Abstract
Presenilins (PS) provide the catalytic activity for γ-secretase, which cleaves physiologically relevant substrates including Notch, ErbB4, and APP. Recent genetic studies indicated that the contribution of PS1 to mouse development includes γ-secretase-independent functions that cannot be easily explained by any of the demonstrated or hypothesized functions of this protein. To begin a nonbiased analysis of PS1 activity unencumbered by the dominant effect stemming from loss of Notch function, we characterized PS functions in the early land plant Physcomitrella patens, which lacks Notch, ErbB4, and APP. Removal of P. patens PS resulted in phenotypic abnormalities. Further assays performed to delineate the defective pathways in PS-deficient P. patens implicated improper function of the cytoskeletal network. Importantly, this characterization of a nonmetazoan PS uncovered a previously undescribed, evolutionarily conserved function (human PS1 can rescue the growth and light responses) that is γ-secretase-independent (mutants with substitutions of the catalytic aspartyl residues retain the activity). Introduction of PpPS into PS-deficient mouse embryonic fibroblasts rescues normal growth rates, demonstrating that at least some metazoan functions of PS are evolutionarily conserved.
Keywords: evolution, iCLiP, Alzheimer's disease, cytoskeleton, Physcomitrella
Presenilin (PS) proteins are polytopic transmembrane domain (TMD) proteins discovered independently as loci frequently mutated in familial forms of Alzheimer's disease (AD) (1–3) and as modifiers of Notch signaling (4, 5). PS proteins form the catalytic center of γ-secretase, an aspartate protease complex that cleaves type I substrates sequentially within their TMD to release soluble C- and N-terminal peptides (6, 7). Enzymatic activity of PS is unique among aspartyl proteases in the location (within the TMD) and the context of conserved catalytic residues (YD/GxGD) and is among the founders of a growing family of intramembrane cleaving enzymes (8, 9). Mutation of either one of the two conserved aspartate residues results in the loss of γ-secretase activity (10–12). PS proteins must associate with at least three other transmembrane proteins [nicastrin (Nct), Aph-1, and Pen-2] to assemble a functional γ-secretase (13). Whereas PS, Nct, Aph-1, and Pen-2 are essential and sufficient for reconstitution of an active γ-secretase, other cellular factors [e.g., TMP21 and CD147 (14, 15)] may play regulatory roles.
Clinically relevant substrates of γ-secretase include Notch and amyloid precursor protein [APP (16, 17)]. Hydrolysis of APP forms Aβ peptides, which cause AD (18). Notch is receptor in a signaling pathway regulating development and tissue renewal in the adult; mutations in Notch cause developmental syndromes (19) and contribute to adult-onset disease such as CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy) (20), aortic valve disease (21), cancer (22, 23), and possibly multiple sclerosis (24). The recent realization that Notch signaling may be critical in maintaining cancer stem cells revived the interest in γ-secretase as a therapeutic target (25–29).
Interestingly, some defects resulting from loss of PS might not be due to loss of γ-secretase activity. The clearest genetic evidence of γ-secretase-independent PS functions in mice emerged from experiments examining the role of Notch signaling in somite development where formation of discernable somites (anterior and posterior) was completely abrogated in embryos deficient in both PS paralogs, PS1 and PS2. However, mouse embryos lacking other components of γ-secretase or missing Notch pathway components developed anterior somites despite complete loss of Notch signaling (30).
To explain this phenotype, we postulated that the involvement of PS in Wnt signaling might impact the somite. If loss of PS1 affects both Notch and Wnt signaling (30), no somite may form. However, a PS–Wnt connection does not have support from genetic experiments reported in the literature (31) or from mice with compound mutations in Wnt3A and PS1; the compound mutants were not any worse than Wnt3A heterozygotes, 50% of which are born with defective tail vertebra, the product of somites (R.K., unpublished observations). More recently, PS were shown to possess ER-Ca2+ leak channel activity (32). However, several PS mutant proteins lacking ER-Ca2+ channel activity (32) can still rescue the somite defects in mice with limiting amounts of PS1 (33).
Other proposed activities of PS that are independent of γ-secretase include a role in transport of several membrane proteins (34–36), autophagy and protein degradation (37, 38), or act as a scaffold in Erk activation (39, 40). However, the dominant phenotype caused by loss of Notch signaling (41–43) severely limits the ability to identify and interrogate the physiological relevance of the entire spectrum of PS functions. These observations suggest that an essential function provided by PS in the somite and possibly elsewhere may have escaped detection because of dominant Notch phenotypes. A system, which will allow an unbiased investigation, will have to shift to organisms encoding PS but lacking Notch.
Notch receptors and APP proteins are metazoan proteins, i.e., they are only present in genomes of multicellular animals. In contrast, PS and related proteins have been identified in both metazoans and plants (44–48). Similar to the genomes of Arabidopsis (Dicot) and Oryza sativa (monocot), whose complete genome sequences are available, an early land plant, the moss Physcomitrella patens, also harbors all four components of γ-secretase. As a laboratory organism in which to interrogate PS functions, P. patens has many additional advantages. It has few cell types and offers excellent cytology because it is mostly single cell layer thick (see ref. 49). The complete genome of P. patens has been sequenced (www.mossgenome.org), and asexual plants grown in the lab are haploid. Most importantly, tissue from vegetative stages grown in the lab can be disrupted and treated to remove the cell wall generating protoplasts that can be used for transformation and other molecular procedures. P. patens can be used to study gene function, either by RNA interference [RNAi (50, 51)], or, because it can undergo homologous recombination at a high frequency, by specific gene deletion and/or replacement (51–54).
In this article, we report the initial characterization of the null phenotype of P. patens PS (PpΔpsn) and demonstrate, using multiple lines of evidence and approaches, that evolutionarily conserved functions underlying the phenotype associated with loss of PS in this basal organism are independent of γ-secretase and may be placed in a pathway regulating the function of the cytoskeleton. This genetically tractable system adds a powerful new tool to the investigation of PS proteins and their cellular partners.
Results
Characteristics of PS Homologues in P. patens.
A full-length PS cDNA from P. patens (PpPS) was cloned based on sequence information available from the EST database, PHYSCObase (moss.nibb.ac.jp). PpPS cDNA contains a single, intron-less ORF that encodes a polypeptide of 477 aa, with 49% and 46% similarity to human PS1 and PS2, respectively. Based on the length of hydrophilic loop, PpPS is a γ-type PS similar to the rice (O. sativa) and both Arabidopsis homologues (55) (Fig. 1A). Characteristics of PS family, i.e., the catalytic aspartates embedded in motifs YD and GxGD and the PAL motif, are conserved in PpPS [supporting information (SI) Fig. 6]. The complete genome sequence suggests that PpPS exists as a single copy in its haploid genome, which we confirmed by Southern blot analysis (data not shown).
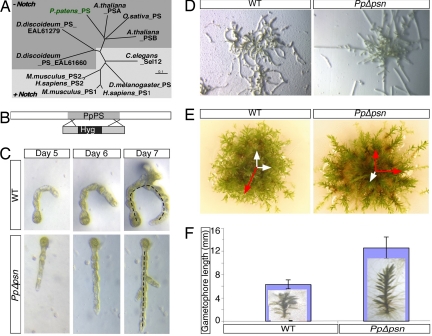
Fig. 1.
Loss of PpPS displays pleiotropic phenotype. (A) ClustalW dendrogram depicting PS homologues from selected species. (B) Schematic representation of the PpPS KO construct used in the study. Gray, PpPS; black, the Hyg cassette used to disrupt the PpPS ORF. (C and D) Filaments regenerating from WT and PpΔpsn protoplasts photographed for several days. Note that WT protonemal filaments curve whereas PpΔpsn colonies are straight. (E) Fifteen-week-old gametophores. PpΔpsn gametophores spread out, whereas WT grows toward the light. Preferred axes of gametophore growth are represented in red. (F) Mean length of each gametophore is graphically represented. (Insets) Pictures of PpΔpsn and WT gametophore.
Mutant Phenotypes Because of the Loss of PS.
To evaluate the role of PS in P. patens, we created a null mutant of PpPS (PpΔpsn) by homologous recombination (Fig. 1C). RT-PCR analysis confirmed that this plant lacked the PpPS transcript (data not shown); genomic PCR using sequences flanking the ORF identified integration into the PpPS locus (data not shown).
P. patens has alternating sexual and asexual stages in its life cycle. Diploid stage plants (sporophyte) generate haploid spores, which germinate into vegetative tissue that further develops into leafy shoots (or gametophores) bearing male and female gametes. Fertilization of these gametes results in diploid sporophytic generation. PpΔpsn plants displayed a unique phenotype early in development: filaments regenerating from PpΔpsn protoplasts grew in straight lines and did not curl like WT filaments (referred to as “coiled” morphology hereafter; Fig. 1C). These differences remained clearly evident throughout filament growth (Fig. 1 D and E). Whereas the number of leaves per unit length was unchanged in mature gametophores, PpΔpsn gametophores exhibited three distinct phenotypes: First, shoots were longer and the total number of leaves was much greater than in WT gametophore (Fig. 1 E and F). Second, the PpΔpsn gametophore did not grow in a predominant upward direction (Fig. 1E, red arrows), suggesting a defect in sensing of light or gravity. Third, WT P. patens became fertile within three months, but PpΔpsn plants were still unable to complete their life cycle (i.e., formation of spores) after a four-month period (data not shown). This inability may indicate a form of sterility, which will not be investigated further here.
Mutant Phenotypes Implicate the Cytoskeleton.
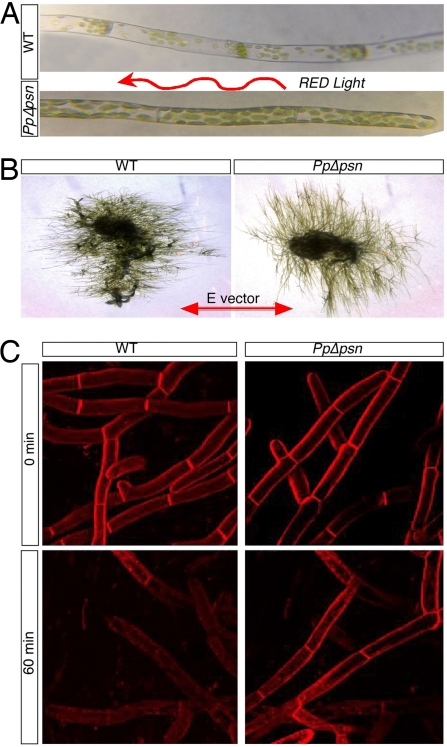
The phenotypes described above could result from a number of defective processes, including the reception and integration of polarizing signals (light, gravity), cytoskeleton organization, membrane and vesicle cycling, and/or cell wall composition and organization. To investigate which processes depend on PpPS, we grew P. patens under different light conditions. When WT filaments were subjected to unidirectional red light, the chloroplasts rearrange and localize primarily to the cell-cell contact zones (56) (Fig. 2A). The redistribution of chloroplasts to contact zones was not observed in PpΔpsn; instead, chloroplasts remained randomly oriented (Fig. 2A). Chloroplast relocalization in P. patens requires an intact photoreceptor complex, signal transduction to downstream effectors, and a dynamic actin and tubulin filament networks (57). To determine which of these pathways have become impaired in PpΔpsn, we subjected WT and PpΔpsn filaments to polarized red light and monitored their growth response. WT filaments responded by aligning themselves parallel to the plane of polarized light, whereas PpΔpsn filaments aligned perpendicular to the plane of polarized light (Fig. 2B). Because filaments did align in response to light (albeit in the incorrect plane), the defect in PpΔpsn is likely downstream of light detection in either a signaling system (which, as in metazoans, may require γ-secretase activity) or in the cytoskeletal network(s) or both.
Fig. 2.
Differential responses of WT and PpΔpsn filaments. (A) Chloroplasts relocalization differs between WT and PpΔpsn cells exposed to unidirectional red light (arrow represents the light vector). (B) PpΔpsn filaments align perpendicular to the E-vector under polarized red light whereas WT filaments align parallel to the E-vector. (C) Clearance of FM4-64 fluorescence from the membrane is delayed in PpΔpsn filaments.
To further define the step altered in PpΔpsn, we performed a light- and gravity-independent assay involving the function of the cytoskeleton. WT and PpΔpsn protoplasts were stained for two minutes with FM4-64 (a lipophilic dye used to stain membranes), washed and chased for one hour in the absence of the dye. Immediately after staining, both WT and PpΔpsn filaments stained bright red, but after the 1-h chase, fluorescence was lost in WT filaments because of surface membranes recycling via endocytosis. In contrast, PpΔpsn filaments retained significant fluorescence (Fig. 2C), most likely reflective of a slower rate of endocytosis and/or membrane recycling. Together, these experiments point to a defect in cytoskeletal related responses in PpΔpsn.
Transient Silencing of PpPS by RNAi Mimics Deletion of PpPS.
An unfortunate consequence of altered membrane dynamics in PpΔpsn protoplasts is that they are refractory to transformation; this phenotype prevented us from performing rescue experiments directly in PpΔpsn. Also, as the knockout was created by disrupting the ORF and not by deletion, an amino-terminal truncated product may be produced, contributing to the observed phenotypes. We therefore asked whether RNAi could mimic faithfully the effects of PS loss. RNAi lines were established in the NLS4 background, which constitutively expresses a nuclear green fluorescent protein (GFP) (50, 51). When this line is used in conjunction with a vector that expresses complementary RNA strands for targeted gene-fragments contiguous with GFP sequences (pUFGi_PS-KD), effective reduction of both GFP and target gene expression is achieved. Protoplasts transformed with the vector alone (harboring RNAi for GFP) lost GFP expression but displayed the WT, coiled filament morphology (Fig. 3A) and chloroplast realignment (data not shown). When transformed with a RNAi vector targeting both GFP and PpPS, filaments lacking GFP also exhibited the mutant, straight filaments morphology observed in PpΔpsn (Fig. 3A). Silencing of an unrelated protein involved in assembly of actin-based structures (58) in P. patens (51) did not result in the straight filament phenotype (data not shown) demonstrating that the phenotype was not a general response to RNAi.
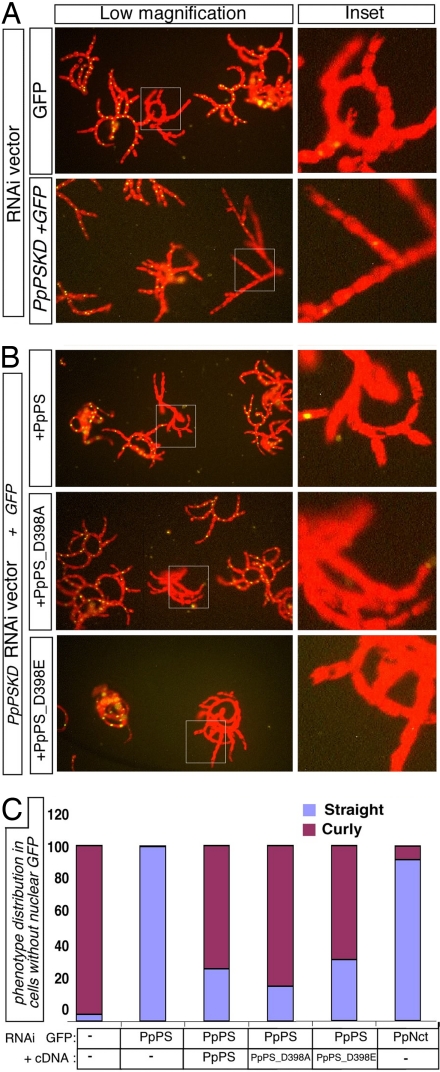
Fig. 3.
Transient silencing of PpPS by RNAi mimics genetic loss of PpPS. (A) Fluorescent images of filaments regenerating from protoplasts transformed with PpPSKD construct or the corresponding vector alone. Nuclear GFP fluorescence overlaps with the red autofluorescence of chlorophyll in untransfected cells; straight filaments in GFP-negative cells are observed with PpPSKD, but not vector controls. (B) PpPSKD filaments cotransformed with WT or aspartate mutants of PpPS display normal phenotype despite loss of GFP expression (reflective of silencing). (C) Quantitative analysis of three independent experiments.
To address whether the other γ-secretase components contributed to straight filament phenotype, we asked whether PpNct is involved in the pathway leading to the PpΔpsn phenotype. PpNct knockdown by RNAi (NctKD) resulted in protoplasts with straight filaments (Fig. 3C), similar to the PpPSKD (Fig. 3D). The similarity between the phenotype observed in PpPS and PpNct knockdown could result from loss of γ-secretase activity or by means other mechanisms, including loss of PS protein in Nct-deficient cells (59–61). To test whether indeed γ-secretase activity was involved in the phenotype, we asked whether the PpPSKD phenotype could be rescued with WT PpPS or PpPS cDNAs containing substitutions of D398, replacing the conserved aspartyl in the essential GxGD motif to glutamate (D398E) or alanine (D398A). Interestingly, the WT as well as either D398 substitution in PpPS could rescue filament morphology in PpPSKD plants (Fig. 3B), presumably by overriding the RNAi effect. The ability of PpPSKD to phenocopy PpΔpsn, and the ability of WT PpPS to rescue this phenotype, confirmed that the straight filament phenotype is caused by loss of PS.
Human PS Can Rescue PpPSKD Independent of γ-Secretase Function.
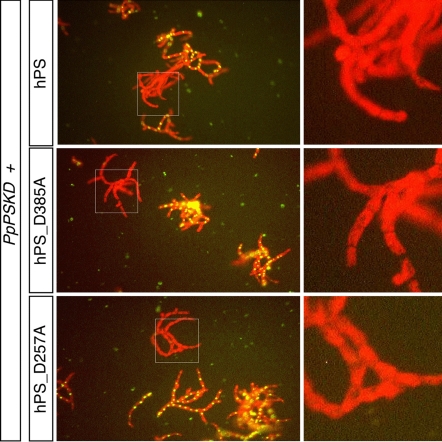
The results described above suggest that the coiled morphology of regenerating filaments is independent of proteolytic activity associated with PS proteins because aspartyl mutations in PS proteins eliminate enzymatic activity; however, we were unable to demonstrate any enzymatic activity for PpPS. To ask whether the loss of proteolytic activity contributed to the PpPS phenotype, we next tested whether the human PS (hPS) could provide an evolutionarily conserved activity and rescue the PpPSKD phenotype. hPS rescued coiled growth (Fig. 4A) and chloroplast alignment (data not shown). This conservation enabled us to test the rescuing ability of PS mutants lacking γ-secretase activity. Both the hPSD257A and hPSD385A substitutions abolish enzymatic activity [they are unable to reconstitute γ-secretase activity in PS-deficient mouse embryonic fibroblasts (MEFs) (7)]. Nonetheless, both aspartyl-substituted hPS proteins were capable of rescuing the growth phenotype of PpPSKD. This observation confirmed that retention of γ-secretase activity was not required for the growth phenotype in P. patens. We conclude that a conserved function of PS proteins is γ-secretase-independent.
Fig. 4.
Mammalian PS retained the activity responsible for PpPSKD phenotype in moss. PpPSKD protoplasts cotransformed with human PS constructs display WT growth morphology.
Plant PS Fail to Rescue γ-Secretase Activity in PSDKO Cells.
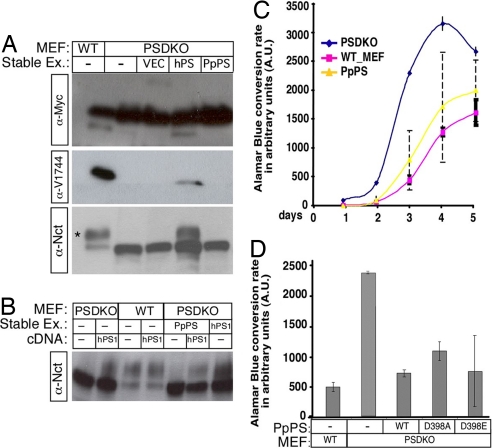
The ability of a mammalian protein (hPS) to rescue a plant PS deficiency prompted us to ask whether PpPS could rescue any of the documented defects in PS1, PS2 deficient mouse fibroblasts (PSDKO MEF; ref. 62). To this possibility, we established several clones of mammalian PSDKO cells stably expressing PpPS, PpPSD398A and PpPSD398E (collectively, PSDKO_PpPS lines). PSDKO_PpPS cell lines were first tested for their ability to cleave Δice (63). Neither α-MYC antibodies, which can recognize cleaved and uncleaved Δice molecules independent of scissile bond selection, nor α-V1744 antibodies, which can recognize the amino terminus of the common γ-secretase-cleaved Notch1-based substrates, detected any significant cleavage in PSDKO_PpPS. Both antibodies readily recognized cleavage of Δice in cell line expressing human PS1 (PSDKO_hPS; Fig. 5A).
Fig. 5.
PpPS can suppress accelerated proliferation in PS-deficient embryonic fibroblasts. (A) Analysis of the Notch cleavage in stable MEF lines expressing WT or mutant PpPS. Stable PpPS expression cannot rescue NCT maturation (*) or Notch substrate cleavage in PSDKO. (B) Nct maturation can be restored by transiently transfecting hPS into the PSDKO_PpPS MEFs. (C) Growth curves of WT (n = 4), PSDKO (n = 4), and PSDKO_PpPS clones (n = 4) were determined by alamarBlue conversion in 1 h as a reflection of increase in cell number. (D) alamarBlue conversion on day 3 is shown for each PSDKO_PpPS clone (average of 8 replicates).
Lack of Δice cleavage in PSDKO_PpPS can be due to either the inability of PpPS to perform proteolysis, or to the possibility that PpPS cannot associate with mammalian partners because of sequence divergence and hence cannot incorporate itself into γ-secretase complex (64). To differentiate between these possibilities, we examined the glycosylation of Nct [a measure of Nct maturation and functional assembly of γ-secretase complex, (65–68)]. Mature Nct was not observed in PSDKO_PpPS lines (Fig. 5 A and B), indicating that PpPS failed to assemble into functional γ-secretase complexes in the mammalian cells. To confirm that cells expressing plant proteins have not lost the ability to assemble γ-secretase, PSDKO_PpPS lines were transiently transfected with hPS; significant amounts of mature Nct were detected (Fig. 5B) indicating recovery of the complex. These results demonstrate that no γ-secretase complex is present in PSDKO_PpPS lines.
PpPS Reverts Accelerated Proliferation in PS-Deficient Mouse Embryonic Fibroblasts.
We next asked whether PpPS could complement a γ-secretase-independent function of PS. The most obvious difference between PSDKO and WT MEF lines derived from heterozygote littermates is their growth rates (69). To evaluate growth, we assayed all our MEF sublines for their ability to convert alamarBlue, a redox dye whose metabolism in live cells leads to increase in fluorescence which correlates well with cell numbers (70). Fluorescence from WT MEFs, PSDKO cell lines and stable lines expressing human or plant PS were recorded over several days and plotted as a function of time. Repeated measures analysis (see SI Text for details) confirmed that growth increased across days 2 to 4 in all clones (P < 0.0001), and that the least square (LS) mean (i.e., adjusted mean) of alamarBlue conversion rate values indicated that the growth of the individual WT_MEF and PSDKO clones did not differ (see SI Fig. 7 and SI Text) and thus could be combined to a single value characteristic for each genotype. PSDKO grow at an identical rate as PSDKO cells transfected with an empty vector, allowing us to combine all four PSDKO clones into a single pool. Consistent with previous reports, PSDKO clones were hyperproliferative (LS mean value of 2,030.4) when compared with the two WT MEFs (LS mean value of 662.1; P < 0.0001).
In contrast, all PpPS-transfected clones differed significantly from each other (P < 0.03); nonetheless, comparing the average across replicates for all clones shows that expression of PpPS and PpPS D398 mutants could significantly reverse hyperproliferation (Fig. 5C). As reflected in SI Fig. 7, all four PpPS clones provided better rescue than hPS. As can be seen in SI Fig. 7, the growth rate (reflected in LS mean) of line PpPS621 (expressing D398E) was lower than that of both the knockout (KO) and WT groups, with the means of each of the remaining PpPS clones falling between the combined rates for PSDKO and WT_MEF.
Discussion
This study provides a genetic and functional investigation of nonmetazoan PS. The function we identified is evolutionarily conserved and γ-secretase independent. P. patens, with well established genetic tools and a sequenced genome, is thus an attractive model system to study Notch- and γ-secretase independent functions of PS.
Loss of PS Results in Pleiotropic Defects at Various Stages of P. patens Life Cycle.
Phenotypic analysis of PpΔpsn suggested involvement of the cytoskeleton as the system most likely to be impaired. Growth of filaments perpendicular to the plane of polarized red light indicates that PpΔpsn can detect directional light. Because the alignment of tip growth relative to the directional light is reversed relative to WT, cytoskeleton-mediated vesicle transport may fail to target the proper site for tip growth. Furthermore, lack of chloroplast movement in response to light may also reflect similar defects in organelle transport along microtubules. In addition, altered membrane recycling observed in PpΔpsn also points to a defective cytoskeletal functions. PS may act as scaffolding protein facilitating interactions between membrane receptors and downstream proteins that direct vesicle transport, similar to the proposed role of PS in mammalian cells (39, 71, 72). Without PS, such interactions may be delayed or fail all together. The phenotypic defects observed in PpΔpsn could be due to the absence of γ-secretase activity, which might be involved in transducing signals akin to its role in Notch and ErbB4 (73) signaling in mammals. However, the ability of both P. patens and Homo sapiens PS proteins lacking critical aspartyl residues to rescue PpPSKD suggests that the observed phenotypes are independent of any proteolytic ability PpPS may possess.
Whereas proteolytic activity may not be required in P. patens, association with Nct may be. Nct knockdown resulted in straight filament growth and failure to localize chloroplasts in a manner reminiscent of PpPS knockdown (Fig. 3). Previous reports documented similar collaboration between PS and Nct in other γ-secretase independent activity. Examples include the organization of spectrin cytoskeleton (60) and tau hyperphosphorylation (74). Because Nct maturation was not restored in PSDKO_PpPS whereas the growth phenotype was rescued, it is possible that a PS/Nct complex has distinct roles in the endoplasmic reticulum (ER). Alternatively, because Nct plays an important role in stability and trafficking of PS in metazoans (60, 75), the Nct phenotypes may simply reflect loss of PS.
In summary, our findings demonstrate unequivocally that PS has an evolutionarily conserved function that is unrelated to γ-secretase. We demonstrate that mammalian PS retained this conserved, proteolysis-independent activity, and that P. patens PS has the ability to rescue a γ-secretase-independent phenotype in PSDKO tissue culture cells. Whereas still far from identifying the activity of PS in the somite, the defects observed in PS-deficient plant point to the cytoskeleton. Mesenchymal to epithelial transition and cleft formation during somitogenesis (missing in PS1, 2 deficient mice) requires coordinated cell motions and intact cytoskeleton (76), suggesting that perhaps this activity, which remains a subject for speculation at this time, is involved in regulating both mammalian segmentation and plant growth patterns. Alternately, PpPS may have conserved other γ-secretase independent functions previously reported in metazoans.
Materials and Methods
Plant Material, Culture Conditions, and Treatments.
P. patens (Gransden WT) tissue was maintained on cellophane overlaid on minimal medium supplemented with di-ammonium tartarate (0.5 g/liter) at 25°C under a 16 h light and 8 h dark cycle. Phenotypic analyses were performed on minimal medium, whereas protoplast regeneration was carried out on minimal medium supplemented with 8.5% mannitol protoplast regeneration medium (PRM).
Cloning of Moss PS.
RNA was isolated from a week-old P. patens filaments by using RNeasy Plant Mini Kit (Qiagen, Valencia, CA) and cDNA was synthesized by using the Thermoscript RT-PCR system (Invitrogen). Specific primers were designed, based on the sequence information from the contigs, and used to amplify and clone PpPS, into pMKUbi.
Homologous Recombination.
The gene deletion vector, HD-PpPS (Fig. 1B) was designed to disrupt the PpPS ORF by inserting the hygromycin (Hyg) cassette. PpPS-specific primers were used to amplify PpPS ORF disrupted with the Hyg cassette, which was purified by gel extraction and used to transform protoplasts as described below. Homologous recombination with this construct will result in replacement of the 1.5-kb genomic copy with the 4-kb disrupted copy, whereas random integration will retain both. The disrupted copy includes insertion of Hyg cassette in PpPS ORF resulting in transcription deletion.
RNAi and Rescue Constructs.
Four-hundred base pairs of the conserved C terminus of PpPS were amplified with gene-specific primers, harboring a CACC sequence at the 5′ primer and cloned into a pENTR/D-Topo kit. The final knockdown construct, pUFGi_PS-KD, was obtained by subcloning into pUFGi, using the protocol described before (50). Similarly, Nct knockdown construct pTUgi_Nct-KD was obtained by amplifying a fragment from 703 bp to 969 bp that is highly conserved across species and cloned in the next generation of RNAi vector as described (50). pUFGi/pTUGi was always used along with the reporter line NLS-4, expressing a nuclear localized GFP:GUS fusion reporter protein, facilitating easy analysis of gene silencing. Silencing of the gene of interest via this plasmid results in simultaneous silencing of nuclear GFP expression, which by itself displays no phenotype (77).
PEG-Mediated Transformation of P. patens.
Protoplasts were isolated from 6- to 7-day-old tissue as described (78). Fifteen micrograms of DNA was used to transform approximately half a million protoplasts in the presence of MMM (d-mannitol/MgCl2/Mes) and PEG as described before (moss.nibb.ac.jp). The transformation mix was incubated at room temperature for 10 min and subjected to heat shock at 45°C for 3 min. The mix was subsequently incubated without shaking at room temperature for 20 min, diluted 10-fold in mannitol, centrifuged at 300 × g for 10 min, resuspended in PRM with Phytagel, and overlaid on PRM with 0.7% agar medium. After 5–7 days of regeneration, transformants were selected on Hyg. To generate stable lines, transformants were alternately grown for a week each in the presence or absence of Hyg.
Mammalian Cell Culture and Transfection.
All cell lines were maintained in Dulbecco's modified Eagle's medium with 10% FBS, 4 mM glutamine, and antibiotics. HEK 293 cells were transfected by calcium phosphate in N, N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid (BES)-buffered saline. PS1−/− PS2−/− dKO mouse embryonic fibroblasts (PSDKO) were transfected by using Lipofectamine (Invitrogen). All mutant PS constructs were generated by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and were subsequently sequenced to confirm that only the desired mutations were introduced. To generate PSDKO cell lines expressing mPS1, WT PpPS, D398E PpPS, or D398A PpPS, appropriate constructs were cotransfected with the plasmid, pBabePuro, conferring antibiotic resistance, at a ratio of 10:1. The next day, cells were trypsinized and plated on P150 plates, and, after 24 h, 4 μg/ml puromycin was added to the medium. Stable lines were subjected to several rounds of antibiotic selection and tested for expression of appropriate PS proteins by Western analysis.
Western Blotting.
Western Blotting was performed by fractionating cellular lysates on SDS/PAGE and transferring them onto nitrocellulose membranes. Membranes were then blocked in 10% milk (PBS and 0.1% Tween 20) and incubated in primary antibody (1:1,000 in 5% milk, PBS, and 0.1% Tween 20) followed by HRP-conjugated secondary antibody (1:5,000 in 5% milk, PBS, and 0.1% Tween 20; Pierce).
Microscopic Observations.
Microscopic images of moss were taken with a Spot RT Slider camera (Diagnostic Instruments, Sterling Heights, MI) on either a Zeiss inverted microscope or an Olympus dissecting microscope. Images were then processed by using Canvas X and Adobe Photoshop CS software.
Statistical Analysis of Growth.
For statistical analysis of growth, see SI Text.
Supplementary Material
Acknowledgments
We thank Drs. Greg Longmore and Ma. Xenia Garcia Ilagan for insightful comments, members of the R.K. and R.S.Q. laboratories for valuable discussions and encouragement, Dr. Michael Neff and laboratory for sharing the light chamber for phototropic assays, and Dr. Magdalena Bezanilla (University of Massachusetts, Amherst, MA) for providing us with capping protein constructs. This work was supported by National Institutes of Health Grant GM55479 (to R.K.) and Alzheimer's Association Grant IRG-03-5283 (to D.C.). National Science Foundation Grant IBN 0112461 (to R.S.Q.) supported A.K and R.S.Q. At the time of writing, A.K. was supported by National Science Foundation Grant EF 0425749-1.
Abbreviations
- AD
Alzheimer's disease
- APP
amyloid precursor protein
- MEF
mouse embryonic fibroblasts
- PS
presenilin
- TMD
transmembrane domain
- PpΔpsn
null phenotype of P. patens PS
- Nct
nicastrin
- LS
least square
- Hyg
hygromycin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702038104/DC1.
References
- 1.Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K, et al. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 2.Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, et al. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 3.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature. 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 4.Li XJ, Greenwald I. Proc Natl Acad Sci USA. 1997;94:12204–12209. doi: 10.1073/pnas.94.22.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levitan D, Greenwald I. Nature. 1995;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- 6.Chandu D, Huppert S, Kopan R. J Neurochem. 2006;96:228–235. doi: 10.1111/j.1471-4159.2005.03547.x. [DOI] [PubMed] [Google Scholar]
- 7.Schroeter EH, Ilagan MXG, Brunkan AL, Hecimovic S, Li Y-m, Xu M, Lewis HD, Saxena MT, De Strooper B, Coonrod A, et al. Proc Natl Acad Sci USA. 2003;100:13075–13080. doi: 10.1073/pnas.1735338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haass C, Steiner H. Trends Cell Biol. 2002;12:556–562. doi: 10.1016/s0962-8924(02)02394-2. [DOI] [PubMed] [Google Scholar]
- 9.Martoglio B, Golde TE. Hum Mol Genet. 2003;12(Spec No 2):R201–R206. doi: 10.1093/hmg/ddg303. [DOI] [PubMed] [Google Scholar]
- 10.Kimberly WT, Xia W, Rahmati T, Wolfe MS, Selkoe DJ. J Biol Chem. 2000;275:3173–3178. doi: 10.1074/jbc.275.5.3173. [DOI] [PubMed] [Google Scholar]
- 11.Steiner H, Duff K, Capell A, Romig H, Grim MG, Lincoln S, Hardy J, Yu X, Picciano M, Fechteler K, et al. J Biol Chem. 1999;274:28669–28673. doi: 10.1074/jbc.274.40.28669. [DOI] [PubMed] [Google Scholar]
- 12.Wolfe MS, Xia WM, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 13.Wolfe MS, Kopan R. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- 14.Chen F, Hasegawa H, Schmitt-Ulms G, Kawarai T, Bohm C, Katayama T, Gu Y, Sanjo N, Glista M, Rogaeva E, et al. Nature. 2006;440:1208–1212. doi: 10.1038/nature04667. [DOI] [PubMed] [Google Scholar]
- 15.Zhou S, Zhou H, Walian PJ, Jap BK. Proc Natl Acad Sci USA. 2005;102:7499–7504. doi: 10.1073/pnas.0502768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baulac S, LaVoie MJ, Kimberly WT, Strahle J, Wolfe MS, Selkoe DJ, Xia W. Neurobiol Dis. 2003;14:194–204. doi: 10.1016/s0969-9961(03)00123-2. [DOI] [PubMed] [Google Scholar]
- 17.Kopan R, Ilagan MX. Nat Rev Mol Cell Biol. 2004;5:499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- 18.Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Nat Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 19.Gridley T. Hum Mol Genet. 2003;12(Suppl 1):R9–R13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- 20.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, et al. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 21.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Nature. 2005;21:180–184. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 22.Radtke F, Raj K. Nat Rev Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 23.Weng AP, Ferrando AA, Lee W, Morris JP, IV, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 24.John GR, Shankar SL, Shafit-Zagardo B, Massimi A, Lee SC, Raine CS, Brosnan CF. Nat Med. 2002;8:1115–1121. doi: 10.1038/nm781. [DOI] [PubMed] [Google Scholar]
- 25.Radtke F, Clevers H. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 26.Pinto D, Clevers H. Biol Cell. 2005;97:185–196. doi: 10.1042/BC20040094. [DOI] [PubMed] [Google Scholar]
- 27.Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Breast Cancer Res. 2004;6:R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Hajj M, Becker MW, Wicha M, Weissman I, Clarke MF. Curr Opin Genet Dev. 2004;14:43–47. doi: 10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li YM, Eberhart CG. Cancer Res. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 30.Huppert S, Ilagan MXG, De Strooper B, Kopan R. Dev Cell. 2005;8:677–688. doi: 10.1016/j.devcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 31.McManus EJ, Sakamoto K, Armit LJ, Ronaldson L, Shpiro N, Marquez R, Alessi DR. EMBO J. 2005;24:1571–1583. doi: 10.1038/sj.emboj.7600633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tu H, Nelson O, Bezprozvanny A, Wang Z, Lee SF, Hao YH, Serneels L, De Strooper B, Yu G, Bezprozvanny I. Cell. 2006;126:981–993. doi: 10.1016/j.cell.2006.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mastrangelo P, Mathews PM, Chishti MA, Schmidt SD, Gu Y, Yang J, Mazzella MJ, Coomaraswamy J, Horne P, Strome B, et al. Proc Natl Acad Sci USA. 2005;102:8972–8977. doi: 10.1073/pnas.0500940102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, Tang P, Wang P, Boissy RE, Zheng H. Proc Natl Acad Sci USA. 2006;103:353–358. doi: 10.1073/pnas.0509822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naruse S, Thinakaran G, Luo JJ, Kusiak JW, Tomita T, Iwatsubo T, Qian XZ, Ginty DD, Price DL, Borchelt DR, et al. Neuron. 1998;21:1213–1221. doi: 10.1016/s0896-6273(00)80637-6. [DOI] [PubMed] [Google Scholar]
- 36.Wrigley JD, Schurov I, Nunn EJ, Martin AC, Clarke EE, Ellis S, Bonnert TP, Shearman MS, Beher D. J Biol Chem. 2004;280:12523–12535. doi: 10.1074/jbc.M413086200. [DOI] [PubMed] [Google Scholar]
- 37.Raemaekers T, Esselens C, Annaert W. Biochem Soc Trans. 2005;33:559–562. doi: 10.1042/BST0330559. [DOI] [PubMed] [Google Scholar]
- 38.Esselens C, Oorschot V, Baert V, Raemaekers T, Spittaels K, Serneels L, Zheng H, Saftig P, De Strooper B, Klumperman J, Annaert W. J Cell Biol. 2004;166:1041–1054. doi: 10.1083/jcb.200406060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang DE, Yoon IS, Repetto E, Busse T, Yermian N, Ie L, Koo EH. J Biol Chem. 2005;280:31537–31547. doi: 10.1074/jbc.M500833200. [DOI] [PubMed] [Google Scholar]
- 40.Kim MY, Park JH, Choi EJ, Park HS. Biochem Biophys Res Commun. 2005;332:609–613. doi: 10.1016/j.bbrc.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Pan Y, Lin M, Tian X, Cheng H, Gridley T, Shen J, Kopan R. Dev Cell. 2004;7:731–743. doi: 10.1016/j.devcel.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Pan Y, Liu Z, Shen J, Kopan R. Dev Biol. 2005;286:472–482. doi: 10.1016/j.ydbio.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 43.Yang X, Tomita T, Wines-Samuelson M, Beglopoulos V, Tansey MG, Kopan R, Shen J. Dev Neurosci. 2006;28:102–117. doi: 10.1159/000090757. [DOI] [PubMed] [Google Scholar]
- 44.Wigge PA, Weigel D. Curr Biol. 2001;11:R112–R114. doi: 10.1016/s0960-9822(01)00043-4. [DOI] [PubMed] [Google Scholar]
- 45.Kimberly WT, Wolfe MS. J Neurosci Res. 2003;74:353–360. doi: 10.1002/jnr.10736. [DOI] [PubMed] [Google Scholar]
- 46.Ponting CP, Hutton M, Nyborg A, Baker M, Jansen K, Golde TE. Hum Mol Genet. 2002;11:1037–1044. doi: 10.1093/hmg/11.9.1037. [DOI] [PubMed] [Google Scholar]
- 47.Grigorenko AP, Moliaka YK, Korovaitseva GI, Rogaev EI. Biochem Russia. 2002;67:826–835. doi: 10.1023/a:1016365227942. [DOI] [PubMed] [Google Scholar]
- 48.Moliaka YK, Grigorenko A, Madera D, Rogaev EI. FEBS Lett. 2004;557:185–192. doi: 10.1016/s0014-5793(03)01489-3. [DOI] [PubMed] [Google Scholar]
- 49.Cove D, Bezanilla M, Harries P, Quatrano R. Annu Rev Plant Biol. 2006;57:497–520. doi: 10.1146/annurev.arplant.57.032905.105338. [DOI] [PubMed] [Google Scholar]
- 50.Bezanilla M, Perroud PF, Pan A, Klueh P, Quatrano RS. Plant Biol (Stuttgart) 2005;7:251–257. doi: 10.1055/s-2005-837597. [DOI] [PubMed] [Google Scholar]
- 51.Harries PA, Pan A, Quatrano RS. Plant Cell. 2005;17:2327–2339. doi: 10.1105/tpc.105.033266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perroud PF, Quatrano RS. Cell Motil Cytoskeleton. 2006;63:162–171. doi: 10.1002/cm.20114. [DOI] [PubMed] [Google Scholar]
- 53.Cove D. Annu Rev Genet. 2005;39:339–358. doi: 10.1146/annurev.genet.39.073003.110214. [DOI] [PubMed] [Google Scholar]
- 54.Quatrano RS, McDaniel SF, Khandelwal A, Perroud PF, Cove DJ. Curr Opin Plant Biol. 2007;10:182–189. doi: 10.1016/j.pbi.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 55.Hashimoto-Gotoh T, Tsujimura A, Watanabe Y, Iwabe N, Miyata T, Tabira T. Gene. 2003;323:115–123. doi: 10.1016/j.gene.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 56.Kasahara M, Kagawa T, Sato Y, Kiyosue T, Wada M. Plant Physiol. 2004;135:1388–1397. doi: 10.1104/pp.104.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sato Y, Wada M, Kadota A. J Cell Sci. 2001;114:269–279. doi: 10.1242/jcs.114.2.269. [DOI] [PubMed] [Google Scholar]
- 58.Wear MA, Cooper JA. Trends Biochem Sci. 2004;29:418–428. doi: 10.1016/j.tibs.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 59.Edbauer D, Winkler E, Haass C, Steiner H. Proc Natl Acad Sci USA. 2002;99:8666–8671. doi: 10.1073/pnas.132277899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopez-Schier H, Johnston DS. Dev Cell. 2002;2:79–89. doi: 10.1016/s1534-5807(01)00109-5. [DOI] [PubMed] [Google Scholar]
- 61.Hu Y, Ye Y, Fortini ME. Dev Cell. 2002;2:69–78. doi: 10.1016/s1534-5807(01)00105-8. [DOI] [PubMed] [Google Scholar]
- 62.Herreman A, Hartmann D, Annaert W, Saftig P, Craessaerts K, Serneels L, Umans L, Schrijvers V, Checler F, Vanderstichele H, et al. Proc Natl Acad Sci USA. 1999;96:11872–11877. doi: 10.1073/pnas.96.21.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vooijs M, Schroeter EH, Pan Y, Blandford M, Kopan R. J Biol Chem. 2004;279:50864–50873. doi: 10.1074/jbc.M409430200. [DOI] [PubMed] [Google Scholar]
- 64.Francis R, McGrath G, Zhang J, Ruddy DA, Sym M, Apfeld J, Nicoll M, Maxwell M, Hai B, Ellis MC, et al. Dev Cell. 2002;3:85–97. doi: 10.1016/s1534-5807(02)00189-2. [DOI] [PubMed] [Google Scholar]
- 65.Yang DS, Tandon A, Chen F, Yu G, Yu H, Arawaka S, Hasegawa H, Duthie M, Schmidt SD, Ramabhadran TV, et al. J Biol Chem. 2002;277:28135–28142. doi: 10.1074/jbc.M110871200. [DOI] [PubMed] [Google Scholar]
- 66.Tomita T, Katayama R, Takikawa R, Iwatsubo T. FEBS Lett. 2002;520:117–121. doi: 10.1016/s0014-5793(02)02802-8. [DOI] [PubMed] [Google Scholar]
- 67.Kimberly WT, LaVoie MJ, Ostaszewski BL, Ye WJ, Wolfe MS, Selkoe DJ. J Biol Chem. 2002;277:35113–35117. doi: 10.1074/jbc.M204446200. [DOI] [PubMed] [Google Scholar]
- 68.Leem JY, Vijayan S, Han P, Cai D, Machura M, Lopes KO, Veselits ML, Xu H, Thinakaran G. J Biol Chem. 2002;277:19236–19240. doi: 10.1074/jbc.C200148200. [DOI] [PubMed] [Google Scholar]
- 69.Soriano S, Kang DE, Fu MF, Pestell R, Chevallier N, Zheng H, Koo EH. J Cell Biol. 2001;152:785–794. doi: 10.1083/jcb.152.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ahmed SA, Gogal RM, Jr, Walsh JE. J Immunol Methods. 1994;170:211–224. doi: 10.1016/0022-1759(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 71.Baki L, Marambaud P, Efthimiopoulos S, Georgakopoulos A, Wen P, Cui W, Shioi J, Koo E, Ozawa M, Friedrich VL, Jr, Robakis NK. Proc Natl Acad Sci USA. 2001;98:2381–2386. doi: 10.1073/pnas.041603398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kang D, Soriano S, Xia X, Eberhart CG, De Strooper B, Zheng H, Koo EH. Cell. 2002;110:751–762. doi: 10.1016/s0092-8674(02)00970-4. [DOI] [PubMed] [Google Scholar]
- 73.Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G. Cell. 2006;127:185–197. doi: 10.1016/j.cell.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 74.Doglio LE, Kanwar R, Jackson GR, Perez M, Avila J, Dingwall C, Dotti CG, Fortini ME, Feiguin F. Neuron. 2006;50:359–375. doi: 10.1016/j.neuron.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 75.Zhang YW, Luo WJ, Wang H, Lin P, Vetrivel KS, Liao F, Li F, Wong PC, Farquhar MG, Thinakaran G, Xu H. J Biol Chem. 2005;280:17020–17026. doi: 10.1074/jbc.M409467200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kulesa PM, Fraser SE. Science. 2002;298:991–995. doi: 10.1126/science.1075544. [DOI] [PubMed] [Google Scholar]
- 77.Bezanilla M, Pan A, Quatrano RS. Plant Physiol. 2003;133:470–474. doi: 10.1104/pp.103.024901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schaefer DG, Zryd JP. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.