Abstract
Cells contain numerous enzymes that use molecular oxygen for their reactions. Often, their active sites are buried deeply inside the protein, which raises the question whether there are specific access channels guiding oxygen to the site of catalysis. Choosing 12/15-lipoxygenase as a typical example for such oxygen-dependent enzymes, we determined the oxygen distribution within the protein and defined potential routes for oxygen access. For this purpose, we have applied an integrated strategy of structural modeling, molecular dynamics simulations, site-directed mutagenesis, and kinetic measurements. First, we computed the 3D free-energy distribution for oxygen, which led to identification of four oxygen channels in the protein. All channels connect the protein surface with a region of high oxygen affinity at the active site. This region is localized opposite to the nonheme iron providing a structural explanation for the reaction specificity of this lipoxygenase isoform. The catalytically most relevant path can be obstructed by L367F exchange, which leads to a strongly increased Michaelis constant for oxygen. The blocking mechanism is explained in detail by reordering the hydrogen-bonding network of water molecules. Our results provide strong evidence that the main route for oxygen access to the active site of the enzyme follows a channel formed by transiently interconnected cavities whereby the opening and closure are governed by side chain dynamics.
Keywords: free energy, lipid peroxidation, oxygen channel, oxygen diffusion, oxygenases
Molecular oxygen participates in numerous cellular processes but, except for a few examples like myoglobin, little is known about the mechanisms of how oxygen reaches the reaction sites of the related proteins. Formerly, the hypothesis of unhampered oxygen diffusion through proteins prevailed (1). However, this picture is changing because a number of studies have shown the existence of specific oxygen diffusion routes (e.g., refs. 2–4).
Localizing dioxygen in proteins is difficult. Oxygen is very mobile and usually is not resolved in crystal structures, although mimicking O2 with xenon, which has a higher electron density, in some cases enabled the detection of well defined oxygen binding sites (e.g., ref. 5). Other techniques, like tryptophane fluorescence quenching, suffer from insufficient resolution. However, an alternative and promising approach to determine the occupation probability of oxygen in a protein consists of the application of computational methods. The equilibrium distribution of oxygen is determined by the Gibbs free-energy cost ΔGS(O2) to transfer an oxygen molecule from the solvent to a given position in the protein. If ΔGS(O2) is negative for a certain region of the protein, then the O2 concentration in that area is increased. Unfortunately the free-energy distribution cannot be measured directly, and computation based on molecular dynamics (MD) simulations is cumbersome because a large number of different protein conformations have to be taken into account. Recently, implicit ligand sampling was introduced (6), a method that allows efficient computation of 3D free-energy maps for small gas molecules in proteins. Here we have applied this technique to the rabbit 12/15-lipoxygenase (LOX).
LOXs form a heterogeneous family of nonheme iron-containing fatty acid dioxygenases (7, 8). A number of mammalian LOXs are implicated in (patho-) physiological processes (9–12). They represent interesting candidates for the detection of specific oxygen access channels. The existence of such a channel in LOXs initially was postulated when the first crystal structure for soybean LOX-1 was solved (13). An alternative oxygen access path later was suggested (14), and recent mutagenesis studies indicate its functionality (15, 16). Inferring from structural comparison that the same oxygen channel also is present in rabbit 12/15-LOX (17) is intricate because of substantial differences in the relevant parts. Furthermore, the dynamics of oxygen access remains an interesting open question.
The stereochemistry of the reaction is well defined. There is an antarafacial relation between initial hydrogen abstraction and subsequent oxygen insertion (8). In principle, there are different ways to explain the stereocontrol of LOX-catalyzed oxygen insertion. (i) The enzyme-bound fatty acid radical is stabilized at the active site in one of its mesomeric limit structures. Such stabilization may be accomplished by electrophilic amino acid side chains, which focus the electron density at a well defined position or space-filling residues that force the fatty acid radical into a steric configuration with a more defined localization of the radical electron (18). (ii) Molecular dioxygen may not be randomly available at the active site but might be targeted to the fatty acid radical via defined channels.
In this work, we present a detailed atomistic model for the oxygen access mechanism in 12/15-LOX based on MD simulations and implicit ligand sampling. We confirmed the computational findings by multiple site-directed mutagenesis and kinetic measurements. Our results reveal an area of high oxygen affinity at the catalytic center and specific pathways that can be used by oxygen for diffusion into the active site.
Results
Dioxygen Is Concentrated at the Catalytic Center of Rabbit 12/15-LOX.
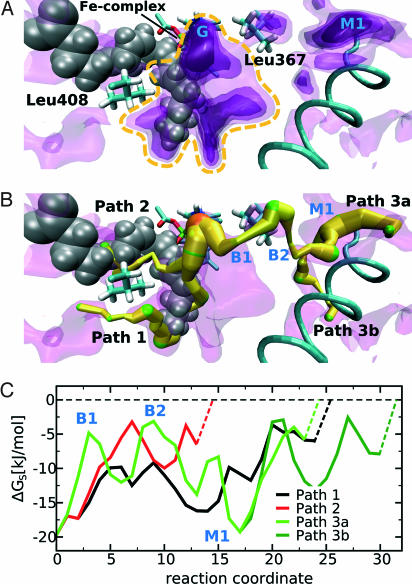
Using implicit ligand sampling (6), we calculated a 3D distribution map of the Gibbs free energy ΔG(O2) for placing one molecule of dioxygen from vacuum into any 1-Å3 volume element. In regions where ΔG(O2) is low, the probability for finding oxygen (i.e., the oxygen affinity) is high. In water this free-energy distribution is uniform (8.2 kJ/mol). In this article, we use the free-energy difference with respect to the solvent instead of to vacuum ΔGS(O2). In Fig. 1A, the free-energy distribution in the environment of the empty substrate-binding pocket is displayed in the form of four nested energy isosurfaces. The inner surfaces enclose regions with lower ΔGS(O2). Localization of the substrate-binding pocket was visualized by an arachidonic acid molecule, albeit the structural model used for these simulations did not contain fatty acid substrate. The free-energy maps indicate that the innermost regions of the substrate-binding pocket exhibit a higher probability for finding oxygen than other parts of the enzyme. The global free-energy minimum (−19.8 kJ/mol) is located deep inside the substrate-binding pocket opposite to the nonheme iron. The chance of finding oxygen in the high-affinity region around the active site (marked by a yellow dashed line in Fig. 1A) was 50- to 100-fold higher than in an equivalent volume of solvent. It has to be noted that these values represent statistical averages sampled from the oxygen distribution pattern of many 12/15-LOX conformations.
Fig. 1.
Free-energy distribution for oxygen inside the substrate-free rabbit 12/15-LOX. (A) Four nested free-energy isosurfaces with energy levels of −12.1, −9.5, −7.0, and −3.0 kJ/mol (from dark to light violet) are superimposed. The dark regions represent areas with a high probability of finding oxygen (low energy). Although this model represents the substrate-free enzyme, arachidonic acid (gray) was projected in to allow better orientation. Parts of the iron complex can be seen behind the substrate molecule. The global maximum of the probability of oxygen occupation (G) is located in front of arachidonic acid. The central high-affinity region close to the active site that is referred to in the text is indicated by a yellow dashed line. (B) Optimal routes for oxygen movement determined by the “flooding” method. The depicted paths start at the global energy minimum (orange) and end at local minima at the protein surface, which are in contact with the solvent. The diameter of the paths is related to the free-energy level and thus the oxygen affinity. A thick path symbolizes high oxygen affinity. Local energy minima are marked in green. The inner part of path 1 coincides with the deepest section of the substrate-binding pocket. The protein surface is intersected in the surrounding of L408. Path 2 is identical with the substrate entrance, whereas path 3 leads to the opposite side of the protein. Just below the protein surface, we identified a deep local energetic minimum (M1) that may act as an initial scavenger for oxygen molecules freely diffusing in the solvent. This initial sink can be accessed from either side of a helix formed by residues 426–438. (C) Energy profiles for oxygen movement along the different paths. The dashed lines connecting the profiles with the energy level of oxygen in the solvent represent the movement of oxygen molecules from the solvent into the initial oxygen traps at the protein surface. The reaction coordinate on the abscissa indicates the length of the oxygen paths in arbitrary units. In the surrounding of L367, path 3 has two characteristic energy barriers (B1 and B2), suggesting that this amino acid is critical for oxygen movement along this path.
Implicit Ligand Sampling Simulations of Substrate-Free 12/15-LOX Revealed Three Major Routes for Intraprotein Oxygen Movement.
Inspection of the highest-energy isosurface in Fig. 1A at −3.0 kJ/mol provided an impression of possible oxygen pathways. However, to identify the most likely routes for oxygen diffusion from the protein surface to the high-affinity area at the active center, we developed an algorithm to search for low-energy paths in the 3D energy map. This task was accomplished by gradually flooding the energy landscape. Whenever an outlet from the current basin was found, a steepest descent path through this saddle point connecting the current with the neighboring basin could be defined. Following this procedure for all newly found local minima in an iterative manner allowed us to construct pathways in the free-energy landscape that could be considered energetically preferred routes of oxygen diffusion.
Using the substrate-free model of 12/15-LOX, we identified three major channels with high probability of oxygen occupancy interconnecting the protein surface with the high-affinity area at the active site (Fig. 1B). The energy profiles along these paths are shown in Fig. 1C. The route of oxygen movement with the lowest barrier (path 1) starts at the bottom of the substrate-binding pocket and reaches the protein surface close to L408 and I414. The inner part of this path corresponds to the oxygen channel postulated for the soybean LOX-1. The second route (path 2) follows the fatty acid-binding pocket. In the absence of the lipid substrate, this route is free of major energetic or steric constraints for oxygen movement. The third channel (path 3) connects the opposite side of the protein molecule with the active site. The energy profile along this path is characterized by two barriers related to the side chain of L367 and a deep local energy minimum (M1) between I150, M368, L437, and L508 just below the protein surface. This minimum can be accessed from both sides of the surface helix formed by residues 425–436. In contrast to the substrate-binding pocket, path 3 is not continuously open and thus escapes detection when the crystal structure is inspected. It rather resembles a chain of separate, mostly hydrophobic cavities that are transiently interconnected because of side chain flexibility of lining amino acids.
Arachidonic Acid Closes the Substrate-Binding Pocket for Oxygen Diffusion but Opens a Fourth Oxygen Access Channel.
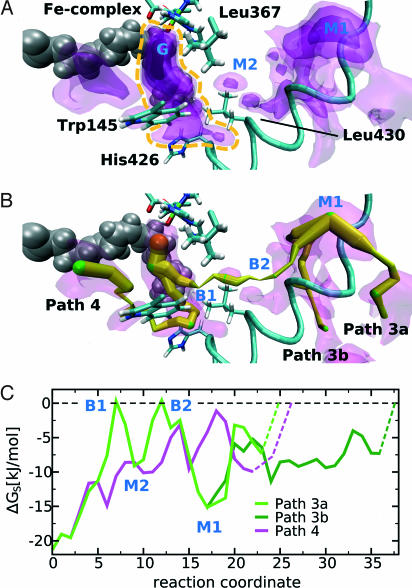
When arachidonic acid was added to the system (Fig. 2A), the regions of highest oxygen affinity at the active site were not altered significantly, and the location of the global energy minimum remained the same. Arachidonic acid does not completely fill the volume of the deeper parts of the binding pocket, leaving space for oxygen. This result indicates that dioxygen could be stored at the active site independent of bound substrate. However, toward the substrate entrance where the binding pocket is narrow, fatty acid substrate displaces oxygen from the pocket. Thus, diffusion of oxygen through the substrate-binding pocket is interrupted in the presence of fatty acid substrate. Fig. 2B shows that path 1 also disappeared. On the other hand, a fourth oxygen path was opened that reached the protein surface between W145 and H426, whereas path 3 was persistent.
Fig. 2.
Free-energy distribution of oxygen inside the 12/15-LOX-arachidonic acid complex. (A) Four nested isosurfaces characterizing the energy levels for oxygen distribution at levels −1.0, −4.9, −10.8, and −16.0 kJ/mol (see legend to Fig. 1A) are shown. Arachidonic acid is shown in gray, and the high-affinity region at the active site is marked by a yellow dashed line. To visualize the steric relations in a better way, this view is rotated right and up when compared with Fig. 1A. (B) Paths 1 and 2 from the substrate-free map disappeared but a new channel (path 4) reaching the protein surface between W145 and H426 becomes apparent. (C) Energy profiles for oxygen movement along paths 3 and 4. In path 3, the two energy barriers are somewhat higher when compared with the substrate-free enzyme.
Directed Oxygen Access Fosters Positional and Stereospecificity of the Reaction.
Because the fatty acid radical intermediates might adopt a different conformation in the pocket, some care has to be taken relating the oxygen distribution to the specificity of the oxygenase reaction. However, the region in which intraenzyme oxygen concentration is maximal and the iron complex is located are roughly at opposite sides of the fatty acid backbone. This spatial arrangement is consistent with the antarafacial character of the LOX reaction, which means hydrogen abstraction and oxygen insertion are taking place from opposite directions of the fatty acid molecule. Furthermore, it can be seen that the oxygen occupation probability within a sphere with 2-Å radius around C15 of the arachidonic acid backbone is 7-fold higher than around C11. Thus, oxygen insertion at C15 appears to be favored over C11, which is consistent with the positional specificity of the enzyme. However, because of the high degree of structural flexibility of arachidonic acid, our calculations do not exclude oxygen insertion at C11 or at the pro-R side of C15 so that additional mechanisms may contribute to determine the stereochemistry of the oxygenation process (19).
High Oxygen Affinity Is Important for Effective Catalysis.
A saturation concentration of 280 μM corresponds to one oxygen molecule in 5.9 × 106 Å3 (a box of 181-Å side length). The oxygen high-affinity region at the active site of the substrate-loaded 12/15-LOX has a volume of 246 Å3. If this region had the same oxygen affinity as the solvent, only every 20,000th enzyme molecule would be carrying oxygen. Integrating the occupation probability over all volume elements of the high-affinity region yields an O2 occupancy at the active center that is 100-fold higher than the average occupancy of the same volume in the solvent. It follows that in equilibrium approximately every 200th enzyme molecule is loaded with oxygen. These numbers illustrate that, to maintain a rate of 10 s−1 with only 1/200 enzymes in action at any instant, the product formation could last no longer than 1/(200 × 10) s = 500 μs.
Channel Conductivity Depends on the Energy Profile Along the Path.
The energy profiles for oxygen movement along the four major oxygen access channels found in the free-energy landscape are shown in Figs. 1C and 2C. The entrances to all channels are formed by solvent-exposed invaginations of the protein surface that exhibit a much higher oxygen affinity when compared with the solvent. The fact that oxygen falls from the solvent energy level into the entrance is symbolized by the dashed lines in Figs. 1C and 2C. Consequently, these hydrophobic invaginations may serve as sinks for initial oxygen enrichment at the protein surface. On their way to the global energy minimum (−19.8 kJ/mol) at the catalytic center, oxygen molecules must overcome several energy barriers. Oxygen conductivity along the putative channels depends on both the extent of the energy barriers to be overcome and the local increase of oxygen density near the surface invaginations. For instance, path 3 involves high-energy barriers that impair oxygen conductivity. On the other hand, the local energy minimum M1 at the protein surface is very low (−19.2 kJ/mol), suggesting high local oxygen concentrations.
Comparing the energy profiles along path 3 of the substrate-free enzyme and substrate containing 12/15-LOX, we found that the internal energy barriers of the enzyme–substrate complex are more pronounced. The molecular changes accounting for this effect have not been studied in detail but may be related to reduced side chain flexibilities in the surrounding of the substrate-binding pocket.
L367F Exchange Corrupts Oxygen Diffusion as Indicated by an Increased Michaelis Constant for Oxygen.
Because in the MD calculations only path 3 appeared to be functional for both substrate-free 12/15-LOX and the enzyme–substrate complex, we attempted to reduce its conductivity by site-directed mutagenesis. For this purpose, we mutated L367, which lines the channel at a critical position, to residues carrying more space-filling or charged side chains (L367F, L367W, L367K, and L367E). All mutants exhibited a somewhat reduced catalytic activity, but other enzymatic characteristics were not altered [see supporting information (SI) Appendix].
To assess the effect of the mutations on the oxygen conductivity of path 3, we determined kcat and KMO2 (Table 1) and calculated kcat/KMO2, the latter revealing specifically how mutants affect oxygen diffusivity. For the wild-type enzyme, a low KMO2 value of 5.2 ± 2.4 μM was determined, indicating a high oxygen affinity in line with previous results (20). Mutant L367K exhibited a notable drop in kcat and thus was ruled out from further considerations. For the mutants L367F, L367E, and L367W, the catalytic efficiency was in the order of the wild type, but kcat/KMO2 was lowered significantly. The largest changes were observed for L367F, showing a 20-fold lower kcat/KMO2 and a 10-fold higher Michaelis constant for oxygen. To interpret these findings with respect to oxygen diffusivity, we used the kinetic expressions 3 and 4 derived for the reaction scheme 2. As our calculations yielded very similar Gibbs free-energy distributions in the high-affinity region of wild-type and mutant L367F, a significant change of the association constant K0 for oxygen channeling is unlikely. We also assume that the rate constant kP, for oxygen insertion into the fatty acid radical, stays approximately the same. Under these premises, it follows from expression 4 that the ratio of the on-rate constants k+0wt and k+0mut for oxygen uptake in the mutant and the wild-type enzyme is given by
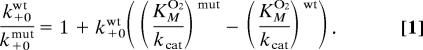 |
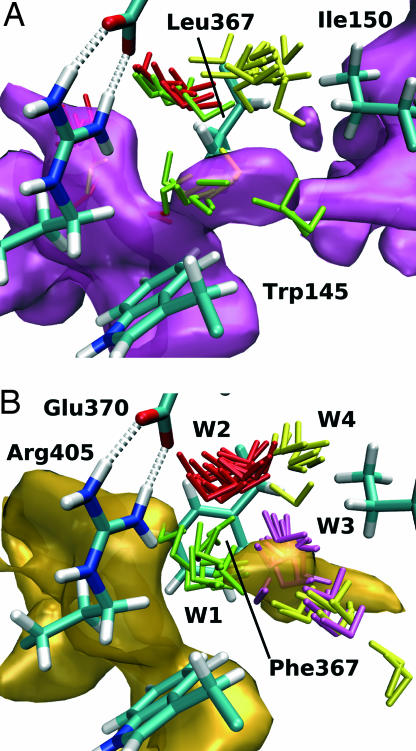
With a typical value of 100 μM−1s−1 for the diffusion on-rate constant we get k+0mut/k+0wt ratios of 677, 390, and 188 for the mutants L367F, L367E, and L367W, respectively. Thus, in the mutant enzymes, the conductivity of the oxygen channel should be reduced by at least two orders of magnitude. The charge of the glutamate mutant might account for a reorientation of neighboring side chains disturbing the architecture of the channel. The effect of the bulky mutants L367F and L367W might to some extent be explained by the reduced channel volume. Mechanistic details accounting for the impact of L367F exchange on oxygen conductivity of channel 3 can be explained when one considers the protein dynamics and the free-energy distribution: In Fig. 3, the oxygen-accessible areas of the substrate-free 12/15-LOX and its L367F mutant are compared in terms of the isosurface at −1.0 kJ/mol. Because the energy level of this isosurface is higher than the largest barrier in the wild-type enzyme, a continuous channel can be seen (Fig. 3A). For the mutant, however, the channel clearly is interrupted, and it is not the F367 side chain itself that is blocking the pathway (Fig. 3B). Instead, the effect is more subtle. We identified several water molecules in the vicinity of L367 that contribute to opening and closure of the oxygen channel. In the wild-type enzyme, these water molecules are not constrained to defined hydration sites but may move around and change their orientations (molecules are colored in red, yellow, and green in Fig. 3). In contrast, for the mutant enzyme (Fig. 3B), the water molecules W1 and W2 are immobilized, forming a stable hydrogen-bonding network that involves E370 and R405. Fig. 3B clearly shows that water molecule W1 is blocking the channel. The decreased water mobility is accompanied with the lack of conformational flexibility of the phenylalanine side chain. Although L367 is able to rotate more or less freely, F367 remained in the same conformation throughout our simulations.
Table 1.
Enzyme characteristics of different mutants
| kcat, s−1 | kcat/KMS*, s−1·μM−1 | KMO2, μM | kcat/KMO2, s−1·μM−1 | |
|---|---|---|---|---|
| WT | 13.7 ± 0.4 | 0.74 ± 0.08 | 5.2 ± 2.4 | 2.63 ± 1.22 |
| L367K | 0.3 ± 0.01 | 0.08 ± 0.01 | 9.0 ± 1.8 | 0.03 ± 0.01 |
| L367E | 2.2 ± 0.2 | 0.26 ± 0.06 | 9.0 ± 2.1 | 0.24 ± 0.06 |
| L367F | 5.6 ± 0.4 | 0.73 ± 0.07 | 40.1 ± 3.8 | 0.14 ± 0.02 |
| L367W | 4.4 ± 0.2 | 0.48 ± 0.05 | 7.0 ± 3.2 | 0.63 ± 0.29 |
*S, linoleic acid.
Fig. 3.
Impact of site-directed mutagenesis of L367F on intraenzyme oxygen movement. (A) Free-energy isosurface area at −1.0 kJ/mol for the wild-type enzyme in the vicinity of L367 (path 3). Three different water molecules (green, red, and yellow) in the surrounding of the mutated residue are displayed. Sixteen conformations from a 4-ns trajectory illustrate the mobility of these water molecules. (B) For the L367F mutant, the energy isosurface is disrupted at the site of mutation. The water molecules are immobilized, and W1 appears to interrupt the flow of oxygen. E370 and R405 participate in the hydrogen-bonding network arresting the water molecules.
Simulations of Oxygen Diffusion Reveal the Dynamic Character of the Oxygen Channels.
To estimate the rate of oxygen movement through the postulated channels, we performed additional MD simulations. The simulations were initialized by placing two oxygen molecules into the high oxygen affinity area of the wild-type 12/15-LOX with and without bound substrate and the arachidonic acid containing L367F mutant. During the 2-ns simulations, one of the introduced O2 molecules escaped via path 3 from the wild-type enzyme into the surrounding solvent.
Discussion
Oxygen Access Channels in LOXs.
For the soybean LOX-1, the presence of a putative oxygen access channel has been suggested, but for mammalian LOX isoforms, similar data currently are not available. Based on extensive MD simulations and implicit ligand sampling calculations, we predict four distinct oxygen access channels joining different sites of the protein surface with a region of high oxygen affinity around the catalytic center. Reliability of our theoretical predictions is supported by the following:
Oxygen access channel(s) have been reported before for the soybean LOX-1 (15, 16), and we compared the localization of these cavities with those of the rabbit enzyme. The inner oxygen cavity of the soybean enzyme channel colocalizes with the lower part of the substrate-binding cage and with parts of the high oxygen affinity region identified for the rabbit enzyme. Comparison of the more distal regions of the oxygen channels is problematic because these regions do not exhibit a high degree of structural similarity.
Our mutagenesis experiments on a critical Leu residue in channel 3 led to a mutant with significantly reduced oxygen conductivity, and the molecular reasons for the impaired oxygen permeability have been identified (see Fig. 3).
According to our structural model, localization of the high-affinity region is consistent with the antarafacial character of the LOX reaction. However, the positional specificity of oxygen insertion cannot alone be explained by the distribution of oxygen occupancy probability at C11 and C15 of the arachidonic acid backbone, mainly because of the high degree of motional flexibility of the substrate. Thus, alternative mechanisms appear to contribute to the stereocontrol of the LOX reaction (19). Similar conclusions also have been drawn for cyclooxygenase (21).
A central finding of our computations is the dynamic character of the predicted oxygen channels, some of which transiently open and close because of fluctuations of the protein conformation and thus are not detectable when inspecting the rigid crystallographic protein structure. Furthermore, our calculations show that two of the oxygen channels (paths 1 and 2) detected in the substrate-free enzyme are blocked upon fatty acid binding. One of them coincides with the substrate-binding pocket (path 2). Because hydrogen abstraction is the rate-limiting step (22), we can assume an equilibrium of oxygen exchange that is fast compared with the sojourn time of fatty acid in the pocket. Hence, it is unlikely that these two paths play a role in catalysis. In fact, mutation of a critical position along path 1 (L408F and L408W) did not impair oxygen affinity of the enzyme (data not shown). The equilibrium of oxygen exchange in the wild type being much faster than the turnover rate has further implications: An increase of KMO2 for the mutant indicates that oxygen access is impeded to an extent that oxygen becomes rate-limiting.
Occurrence of Oxygen Channels in Other Enzymes.
Presence of distinct molecular pathways for oxygen transport in proteins has been demonstrated for other enzymes. In water, oxygen can freely diffuse, and early fluorescence quenching studies suggested that this also may be the case in proteins (1). In contrast, more recent studies on oxygen metabolizing enzymes provide examples for targeted oxygen movement:
For copper amine oxidase reducing O2 to H2O2 when oxidizing primary amines a special channel for oxygen and hydrogen peroxide that is different from the substrate path has been proposed (23).
Cholesterol oxidase is a bacterial enzyme that oxidizes 3β-hydroxysteroids. In the crystal structure, which is highly resolved (0.95 Å), multiple conformations exist for many of the side chains (2, 24). Based on steric considerations, two conformations for the entire protein could be constructed. In one conformation, no channel can be seen, whereas in the other, there is a hydrophobic tunnel from the protein surface to the active center that is distinct from the substrate-binding cleft. This finding leads to the conclusion that the oxygen channel transiently opens and closes because of side chain dynamics.
Cytochrome c oxidase, which catalyzes the final step of the respiratory chain, requires oxygen as electron acceptor at the catalytic center located deeply inside the protein. A preformed channel that is supposed to serve as oxygen access path was detected (3, 25) and was confirmed by MD simulations of oxygen diffusion inside the protein (26).
For cyclooxygenases, solvent-accessible surface calculations and MD studies of oxygen diffusion inside the binding pocket revealed four access channels to the active site. Interestingly, the channel dynamics suggest that only the route also serving as substrate entrance efficiently functions as oxygen access pathway. This channel provides direct access to the stereochemically correct site of insertion, whereas other possible sites are sterically shielded (21).
These reported data and our MD simulations indicate that specialized oxygen access channels exist and that they are not necessarily empty or solvent-filled tubes inside the protein. They can be formed of neighboring, mostly hydrophobic cavities that partition oxygen away from water. These cavities temporarily are interconnected due to side chain flexibility. Because of their dynamic character, such channels might escape detection if static structural models are used.
Outlook.
The results of our MD simulations in connection with the mutagenesis data provide a detailed mechanistic picture and direct experimental evidence for the existence of functional oxygen access channels in the rabbit 12/15-LOX. They prompt the conclusion that similar structures might exist for many other oxygen-consuming enzymes. The methodology also can be applied to other small hydrophobic molecules (6). Hence, similar studies assessing the energetics and dynamics of gas diffusion in enzymes using NO, CO, or CO2 appear promising.
Methods
Structural Modeling and in Silico Mutagenesis.
Using the VMD software package (27), we set up structural models of the substrate-free enzyme and of the 12/15-LOX–arachidonic acid complex based on the x-ray coordinates of rabbit 12/15-LOX (17). These models, fully solvated with water at physiological salt concentrations, were used for MD simulations. In the enzyme–substrate complex, arachidonic acid was positioned in the substrate-binding pocket with its methyl terminus placed in proximity to the sequence determinants of the positional specificity (28). The fatty acid carboxylate was positioned in bonding distance to R403 (29), and the pro-S hydrogen at carbon 13 was placed in close proximity to the hydroxide ion ligand of the iron complex, the putative reaction partner (18). Details of the modeling procedure are described in SI Appendix. For in silico mutagenesis (L367F), the amino acid side chain was replaced in the equilibrated structure. A sterically favorable side chain conformation was found by stepwise rotation, then the system was reequilibrated for 10 ps to reach energetic optimization.
MD Simulations of Intraenzyme Oxygen Movement.
All MD simulations were carried out with the program NAMD (30). For protein, water, and ions, the CHARMM force field parameters (31) were used. For arachidonic acid, the corresponding values were compiled from molecular building blocks of lipids taken from the same force field database. The parameter set for the 12/15-LOX iron complex was calculated by using the PARATOOL program. PARATOOL is a plug-in for the molecular viewer VMD and is distributed with VMD version 1.8.5 or greater. Both can be obtained free of charge. Further details are given in SI Appendix. After equilibration, we tested intraenzyme oxygen movement by two independent approaches. To assess oxygen movement within the 12/15-LOX protein, one molecule of dioxygen was placed inside the substrate-binding pocket in close proximity to the site of oxygen insertion during catalysis (C15 of arachidonic acid). Another oxygen molecule was added next to the side chains of H426, V427, and K146, a site homologous to the internal part of the soybean LOX-1 oxygen channel (16). Oxygen diffusion was monitored over a 2-ns simulation period for the wild-type 12/15-LOX and its L367F mutant.
In addition to explicit simulations of oxygen diffusion, we also used implicit ligand sampling (6). With this method, it is possible to compute a 3D grid of the Gibbs free-energy cost ΔG for placing a molecule from vacuum into a certain volume element (voxel). In a first step, the system was equilibrated for a certain time period without oxygen. Next, oxygen molecules were placed in every trajectory frame on test points of a fine grid throughout the entire system. Averaging over many conformations, the free energy can be computed at every grid position considering the interaction energy of oxygen with the other molecules of the system. We calculated free-energy maps for the wild-type 12/15-LOX and its L367F mutant. For this purpose, we sampled 8,000 protein conformations from a 4-ns trajectory, which was obtained under the same general conditions used in the explicit oxygen MD simulations except that no oxygen was present in that system. Each volume element of the free-energy map comprised 1 Å3, and within each of these voxels oxygen was placed in 40 different rotational orientations at 27 different positions. The 3D free-energy distribution mirrors the probability of oxygen to occupy a given spatial region of the enzyme.
Kinetic Measurements and Quantification of Oxygen Affinity.
Oxygenation kinetics were assayed spectrophotometrically recording the time-dependent increase in absorbance at 235 nm (UV2100 spectrophotometer; Shimadzu, Kyoto, Japan). The enzyme initially was activated by incubation with 13S-HpODE (2 μM) in 0.1 M potassium phosphate buffer, pH 7.4. The reaction was started by addition of linoleic acid (500 μM), and all spectrophotometric measurements were carried out at room temperature. Different oxygen concentrations were adjusted by mixing hypoxic and hyperoxic assay buffers, and the oxygen concentration in the assay system was determined by using a Clark electrode. Fatty acid substrate never did become rate-limiting during the time course of oxygenation (pseudo-first-order conditions). Quantitative analysis of the kinetic data was based on the reaction scheme
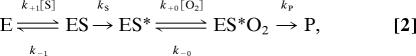 |
where ES* is the enzyme–fatty acid radical complex and ES*O2 is the complex with channeled oxygen. This scheme is identical with the core of a more detailed reaction scheme developed in our previous work (32). Stationary kinetic treatment results approximately in the rate law
where [Etot] is the total enzyme concentration. The catalytic rate constant is identical with the rate constant for hydrogen abstraction, i.e., kcat = kS. The ratio between kcat and the apparent Michaelis constant for oxygen is given by
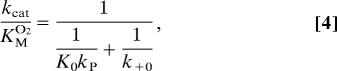 |
where kP denotes the rate constant for oxygen insertion into the fatty acid radical. K0 = k+0/k−0 is the equilibrium constant for oxygen and k+0 and k−0 mean the rate constants for uptake and release of oxygen. Numerical values for the parameters Vmax = kcat[Etot] and KMO2 were determined by minimizing the sum of squared distances between the rate Eq. 3 and experimentally determined initial rates taken from the slope of photometric progress curves.
Supplementary Material
Acknowledgments
We thank Jesper Haeggstrom for fruitful discussions and critical reading of our manuscript. Computer time was granted by Norddeutscher Verbund für Hoch-und Höchstleistungsrechnen. Financial support for this study was provided by the European Commission (FP6, LSHM-CT-2004–0050333, and MIFI-CT-2006–021230) and Deutsche Forschungegemeinschaft Graduiertenkolleg 268.
Abbreviations
- MD
molecular dynamics
- LOX
lipoxygenase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702401104/DC1.
References
- 1.Calhoun DB, Vanderkooi JM, Woodrow GW, III, Englander SW. Biochemistry. 1983;22:1526–1532. doi: 10.1021/bi00276a002. [DOI] [PubMed] [Google Scholar]
- 2.Lario PI, Sampson N, Vrielink A. J Mol Biol. 2003;326:1635–1650. doi: 10.1016/s0022-2836(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 3.Koutsoupakis K, Stavrakis S, Soulimane T, Varotsis C. J Biol Chem. 2003;278:14893–14896. doi: 10.1074/jbc.M210293200. [DOI] [PubMed] [Google Scholar]
- 4.Brunori M, Cutruzzola F, Savino C, Travaglini-Allocatelli C, Vallone B, Gibson QH. Biophys J. 1999;76:1259–1269. doi: 10.1016/S0006-3495(99)77289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tilton RF, Kuntz ID, Petsko G. Biol Cyber. 1984;23:2849–2857. doi: 10.1021/bi00308a002. [DOI] [PubMed] [Google Scholar]
- 6.Cohen J, Arkhipov A, Braun R, Schulten K. Biophys J. 2006;91:1844–1857. doi: 10.1529/biophysj.106.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brash AR. J Biol Chem. 1999;274:23679–23682. doi: 10.1074/jbc.274.34.23679. [DOI] [PubMed] [Google Scholar]
- 8.Feussner I, Wasternack K. Annu Rev Plant Biol. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- 9.Funk CD. Science. 2001;294:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- 10.van Leyen K, Duvoisin RM, Engelhardt H, Wiedmann M. Nature. 1998;395:392–395. doi: 10.1038/26500. [DOI] [PubMed] [Google Scholar]
- 11.Kuhn H, Römisch I, Belkner J. Mol Nutr Food Res. 2005;49:1014–1029. doi: 10.1002/mnfr.200500131. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn H, O'Donnel V. Prog Lipid Res. 2006;45:334–356. doi: 10.1016/j.plipres.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Boyington JC, Gaffney BC, Amzel LM. Science. 1993;260:1482–1486. doi: 10.1126/science.8502991. [DOI] [PubMed] [Google Scholar]
- 14.Minor W, Steczko J, Stec B, Otwinowski Z, Bolin JT, Walter R, Axelrod B. Biochemistry. 1996;35:10687–10701. doi: 10.1021/bi960576u. [DOI] [PubMed] [Google Scholar]
- 15.Knapp MJ, Seebeck FP, Klinman JP. J Am Chem Soc. 2001;123:2931–2932. doi: 10.1021/ja003855k. [DOI] [PubMed] [Google Scholar]
- 16.Knapp MJ, Klinman JP. Biochemistry. 2003;42:11466–11475. doi: 10.1021/bi0300884. [DOI] [PubMed] [Google Scholar]
- 17.Gillmor SA, Villasenor A, Fletterick R, Sigal E, Browner MF. Nat Struct Biol. 1997;4:1003–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 18.Nelson MJ, Seitz SP. Curr Opin Struct Biol. 1994;4:878–884. doi: 10.1016/0959-440x(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 19.Schneider C, Pratt DA, Porter NA, Brash AR. Chem Biol. 2007;14:473–488. doi: 10.1016/j.chembiol.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juranek I, Suzuki H, Yamamoto S. Biochim Biophys Acta. 1999;1436:509–518. doi: 10.1016/s0005-2760(98)00159-3. [DOI] [PubMed] [Google Scholar]
- 21.Furse KE, Pratt DA, Schneider C, Brash AR, Porter NA, Lybrand TP. Biochemistry. 2006;45:3206–3218. doi: 10.1021/bi052338h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egmond MR, Veldinkand GA, Vliegenthart JF, Boldingh J. Biochem Biophys Res Commun. 1973;54:1178–1184. doi: 10.1016/0006-291x(73)90816-4. [DOI] [PubMed] [Google Scholar]
- 23.Li R, Klinman JP, Matthews FS. Structure (London) 1998;6:293–307. doi: 10.1016/s0969-2126(98)00033-1. [DOI] [PubMed] [Google Scholar]
- 24.Coulombe R, Yue KQ, Ghisla S, Vrielink A. J Biol Chem. 2001;276:30435–30441. doi: 10.1074/jbc.M104103200. [DOI] [PubMed] [Google Scholar]
- 25.Riistama S, Puustinen A, García-Horsman A, Iwata S, Michel H, Wikström M. Biochim Biophys Acta. 1996;1275:1–4. doi: 10.1016/0005-2728(96)00040-0. [DOI] [PubMed] [Google Scholar]
- 26.Hofacker I, Schulten K. Proteins Struct Funct Gen. 1998;30:100–107. [PubMed] [Google Scholar]
- 27.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 28.Kuhn H, Saam J, Eibach S, Holzhütter HG, Ivanov I, Walther M. Biochem Biophys Res Commun. 2005;338:93–101. doi: 10.1016/j.bbrc.2005.08.238. [DOI] [PubMed] [Google Scholar]
- 29.Gan QF, Browner MF, Sloane DL, Sigal E. J Biol Chem. 1996;271:25412–25418. doi: 10.1074/jbc.271.41.25412. [DOI] [PubMed] [Google Scholar]
- 30.Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J Comp Phys. 1999;151:283–312. [Google Scholar]
- 31.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 32.Ivanov I, Saam J, Kuhn H, Holzhütter H. FEBS J. 2005;272:2523–2535. doi: 10.1111/j.1742-4658.2005.04673.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.