were transfected as negative controls. The transfected WT and mutant cells were grown in culture medium RL with 10% FBS and 1% penicillin/streptomycin. Thirty-six hours after transfection, cell pellets were collected and cell lysates were prepared. DHFR and α-tubulin protein levels were determined by using monoclonal anti-DHFR antibody and anti-α-tubulin antibody by SDS/PAGE followed by Western blot analysis.
DHFR mRNA Half-Life.
Quantitative RT-PCR analysis was used to measure DHFR mRNA decay (45). A total of 1 × 106 cells are plated per 10-cm cell culture dish. Actinomycin D (5 μg/ml) was added after 24 h. After the addition of actinomycin D, at time points 0, 4, 8, 12, 19, 24, and 36 h, cells were lysed by using TRIzol. Total RNA was extracted and treated with DNase to digest residual DNA in the RNA preparation. The RNA was quantitated with fluorescent dye RiboGreen by using Mx4000 PCR system (Stratagene) according to manufacturer's protocol. Total RNA (200 ng per well) was used in a real-time RT-PCR by using TaqMan 7000 as described below.
DHFR mRNA Levels.
DHFR and hamster β-actin (control) mRNA levels were examined by quantitative RT-PCR. Total RNA was isolated from the cell pellets by using TRIzol reagent (Invitrogen). Quantitative RT-PCR was conducted with the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). The following primers and probes were designed to amplify DHFR and hamster β-actin cDNA: DHFR forward primer 5′-TAAACTGCATCGTCGCTGTGT-3′, reverse primer 5′-AGGTTGTGGTCATTCTCTGGAAA-3′ and probe 5′-56-FAM/CCCGTTCTTGCCGATGCCCA/36-TAMTph-3′. Hamster β-actin forward primer 5′-TGAACCGTATCTGCCCACCT-3′, reverse primer 5′-GCCACCAAGGCATGATCG-3′ and probe 5′-56-FAM/ACACAAGGTCGGGAGCCAAACGC/36-TAMTph-3′. Briefly, each 25-μl reaction contained 300 nM of each primer, 100 nM probe, and quantitative RT-PCR master mix (Eurogentec, San Diego, CA). Quantitative RT-PCR was performed with the ABI Prism 7000 sequence detection system. The cycling conditions were: initial step 48°C for 30 min, activation step 95°C for 10 min, followed by 40 cycles at 94°C for 15 s and 60°C for 1 min. Each sample was run using eight different wells of the 96-well plate, and each experiment was repeated three times.
Western Blot Analysis.
The protein level of the clones expressing WT and mutant DHFR was determined by using SDS/PAGE followed by Western blot analysis using anti-DHFR and anti-tubulin antibody. Equal loading was determined by using Ponceau S and anti-α-tubulin antibody. The bands were quantitated by using the Kodak Image Station 2000MM multimodal imaging system (IS2000MM; Kodak, Rochester, NY), and fold change in protein level between WT and mutant clones was determined.
DHFR Genomic DNA Levels.
Quantitative PCR was used to determine DHFR genomic DNA levels. Endogenous hamster β-actin DNA was the control. The following primers and probes were used to detect DHFR and hamster β-actin genomic DNA level: DHFR forward primer 5′-TAAACTGCATCGTCGCTGTGT-3′, reverse primer 5′-AGGTTGTGGTCATTCTCTGGAAA-3′ and probe 5′-56-FAM/CCCGTTCTTGCCGATGCCCA/36-TAMTph-3′. Hamster β-actin forward primer 5′-GGCCAACCGTGAAAAGATGA-3′, reverse primer 5′-CGAGAAGTTGGCAAAGATGGA-3′, and probe 5′-56-FAM/CCAGGTCAGCAGCCAGGGTGG/36-TAMTph-3′. Genomic DNA was extracted from the cell pellets by using the phenol-chloroform-isoamylalcohol method. Each 25-μl quantitative PCR mixture contained 200 nM of each primer and probe, 200 ng of genomic DNA, and TaqMan universal PCR master mix (Applied Biosystems). The cycling conditions were 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 94°C for 15 s and 60°C for 1 min. Each sample was run using eight different wells of the 96-well plate, and each experiment was repeated three times.
MTX Cytotoxicity.
MTX cytotoxicity was carried out in RPMI medium 1640 with 10% dialyzed FBS (dialysis removes thymidine), penicillin/streptomycin, and l-glutamine. The clones were acclimatized in the RPMI media for few passages before plating for the cytotoxicity assay. The [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] (MTS) (Promega, Madison, WI) was used to assay the cytotoxicity according to the Cell-Titer 96 aqueous one-solution protocol. Two thousand cells were plated in each well of 96-well plates in 180 μl of the medium. After 24 h, varying concentrations of MTX in 20 μl of media were added and cells were incubated for 96 h. After 96 h of incubation, the MTS assay was performed and the absorbance in each well was determined at 490 nm by using a plate reader. Wells containing cells with no MTX and wells with medium alone were used as positive and negative controls, respectively. All experimental points were set up in replicate wells, and all experiments were repeated four times.
Statistical Analysis.
Student's t test was performed to assay the statistical significance.
Acknowledgments
This work was supported by National Cancer Institute Grant CA0810.
Abbreviations
- MTX
methotrexate
- miRNA
microRNA
- RISC
RNA-induced silencing complex
- DHFR
dihydrofolate reductase
- Ago
Argonaute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 3.Lau NC, Lim LP, Weinstein EG, Bartel DP. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Ambros V. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 5.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 6.Sevignani C, Calin GA, Siracusa LD, Croce CM. Mamm Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai RS. RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CZ. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 15.Mazumder B, Seshadri V, Fox PL. Trends Biochem Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 16.Banerjee D, Mayer-Kuckuk P, Capiaux G, Budak-Alpdogan T, Gorlick R, Bertino JR. Biochim Biophys Acta. 2002;1587:164–173. doi: 10.1016/s0925-4439(02)00079-0. [DOI] [PubMed] [Google Scholar]
- 17.Bertino JR, Booth BA, Bieber AL, Cashmore A, Sartorelli AC. J Biol Chem. 1964;239:479–485. [PubMed] [Google Scholar]
- 18.Gorlick R, Goker E, Trippett T, Waltham M, Banerjee D, Bertino JR. N Engl J Med. 1996;335:1041–1048. doi: 10.1056/NEJM199610033351408. [DOI] [PubMed] [Google Scholar]
- 19.Alt FW, Kellems RE, Bertino JR, Schimke RT. J Biol Chem. 1978;253:1357–1370. [PubMed] [Google Scholar]
- 20.Dicker AP, Waltham MC, Volkenandt M, Schweitzer BI, Otter GM, Schmid FA, Sirotnak FM, Bertino JR. Proc Natl Acad Sci USA. 1993;90:11797–11801. doi: 10.1073/pnas.90.24.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorlick R, Goker E, Trippett T, Steinherz P, Elisseyeff Y, Mazumdar M, Flintoff WF, Bertino JR. Blood. 1997;89:1013–1018. [PubMed] [Google Scholar]
- 22.McCloskey DE, McGuire JJ, Russell CA, Rowan BG, Bertino JR, Pizzorno G, Mini E. J Biol Chem. 1991;266:6181–6187. [PubMed] [Google Scholar]
- 23.Mesner LD, Hamlin JL. Genes Dev. 2005;19:1053–1066. doi: 10.1101/gad.1307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goto Y, Yue L, Yokoi A, Nishimura R, Uehara T, Koizumi S, Saikawa Y. Clin Cancer Res. 2001;7:1952–1956. [PubMed] [Google Scholar]
- 25.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 27.Urlaub G, Mitchell PJ, Kas E, Chasin LA, Funanage VL, Myoda TT, Hamlin J. Somatic Cell Mol Genet. 1986;12:555–566. doi: 10.1007/BF01671941. [DOI] [PubMed] [Google Scholar]
- 28.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 29.Leys EJ, Crouse GF, Kellems RE. J Cell Biol. 1984;99:180–187. doi: 10.1083/jcb.99.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendrickson SL, Wu JS, Johnson LF. Proc Natl Acad Sci USA. 1980;77:5140–5144. doi: 10.1073/pnas.77.9.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, Hussain A, Melera PW. Gene. 1995;163:185–191. doi: 10.1016/0378-1119(95)00381-f. [DOI] [PubMed] [Google Scholar]
- 32.Gehring NH, Frede U, Neu-Yilik G, Hundsdoerfer P, Vetter B, Hentze MW, Kulozik AE. Nat Genet. 2001;28:389–392. doi: 10.1038/ng578. [DOI] [PubMed] [Google Scholar]
- 33.Conne B, Stutz A, Vassalli JD. Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 34.Pizzuti A, Argiolas A, Di Paola R, Baratta R, Rauseo A, Bozzali M, Vigneri R, Dallapiccola B, Trischitta V, Frittitta L. J Clin Endocrinol Metab. 2002;87:4403–4406. doi: 10.1210/jc.2002-020096. [DOI] [PubMed] [Google Scholar]
- 35.Tsai FJ, Lin CC, Lu HF, Chen HY, Chen WC. Urology. 2002;59:458–461. doi: 10.1016/s0090-4295(01)01576-x. [DOI] [PubMed] [Google Scholar]
- 36.Jeon S, Lambert PF. Proc Natl Acad Sci USA. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandola MV, Stoehlmacher J, Zhang W, Groshen S, Yu MC, Iqbal S, Lenz HJ, Ladner RD. Pharmacogenetics. 2004;14:319–327. doi: 10.1097/00008571-200405000-00007. [DOI] [PubMed] [Google Scholar]
- 38.Lu JW, Gao CM, Wu JZ, Cao HX, Tajima K, Feng JF. J Hum Genet. 2006;51:155–160. doi: 10.1007/s10038-005-0339-4. [DOI] [PubMed] [Google Scholar]
- 39.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, et al. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 40.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 41.Ross J. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker JS, Roe SM, Barford D. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song JJ, Joshua-Tor L. Curr Opin Struct Biol. 2006;16:5–11. doi: 10.1016/j.sbi.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 44.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leclerc GJ, Leclerc GM, Barredo JC. Cancer Cell Int. 2002;2:1. doi: 10.1186/1475-2867-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]