Abstract
Damage to the genome is unavoidable in living creatures, because of sunlight exposure as well as environmental chemicals present in food and drinking water. There is a need to monitor and purify the drinking water; therefore, several methods of detection have been developed. A very promising model system for this purpose is the zebrafish (Danio rerio), which is endowed with special qualities for detecting external as well as internal abnormalities. Grossman and Wei's assay [Grossman L, Wei Q (1995) Clin Chem 12:1854–1863], which measures the expression level of a nonreplicating recombinant plasmid DNA containing a UV-damaged luciferase reporter gene, shows that zebrafish can repair chromosomal lesions to a much greater extent than the human population. This vertebrate model is still very promising after possible down-regulation of the DNA repair enzymes.
Keywords: cancer, genotoxic agents, DNA photoproducts, toxicology
The necessity of determining the DNA repair capacity (DRC) of Danio rerio became evident when several laboratories chose this vertebrate as a model system to detect genotoxic agents in the water (1–3). The advantages of using zebrafish over other assays were the following. (i) The Ames test (4), based on the determination of mutation rate in bacteria, although inexpensive and fast, is insensitive for those agents that need to be metabolized in the liver to become biologically active. (ii) Mice are very expensive and labor intensive. (iii) Tissue culture is optimal for quantifying DRC but requires dedicated laboratory facilities and trained personnel. In contrast, zebrafish can be maintained in large quantities using inexpensive tanks. The fish are easy to grow and exceptional for being transparent and small enough to reveal internal abnormalities just by looking under a dissecting microscope. Moreover, zebrafish have proven to be a good model system to study toxicology and carcinogenesis (5–10).
The incidence of cancer not only has been correlated to the exposure of chemical agents in the environment that inflict lesions on DNA and the chromosomes but, to a significant extent, has also been determined by the capacity of the organism to repair lesions (11). The DNA excision repair system (12, 13) is the major cellular enzymatic complex that detects and removes the abnormal bases that are constantly being inflicted on the chromosomes [nuclear or mitochondrial (14)]. Without this vigilant set of enzymes, the cells would accumulate mutated genes to such a large extent that the organism would not survive long. These phenomena have been demonstrated in xeroderma pigmentosum patients (15) and other people with inherited mutations in the DNA repair enzymes (16). Studying the altered genes in this group of patients helped identify the specific enzymes that are essential in performing the repair (17). This global system was studied by using DNA photoproducts caused by UV light (UV), but it turned out that most of these enzymes also correct damage to DNA caused by reactive oxygen species, which are constantly produced in mitochondria (18–20) and by environmental pollutants (21). Therefore, the best sentinel for detecting carcinogens would be an organism that has the lowest DRC possible.
The work of Grossman and Wei on the host-cell reactivation (HCR) assay was pivotal for establishing the correlation of DRC to carcinogenesis (22). Using the quantitative HCR assay with stimulated T lymphocytes obtained from peripheral blood opened up the possibility to determine DRC in humans and extend it to epidemiological studies (23–25). First, Grossman and coworkers established the distribution of DRC in the normal population and compared it with that of people born with xeroderma pigmentosum (XP) mutations who die of cancer at an early age (24, 26). They found that XP patients displayed the lowest DRC observed in the study population. Later studies have shown that DRC diminishes with age in the human population, providing an explanation why cancer is a disease of old age (27, 28) and contributed to a large extent in establishing which enzymes are induced in the process and their specific activities (29). The HCR assay has been used to determine the correlation of DRC to the risk of developing tumors, such as breast cancer (30), prostate cancer (31), and to the onset of neurological abnormalities (32).
This report assesses DRC in living zebrafish embryos using this HCR assay. The present study reveals that WT D. rerio is highly competent to repair UV radiation-induced damage and consequently would not be effective as a sentinel for environmental agents. Nevertheless, it is possible to obtain sensitive transgenic lines by knocking out or down-regulating the DRC enzymes, which have been identified and characterized by the work of L. Grossman and others.
Results
The HCR assay developed to test for DRC used peripheral blood lymphoblasts obtained from human populations (23). Each assay determination required 2 × 106 cells for transfection with undamaged or UV-damaged plasmid DNA carrying a reporter enzyme. Applying this protocol to zebrafish was impossible, because of the low quantity of circulating blood. We reasoned that very young embryos that produce thousands of dividing cells in the first hours of development should be able to provide significant results. To test this assumption, we injected two to four cell embryos with the pCMVluc plasmid, in equal concentration with the pCMVren control plasmid, and let the embryos develop normally for 24 h or more. We dechorionated the surviving embryos, to expose the cells to the substrate, and tested for the expression of the reporter genes. In Assay A, we chose substrates that could be used in vivo to be able to follow the luciferases activities during development. Table 1 shows the results obtained by transfecting undamaged pCMVluc and control pCMVren in equal proportion. Seventy percent of the surviving embryos had luciferase activity, with 50% of those due to firefly enzyme and 50% to renilla enzyme. Thus, the proportion of luc/ren luciferases was 1.0. Thirty percent of the transfected embryos had activity for both plasmids.
Table 1.
Assay I, determination of luciferase activity in transfected live embryos using Luciferin and Viviren as substrates for firefly and renilla luciferases, respectively
| Embryo number | Luciferin AM | Luciferin PM | Washed, no substrate | Viviren AM | Viviren PM |
|---|---|---|---|---|---|
| 1 | 475 ± 7 | 812 ± 19 | 266 | 509 ± 29 | 1,398 ± 20 |
| 2 | — | — | — | — | — |
| 3 | 2,474 ± 29 | 575 ± 3 | 100 | 168 ± 7 | 240 ± 8 |
| 4 | 703 ± 9 | 143 ± 5 | 483 | 511 ± 9 | 844 ± 15 |
| 5 | 109 ± 5 | 897 ± 19 | 118 | — | — |
| 6 | — | — | — | 249 ± 7 | 795 ± 27 |
| 7 | — | — | — | — | — |
| 8 | 302 ± 3 | 159 ± 2 | — | 577 ± 13 | |
| 9 | — | — | — | 147 ± 2 | 255 ± 2 |
| 10 | — | — | — | — | — |
| 11 | — | — | — | — | — |
| 12 | — | — | — | 430 ± 9 | 598 ± 36 |
| 13 | 487 ± 21 | 1,035 ± 23 | — | 222 ± 7 | 730 ± 14 |
| 14 | — | — | — | — | — |
| 15 | 165 ± 3 | 196 ± 2 | — | — | — |
| 16 | 57 ± 2 | 854 ± 15 | — | — | — |
| 17 | — | — | — | — | — |
| 18 | 116 ± 2 | — | — | 388 ± 11 | 678 + 12 |
| 19 | 3,233 ± 36 | 2,284 ± 52 | — | — | — |
| 20 | — | — | 836 | 835 ± 10 | 1,147 ± 18 |
Single embryos were transfected with equal concentrations of pCMVluc and pCMVren. Twenty-four hours later, luciferin was added to single live embryos, and light emission was recorded. After washing embryos and ascertaining that activity was low, viviren was added and renilla enzymatic activity recorded. The same measurements were repeated in the afternoon. The numbers represent the mean RLU of three independent luminometer readings ± SD.
Later, it was found to be easier, less expensive, and still reliable to change to Assay B, which consisted of determining firefly luciferase in vivo with luciferin, as in Assay A, and afterward the embryos were lysed with 20 μl of Passive Lysis Reagent (Promega, Madison, WI) and frozen. The extracts no longer had ATP, which is necessary for the firefly luciferase but does not affect the renilla luciferase activity, thus no light was emitted. After adding 4 μl of Stop & Glow reagent (Promega), which contains the substrate for renilla luciferase, the light emitted by the extracts was determined. Table 2 shows that Assay B is similar to Assay A: 25% of the embryos elicited firefly luminescence, and 35% responded with the renilla substrate. Twenty percent expressed both enzymes. The ratio of firefly luciferase activity to that of renilla is a measure of the proportion of repaired UV-treated plasmids.
Table 2.
Assay B, determination of activity in transfected embryos using Luciferin and Bright & Glo as substrates for firefly and renilla luciferases, respectively
| Embryo number | Luciferin RLU | Bright & Glo RLU |
|---|---|---|
| 1 | — | 1,703 ± 90 |
| 2 | — | — |
| 3 | — | |
| 4 | 180 ± 5 | 14,518 ± 78 |
| 5 | — | — |
| 6 | — | 3,360 ± 95 |
| 7 | 3,955 ± 140 | 762 ± 17 |
| 8 | — | — |
| 9 | — | — |
| 10 | — | — |
| 11 | — | — |
| 12 | 1,016 ± 24 | 434 ± 10 |
| 13 | — | — |
| 14 | — | — |
| 15 | — | — |
| 16 | 1,476 ± 29 | 2,740 + 123 |
| 17 | — | 467 ± 13 |
| 18 | — | — |
| 19 | — | — |
| 20 | — | 592 ± 38 |
| 21 | 224 ± 8 | 628 ± 41 |
| 22 | 3,030 ± 34 | 1,205 ± 50 |
| 23 | — | — |
| 24 | 85,390 ± 1,020 | |
| 25 | 181 ± 8 | 7,353 ± 435 |
| 26 | — | — |
| 27 | — | — |
| 28 | — | — |
| 37 | 6,916 ± 99 | — |
| 38 | — | — |
| 39 | 5,423 ± 160 | — |
| 40 | — |
Single embryos were transfected with equal concentrations of pCMVluc and pCMVren. Twenty-four hours later, luciferin was added and their light emission measured. After washing, the embryos were lysed, frozen, defrosted, and Stop & Glo added to measure renilla luciferase activity. The numbers represent the mean RLU of three independent luminometer readings ± SD.
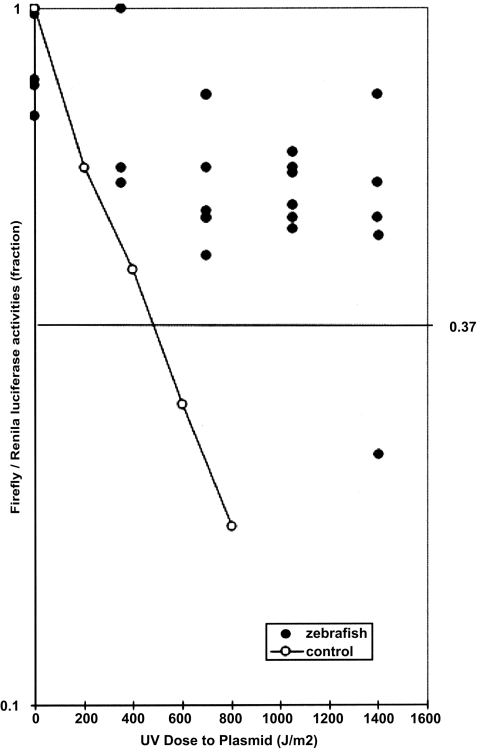
Table 3 shows the results of many transfections performed with pCMVluc, either intact or with increasing numbers of UV lesions, mixed with the same concentration of pCMVren as control of transfection. The ratios obtained, even with the untreated plasmid, show great variability. This is to be expected, because the plasmids have to diffuse from the yolk to the cells; this process may vary for different embryos. Although it is not possible to treat the data statistically, it is possible to compare these results with the DRC obtained for the human population, which estimates the median dose (37% of the lymphoblastoid cells that show repair activity) at a UV dose of ≈400 J/m2 (25). In Fig. 1, which plots the results obtained in all of the experiments performed (Table 3), clearly indicates that the median dose of zebrafish embryos (1,200–1,400 J/m2) is much higher than that of humans.
Table 3.
Transfections of zebrafish embryos with untreated and UV-treated plasmids
| Experiment | UV dose,* J/m2 | No. of embryos | Percent firefly,† RLU | Percent renilla,† RLU | luc/ren‡ |
|---|---|---|---|---|---|
| 1 | 0 | 23 | 30 | 39 | 0.77 |
| 2 | 0 | 66 | 30.5 | 30.5 | 1.00 |
| 3 | 0 | 71 | 12.6 | 16 | 0.79 |
| 4 | 0 | 37 | 27 | 35 | 0.77 |
| 5 | 0 | 35 | 14 | 14 | 1.00 |
| 6 | 0 | 40 | 22.5 | 22.5 | 1.00 |
| 7 | 0 | 33 | 39.4 | 42 | 0.93 |
| 1 | 350 | 52 | 7.7 | 7.7 | 1.00 |
| 2 | 350 | 20 | 24 | 30 | 0.56 |
| 3 | 350 | 35 | 17 | 28.6 | 0.59 |
| 1 | 700 | 42 | 7.1 | 14 | 0.50 |
| 2 | 700 | 32 | 37.5 | 50 | 0.75 |
| 3 | 700 | 70 | 8.6 | 17 | 0.51 |
| 4 | 700 | 11 | 18 | 36 | 0.50 |
| 5 | 700 | 33 | 24 | 54.5 | 0.44 |
| 6 | 700 | 35 | 8.6 | 14.6 | 0.59 |
| 7 | 700 | 39 | 25.6 | 51 | 0.50 |
| 1 | 1,050 | 130 | 10 | 21 | 0.48 |
| 2 | 1,050 | 27 | 3.7 | 7.4 | 0.50 |
| 3 | 1,050 | 21 | 19 | 33 | 0.58 |
| 4 | 1,050 | 42 | 28.6 | 54.8 | 0.52 |
| 5 | 1,050 | 55 | 16 | 27 | 0.59 |
| 6 | 1,050 | 43 | 21.6 | 35 | 0.62 |
| 1 | 1,400 | 33 | 9,1 | 39 | 0.23 |
| 2 | 1,400 | 38 | 26 | 55 | 0.47 |
| 3 | 1,400 | 36 | 30.5 | 61 | 0.50 |
| 4 | 1,400 | 12 | 33 | 58 | 0.56 |
| 5 | 1,400 | 36 | 16.6 | 22 | 0.75 |
Summary of all the transfection experiments performed with either untreated or UV-treated pCMVluc mixed with the control pCMVren used to determine transfection efficiency.
*UV dose of pCMVluc.
†Mean values of three consecutive luminometer readings.
‡Fraction of firefly/renilla activities.
Fig. 1.
Comparison of DRC from zebrafish and human. The zebrafish values represent the proportion of UV lesions repaired, taken from the last column of Table 3. The control human lymphoblastoid cell line is reproduced from published experiments (25), with permission from Qingyi Wei.
Discussion
Susceptibility to cancer is a multibody phenomenon, because many external stimuli, such as sunlight, x-rays used in medical practice, pollutants in water, chemicals applied to food, insecticides, etc., are the most frequent inducers of mutations in animals. Just as important is the complex cellular system that causes the formation of oxygen radicals and reactive oxygen species (ROS), leading to the up-regulation of at least 40 mammalian genes (33). The first scientist to intuitively recognize the correlation of ROS and induction of cancer was Albert Szent-Gyorgyi (34, 35). All these stimuli work by causing specific chromosomal lesions, which are sensed by the cell, inducing the DNA damage response. This signaling mechanism apparently starts by recruiting two protein kinases: ataxia telangiectasia mutated (ATM) and ATM Rad3-related (ATR), which are capable of sensing specific DNA lesions when they are activated by phosphorylation and in turn start a cascade of at least 900 phosphorylation sites in 700 proteins (36–39).
By far the most important parameter among biomarkers for DNA repair in the susceptibility to cancer is DRC, as demonstrated by the work of L. Grossman and coworkers, who devised the quantitative HCR assay (23) that could be used for epidemiology studies.
In the present work, the DRC assay was applied to zebrafish embryos to determine whether the WT species would be sensitive as a detector of carcinogens in water. The data obtained, even with untreated plasmid, are more scattered than the results published for humans. This is because, in those studies, the lymphocytes used are homogeneous suspensions of cells belonging to a single individual and exposed to the plasmid uniformly at a given time. Working with embryos has more variables, such as (i) the number of cells per embryo at the time of transfection; (ii) the stage of development, which is not so well synchronized; and (iii) most important of all, that each embryo has a different genotype. Because the average number of embryos transiently transfected in these experiments was ≈30% compared with 70% in human lymphocytes (40), we should consider the results as Poisson distributions, where the control and the experimental plasmids have independent distributions. This is clearly seen in Tables 1 and 2. Nevertheless, Fig. 1 shows that, even with the large scattering of values obtained, zebrafish embryos have a high capacity for repairing UV photoproducts (D, 37% = 1,400 J/m2) compared with human (D, 37% = ≈400 J/m2). This may be easily explained considering this fish survived for centuries in the Delta region of India, because it adapted to high solar radiation. On the other hand, because zebrafish is transparent to light, exposing its internal organs to radiation may indicate that pollution was probably the selective agent.
Some experimental variation observed could be ameliorated by injecting the plasmids directly into the cell of single-cell embryos or to use some of the highly inbred lines of fish being generated (ZFIN ID: ZDB-LAB-980202-4; www.shigen.nig.ac.jp:6070/zf_info/downloads.html). There is evidence that most of the variability in WT strains may be due to intrinsic factors. For example, when oxygen consumption was measured in single 24–72 h postfertilization zebrafish embryos, it fluctuated between 0.26 and 0.46 μmol O2 (41). Other fish also display variations in size and growth rates in developing embryos (42, 43).
This work shows that the DRC assays can be applied to animals, and that, in the case of WT zebrafish, it proves it would not be sensitive as a detector of carcinogens in water. In addition, by using luciferase as a reporter gene, it is possible to keep the embryos alive (Assay A) for selection purposes. Considering that D. rerio is a vertebrate with oncogenic proteins similar to those of humans (44, 45), it should be relatively easy to engineer transgenic lines that are defective in the major DNA repair enzymes. The sequences of these enzymes are well known and either morpholinos or microRNAs have been successfully used in zebrafish for this purpose. The advantage of having a model animal with higher DRC than humans makes it possible to uncover new parameters for cancer prevention.
Materials and Methods
Cells and Transfection.
Two to eight celled embryos from WT D. rerio were placed in a Petri dish that supported a 1-cm-wide strip of dental wax cemented to the center of the plate. The strip was 1 mm high, which could hold the embryos lined up along its edge. By removing the excess liquid, the embryos attached by capillarity to the strip on the left side and were injected from the right side into the yolk sac. Injections were performed with a glass micropipette attached to an air-pressure-driven micromanipulator introducing 20–25 nl of the plasmids (25 μg/ml each) in Tris-EDTA containing 5% sterile Phenol red (Sigma, St. Louis, MO). The Phenol red served as an estimate of the volume delivered into each embryo. After injection, the embryos were transferred to sterile Petri dishes containing embryo medium (46) with 0.25 μg/ml methylene blue and incubated at 28.5°C. The surviving embryos developed normally, and 27–30 h after injection, they reached the prim 6 to prim 16 embryonic stage (46), displaying spontaneous movements. They were dechorionated manually and distributed singly into Eppendorf tubes. Most of the liquid in the tube was removed, leaving enough to cover the embryo.
Plasmids.
The plasmid expression vector pCMVluc (4,863 bp) (25) containing the firefly luciferase reporter gene under the control of the CMV promoter enhancer was used. The plasmid was purified through CsCl to select the supercoiled species and irradiated with UV of 240 nm at 0, 300, 700, 1,050 and 1,400 J/m2. (This set of plasmids was a gift of L. Grossman and recently of Qingyi Wei.) Because the transfection efficiency varied considerably, we included an untreated control plasmid: pRLluc (Promega), coding for Renilla luciferase, which requires a different substrate to emit light.
Dual Luciferase Assays.
Assay A was designed to keep the embryos alive, while determining the activity of the reporter genes repeatedly for several hours of development: 10 μl of luciferin (0.5 mM) was added to each dechorionated embryo, and three consecutive measurements of the light output in relative light units were recorded by using a luminometer. The activity could be measured again during the next hours of development. After washing the embryos with embryo medium and ascertaining that only background light was emitted, 10 μl of Viviren (Promega) was added to the tube, and triplicate measurements of Renilla luciferase were recorded. Again, the activity of the control reporter enzyme could be repeated during further incubation (Table 1).
Assay B.
Ten microliters of Luciferin was used to measure the firefly luciferase activity, as in assay A, then the washed embryos were lysed with 20 μl of Passive Lysis Reagent(Promega) and frozen. The defrosted embryo extract no longer had ATP, which is necessary for the firefly luciferase but does not affect the renilla luciferase activity; thus, no light was emitted. After adding 4 μl of Stop & Glow reagent (Promega) to each tube, the light emitted by the Renilla luciferase present in the extracts was determined in triplicate (Table 2).
The luminometer used was a Femtomaster FB12 (Zylux Corp., Maryville, TN) programmed for a 3 s measurement delay followed by a 10 s measurement of light emission, and it was checked with a solution of Quantilum Recombinant Luciferin monopotassium salt (Pierce) at 10−10 M in medium containing 1 mg/ml BSA after adding a few microliters of 0.5 mM solution of d-Luciferin monopotassium salt (Pierce).
Acknowledgments
This publication is dedicated in memoriam to Dr. Lawrence Grossman. I thank Qingyi Wei (University of Texas M.D. Anderson Cancer Center, Houston, TX) for providing the plasmids, permitting the addition of previously published data on human DRC, and helpful revision of the manuscript; Dr. Daniel Sussman (National Institutes of Health, Bethesda, MD) for correcting the manuscript; Dr. Kamala Patel (University of Calgary, Calgary, AB, Canada) for helpful comments; Christina Holmes for assistance with the data; and Janice Simmons and Erika Woods for expert maintenance of the zebrafish. I gratefully acknowledge Dr. Steve Roberts for laboratory facilities and the BioCurrents Research Center (Marine Biological Laboratory) for access to injection equipment.
Abbreviations
- DRC
DNA repair capacity
- HCR
host-cell reactivation
- RLU
relative light units.
Footnotes
The authors declare no conflict of interest.
References
- 1.Mizell M, Romig ES. Int J Dev Biol. 1997;41:411–423. [PubMed] [Google Scholar]
- 2.Carvan MJ, III, Sonntag DM, Cmar CB, Cook RS, Curran MA, Miller GL. Sci Total Environ. 2001;274:183–196. doi: 10.1016/s0048-9697(01)00742-2. [DOI] [PubMed] [Google Scholar]
- 3.Langheinrich U. Mol Cell Biol (2006) 2003;26:9083–9093. doi: 10.1128/MCB.01216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ames BN, Mccann J, Yamasaki E. Mutat Res. 1975;31:347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- 5.Amatruda JF, Shepard JL, Stern HM, Zon LI. Mutat Res. 1975;31:347–364. [Google Scholar]
- 6.Bladen CL, Lam WK, Dynan WS, Kozlowski DJ. Nucleic Acids Res. 2005;33:3002–3010. doi: 10.1093/nar/gki613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collodi P, Miranda C, Zhao X, Buhler DR, Barnes DW. Xenobiotica. 1994;24:487–493. doi: 10.3109/00498259409043251. [DOI] [PubMed] [Google Scholar]
- 8.Cheng R, Hendricks JD, Bradford CS, Barnes DW, Bailey GS. Mol Mar Biol Biotechnol. 1997;6:49–47. [PubMed] [Google Scholar]
- 9.Miranda CL, Collodi P, Zhao X, Barnes D, Buhler DR. Arch Biochem Biophys. 1993;301:320–327. doi: 10.1006/abbi.1993.1429. [DOI] [PubMed] [Google Scholar]
- 10.Scata KA, El-Deiry WS. Cancer Biol Ther. 2004;3:501–502. doi: 10.4161/cbt.3.6.946. [DOI] [PubMed] [Google Scholar]
- 11.Sarasin AR, Hanawalt PC. Proc Natl Acad Sci USA. 1978;75:346–350. doi: 10.1073/pnas.75.1.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Setlow RB. Basic Life Sci Rev. 1980;15:45–54. doi: 10.1007/978-1-4684-3842-0_3. [DOI] [PubMed] [Google Scholar]
- 13.Grossman L Mazur SJ, Caron PR, Oh EY. FASEB J. 1988;2:2196–2701. doi: 10.1096/fasebj.2.11.3294078. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen LJ, Stevnsner T, Bohr VA. Ugeskr Laeger. 2006;162:2332–2335. [PubMed] [Google Scholar]
- 15.Kraemer KH. In: Dermatology in General Medicine. Fitzpatrick TB, Eisen AZ, Austen F, Freedberg IM, Wolff K, editors. New York: McGraw–Hill; 1983. pp. 13–142. Update I. [Google Scholar]
- 16.Bootsma D, Kraemer KH, Cleaver JE, Hoeljmakers JLJ. In: The Genetic Basuis of Human Cancer. Vogelstein B, Kinzler KW, editors. New York, NY: McGraw–Hill; 1998. pp. 245–274. [Google Scholar]
- 17.Shiloh Y. Curr Opin Genet Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 18.Davies KJ. IUBMB Life. 2000;50:279–289. doi: 10.1080/713803728. [DOI] [PubMed] [Google Scholar]
- 19.Wood RD, Mitchell M, Sgouros J, Lindahl T. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 20.Barzilai A, Yamamoto KI. DNA Rep. 2004;3:1109–1115. doi: 10.1016/j.dnarep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Stegeman JJ, Lech JJ. Environ Health Perspect. 1991;90:101–109. doi: 10.1289/ehp.90-1519513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossman L, Wei Q. Clin Chem. 1995;12:1854–1863. [PubMed] [Google Scholar]
- 23.Athas WF, Hedayati MA, Matanoski GM, Farmer ER, Grossman L. Cancer Res. 1991;51:5786–5793. [PubMed] [Google Scholar]
- 24.Kraemer KH, Moriwaki SI, Tarone RE, Khan SG, Grossman J. Molecular Biology of Aging. In: Bohr W, Clark B, Stevnsner T, Svejgard A, editors. Alfred Benson Symposium 44; Copenhagen: Munksgaard; 1998. [Google Scholar]
- 25.Qiao Y, Spitz MR, Guo Z, Hadeyati M, Grossman L, Kraemer KH, Wei Q. Mutat Res. 2002;509:165–174. doi: 10.1016/s0027-5107(02)00219-1. [DOI] [PubMed] [Google Scholar]
- 26.Cleaver JE, Kraemer KH. In: The Metabolic Basis of Inherited Diseases. Scrivner CR, Beaudet AL, Sly WS, Valle E, editors. New York: McGraw–Hill; 1989. pp. 2949–2971. [Google Scholar]
- 27.Wei Q, Matanoski GM, Farmer ER, Hedayati MA, Grossman L. Proc Natl Acad Sci USA. 1993;90:1614–1618. doi: 10.1073/pnas.90.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossman L. Aging. 1992;4:252–255. doi: 10.1007/BF03324100. [DOI] [PubMed] [Google Scholar]
- 29.Grossman L, Yenng AT. Mutat Res. 1990;236:213–221. doi: 10.1016/0921-8777(90)90006-q. [DOI] [PubMed] [Google Scholar]
- 30.Ramos JM, Ruiz A, Colen R, Lopez ID, Grossman L, Matta JL. Cancer. 2004;100:1352–1357. doi: 10.1002/cncr.20135. [DOI] [PubMed] [Google Scholar]
- 31.Hu JJ, Hall MC, Grossman L, Hedayati M, McCullough DL, Lohman K, Case LD. Cancer Res. 2004;64:1197–1120. doi: 10.1158/0008-5472.can-03-2670. [DOI] [PubMed] [Google Scholar]
- 32.Emmert S, Slor H, Busch DB, Albert RB, Coleman D, Khan SG, Abu-Libdeh B, DiGiovanna JJ, Cunningham BB, Lee MM, et al. J Invest Dermatol. 2002;118:972–982. doi: 10.1046/j.1523-1747.2002.01782.x. [DOI] [PubMed] [Google Scholar]
- 33.Davis KJ. IUBMB Life. 2000;50:279–289. doi: 10.1080/713803728. [DOI] [PubMed] [Google Scholar]
- 34.Szent-Gyorgyi A. PhysChemPhys. 1980;12:99–110. [PubMed] [Google Scholar]
- 35.Pethig R, Gascoyne PR, McLaughlin JA, Szent-Gyorgyi A. Proc Natl Acad Sci USA. 1983;82:1439–1442. doi: 10.1073/pnas.82.5.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 37.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H, Chen J, Yu X. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 40.Cheng L Bucana CD, Wei Q. BioTechniques. 1996;21:486–491. doi: 10.2144/96213rr01. [DOI] [PubMed] [Google Scholar]
- 41.Bang A, Gronkjar P, Malte H. J Fish Biol. 2004;64:1285–1296. [Google Scholar]
- 42.Chambers RC, Leggett WC. Am Zool. 1996;36:180–196. [Google Scholar]
- 43.Vollestad LA Lillehammer T. Ecol Fresh Water Fish. 2000;9:242–247. [Google Scholar]
- 44.Cheng R, Hendricks JD, Bradford CS, Barnes D, Bailey GS. Mol Marine Biol Biotechnol. 1997;6:40–47. [PubMed] [Google Scholar]
- 45.Cheng R, Ford B, O'Neal P, Mathews CS, Bradford S, Thongtan T, Barnes D, Hendricks J, Bailey G. Mol Marine Biol Biotechnol. 1997;6:88–97. [PubMed] [Google Scholar]
- 46.Westerfield M. The Zebrafish Book, A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio) 2nd Ed. Eugene, OR: Univ of Oregon Press; 1994. [Google Scholar]