Abstract
The unicellular red alga Galdieria sulphuraria is a facultative heterotrophic member of the Cyanidiaceae, a group of evolutionary highly conserved extremophilic red algae. Uptake of various sugars and polyols is accomplished by a large number of distinct plasma membrane transporters. We have cloned three transporters [GsSPT1 (G. sulphuraria sugar and polyol transporter 1), GsSPT2 and GsSPT4], followed their transcriptional regulation and assayed their transport capacities in the heterologous yeast system. SPT1 is a conserved type of sugar/H+ symporter with 12 predicted transmembrane-spanning domains, whereas SPT2 and SPT4 represent monosaccharide transporters, characterized by only nine hydrophobic domains. Surprisingly, all three proteins are functional plasma membrane transporters, as demonstrated by genetic complementation of a sugar uptake-deficient yeast mutant. Substrate specificities were broad and largely redundant, except for glucose, which was only taken up by SPT1. Comparison of SPT1 and truncated SPT1(Δ1–3) indicated that the N-terminus of the protein is not required for sugar transport or substrate recognition. However, its deletion affected substrate affinity as well as maximal transport velocity and released the pH dependency of sugar uptake. In line with these results, uptake by SPT2 and SPT4 was active but not pH-dependent, making a H+ symport mechanism unlikely for the truncated proteins. We postulate SPT2 and SPT4 as functional plasma membrane transporters in G. sulphuraria. Most likely, they originated from genes encoding active monosaccharide/H+ symporters with 12 transmembrane-spanning domains.
Keywords: Cyanidiaceae, Galdieria sulphuraria, polyol transport, red alga, sugar transport, transcriptional regulation
Abbreviations: DEPC, diethyl pyrocarbonate; FW, fresh weight; HUP, hexose uptake; (Gs)SPT, (Galdieria sulphuraria) sugar and polyol transporter
INTRODUCTION
Unicellular algae are ideal candidates to study transport processes across the plasma membrane. The hexose uptake machinery [HUP1 (hexose uptake 1), HUP2 and HUP3] of the green alga Chlorella kessleri was the first to be characterized within the green plant lineage and it still serves as a model for conserved hexose transport in higher plants [1–6]. However, sugar transport within the branch of non-green plants is not well characterized. Cyanidiaceae, a group of evolutionary conserved, unicellular acidophilic red algae, have been described as the oldest extant members of the red lineage and are most likely located at the basis of secondary endosymbiosis [7,8], an event that was dated back some 1260±30 million years and gave rise to the highly diverse group of chromalveolates [9]. Following current taxonomy, three genera are classified as Cyanidiaceae: Cyanidium, Cyanidioschyzon and Galdieria [10,11]. These algae have a cosmopolitan distribution but are restricted to habitats with pH values between 0.05 and 3 and temperatures not above 56°C. Due to a high sensitivity towards desiccation, cells often grow endolithically and, as a result, are subjected to severe light limitation, hampering autotrophic growth [12,13]. Galdieria sulphuraria can survive periods of light limitation by metabolizing carbohydrates (disaccharides, hexoses, pentoses and deoxysugars), polyols (hexitols, pentiols, tetriols and triols), amino acids and organic acids [14–16]. Many sugars are taken up and metabolized in the D- as well as in the L-configuration [16,17]. This large spectrum of substrates for heterotrophic growth is unique among eukaryotes and, therefore, G. sulphuraria is well suited as a model for sugar transport in the non-green plant lineage, comprising such diverse groups as the cryptomonades, haptophytes, heterokonts, dinoflagellates and apicomplexa [18].
In vivo studies with intact cells indicated at least 14 different sugar and polyol transporters with pronounced pH dependency and a partly overlapping spectrum of substrates [17]. However, up to now, a detailed characterization of the uptake machinery for sugars and polyols in G. sulphuraria has been hampered by a lack of genomic sequence information and thus access to individual transporters. The recent initiation of genome projects for G. sulphuraria [19] (http://genomics.msu.edu/galdieria/index.html) and the obligate autotrophic Cyanidioschyzon merolae [20] (http://merolae.biol.s.u-tokyo.ac.jp/) has now made it possible to identify putative sugar transporters encoded by these red algal genomes. Comparative genomics of the physiologically very different Cyanidiaceae revealed surprisingly few differences. One of them is a large number of putative sugar transporters, probably up to 28, encoded only by G. sulphuraria [21].
In the present paper, we report the cloning of three monosaccharide and polyol transporters of G. sulphuraria. We follow their transcriptional regulation and characterize the functions of these proteins in the heterologous yeast system.
EXPERIMENTAL
Cultivation of algae
G. sulphuraria strain 074G (Mount Lawu, Java, Indonesia; [16]) was cultivated at ambient temperature in inorganic salt medium at pH 2 [22]. Autotrophic cultures were illuminated with white incandescent light (120 μE·m−2·s−1) and supplied with sterile air enriched with 2% CO2 from the bottom of the flask. Heterotrophic cultures were kept in the dark on a rotary shaker (120 rev./min) and supplied with 25 mM organic substrate. Cell growth was monitored by following the attenuance (D) of the culture at 800 nm. For quantification, cells were diluted to give a reading between 0.05 and 0.15.
Extraction of RNA
Cells were harvested by centrifugation at 3000 g (5 min, 4°C) and washed twice with 20 mM Tris/HCl (pH 7.5). The cell pellet was either stored at −20°C or immediately used for extraction. Cells [1 g FW (fresh weight)] were disrupted in 4 ml of lysis buffer [100 mM Tris/HCl, pH 8.6, 2% (w/v) Sarkosyl, 25 mM EDTA, 25 mM EGTA and 100 mM 2-mercaptoethanol] using a bead beater (Biospec. Products, Bartlesville, OK, U.S.A.) operated for 1 min at 4°C. Cell debris and membranes were removed at 35000 g (15 min, 4°C). RNA was isolated as described by Sheen and Bogorad [23]. It was resuspended in a suitable volume of DEPC (diethyl pyrocarbonate)/water, precipitated overnight at 4°C with 0.5 vol. of 6 M LiCl and collected at 14000 g (20 min, 4°C), followed by a second precipitation with 2.5 vol. of 96% (v/v) ethanol (1 h, −20°C) and centrifugation at 14000 g (30 min, 4°C). The pellet was washed twice in 70% ethanol, dried and resuspended in DEPC/water.
Northern-blot analysis
RNA (30 μg) was separated on 1.4% (w/v) agarose/formaldehyde gels and transferred on to Hybond-N+ nylon membrane by capillary blotting. Hybridization was carried out in 0.12 M Na2HPO4 (pH 7.2), 0.25 M NaCl, 7% (w/v) SDS and 50% formamide at 42°C. Filters were washed twice for 15 min in 2×SSC (1×SSC is 0.15 M NaCl/0.015 M sodium citrate) and 0.1% SDS at 65°C and then twice for 20 min in 0.1×SSC and 0.1% SDS at 65°C.
Construction of cDNA libraries
cDNA libraries of G. sulphuraria (strain 074G) were constructed as λ ZAP cDNA libraries following the instructions of the provider (Stratagene, La Jolla, CA, U.S.A.). Autotrophic and heterotrophic (grown in the dark with glucose as carbon source) cells were harvested during exponential growth.
Heterologous expression of transporters in yeast
For heterologous expression in Saccharomyces cerevisiae, the coding sequences of the three sugar and polyol transporters GsSPT1, GsSPT2 and GsSPT4 were cloned into the yeast expression vector pAUR123, under the control of the alcohol dehydrogenase promotor (Takara Bio). Resistance against aureobasidine was used as selective marker for transformands. Transporter GsSPT1 was also cloned as a 5′ deletion construct (omitting the first 444 nucleic acids), termed GsSPT1(Δ1–3). As expression system, we used the sugar uptake-deficient yeast strain EBY.VW4000 (MAT a Δhxt1-17 Δgal2 Δstl1 Δagt1 Δmph2 Δmph3 leu2-3,112 ura3-52 trp1-289 his3-Δ1 MAL2-8c SUC2), kindly provided by Professor E. Boles (Johann Wolfgang Goethe University, Frankfurt/Main, Germany) [24]. Cells were routinely cultivated in yeast minimal medium [YNB (yeast nutrient base)], supplemented with leucine, uracil, tryptophan and histidine, containing 2% maltose as carbon source unless indicated otherwise. Transformation of yeast cells was done as described in [25]. After transformation, cells were plated on to minimal selective medium containing 2% maltose and aureobasidine A (3 μg·ml−1) and incubated for 3–4 days at 30°C. Transformation of colonies was verified by PCR with transporter- and vector-specific primers. Transformed cells were used for complementation studies of yeast growth on minimal selective medium containing 2% sugars and polyols respectively and also for uptake of radiolabelled substrates (see below).
Uptake studies with radiolabelled substrates
Yeast cells were harvested during exponential growth at a D between 0.5 and 0.8 (600 nm), washed twice and resuspended in distilled water to give a final concentration of 100 mg FW·ml−1. Cell suspension (20 μl; 2 mg FW) were mixed with 1 vol. of 100 mM Tris/citrate buffer (pH 5.0). After a 2 min pre-incubation at 30°C, uptake of radiolabelled substrate was initiated by rapid addition to give a final volume of 50 μl. As labelled substrates we used L-[C-6-3H]fucose (85.2 Ci·mmol−1), D-[C-1-3H]ribose (20.0 Ci·mmol−1), L-[C-1-3H]arabinose (3.0 Ci·mmol−1), L-[C-1-3H]rhamnose (5.0 Ci·mmol−1), D-[U-14C]glucose (0.29 Ci·mmol−1), D-[U-14C]fructose (0.30 Ci·mmol−1), D-[U-14C]sorbitol (0.26 Ci·mmol−1) and D-[U-14C]mannitol (0.05 Ci·mmol−1) (Moravek Biochemicals and Radiochemicals, Brea, CA, U.S.A.). The final sugar concentration was 0.1 mM with a specific radioactivity of 0.5 mCi·mmol−1 for fucose, rhamnose, ribose, arabinose and glucose and 0.25 mCi·mmol−1 for fructose, mannose and sorbitol. Incubation was done for 20 s at 30°C unless indicated otherwise. Uptake was stopped by the addition of 1 ml of ice-cold distilled water. Cells were harvested on a glass fibre filter (GE Healthcare, Little Chalfont, Bucks., U.K.) and washed four times with 1 ml of cold distilled water. Radioactivity was monitored using 3 ml of scintillation cocktail and a Beckman liquid-scintillation counter (Beckman Coulter, Fullerton, CA, U.S.A.). Assays were performed in duplicates and repeated at least twice in independent experiments. Untransformed cells were used as a blank. Their intake as well as binding of radioactive material to the surface was subtracted from the samples in all experiments.
RESULTS
Cloning of a novel type of sugar transporter
We have cloned three sugar (monosaccharide) and polyol transporters from the acidophilic red alga G. sulphuraria (strain 074G). They were termed GsSPT1, GsSPT2 and GsSPT4 and their coding sequences were deposited into the NCBI (National Center for Biotechnology Information) GenBank® database as EF166070, EF166071 and EF166072 respectively. SPT1 was isolated from cDNA libraries of autotrophic as well as of glucose-grown heterotrophic cells. SPT2 could only be isolated from cDNA of autotrophic cells, whereas SPT4 was specific for cDNA of glucose-grown cells. Attempts to amplify PCR fragments of SPT2 and SPT4 from the respective other types of cDNA yielded no results, suggesting a sugar-dependent expression of transporters. The cDNA of SPT1 comprises a coding sequence of 1671 bp. cDNA sequences of SPT2 and SPT4 were found to contain stop codons upstream of the first ATG. SPT2 contains three stop codons at −120, −147 and −204 bp, leaving a 1206 bp coding sequence of the transporter. SPT4 contains only one stop codon at −381 bp, resulting in a 1236 bp coding sequence. These stop codons were confirmed by multiple and independent RT (reverse transcriptase)–PCR. The genomic DNA sequences of SPT1, SPT2 and SPT4 were identical with the corresponding cDNA sequences and did not contain introns. SPT1 translates into a mature protein of 557 amino acids, SPT2 into 402 amino acids, and SPT4 into 412 amino acids. A protein alignment indicates an overall identity of 25% for all three transporters. Closest homologues to the algal proteins are myo-inositol transporters from yeast. Secondary structures of the deduced protein sequences were predicted using the public DAS, transmembrane prediction server for putative membrane proteins. Apparently, SPT1 follows the highly conserved structure of sugar transporters, exhibiting 12 transmembrane-spanning domains with a large cytosolic loop in the middle of the protein (predicted 6+6 structure). SPT2 and SPT4 were predicted to consist of only nine transmembrane-spanning domains: three at the N-terminus and six at the C-terminus. They are separated by a cytosolic middle loop, resulting in a 3+6 structure of the transporters. Going back into the putatively non-translated 5′ region of the cDNA sequences of SPT2 and SPT4, we found the three ‘missing’ N-terminal domains present in both sequences. However, as stop codons separate the corresponding nucleotides from the putative start of translation, the proteins have to be regarded as N-terminally deleted. The new N-termini of both 3+6 transporters were predicted on the outer side of the plasma membrane, with 8–10 (SPT2) and 10 (SPT4) amino acids reaching into the periplasmatic space.
Transcriptional regulation of transporters
The fact that SPT2 could only be isolated from cDNA of autotrophic algae and SPT4 only from glucose-grown heterotrophic algae indicated sugar-dependent transcriptional regulation of protein expression. We used Northern blot analysis to investigate this regulation in more detail (Figure 1). Algae were grown autotrophically as well as heterotrophically on hexoses (glucose and fructose), pentoses (xylose and L-arabinose), polyols (mannitol, sorbitol, dulcitol, xylitol, arabitol) and deoxysugars (L-fucose). They were harvested and extracted during early exponential growth. Transporter-specific probes were used for RNA hybridization. SPT1 and SPT2 transcripts were detected in autotrophic as well as in heterotrophic cells. Transcript levels, however, were much higher in heterotrophic cells. The induction of SPT1 was independent of the type of substrate provided for heterotrophic growth, while induction of SPT2 was highly specific for dulcitol. Transcript levels of SPT4 were generally very low. In autotrophic cells, no signal was detected. Under heterotrophic conditions, only cells grown in the presence of glucose and arabitol exhibited a faint signal. We conclude that the regulation of monosaccharide and polyol transporters takes place – at least in part – at the transcriptional level in G. sulphuraria. Transcription of all three transporters was up-regulated under heterotrophic conditions. For SPT2 and SPT4, this induction was substrate-specific, while SPT1 transcription was unspecifically up-regulated in the presence of monosaccharides or polyols.
Figure 1. Northern-blot analysis for GsSPT1, GsSPT2, and GsSPT4.
G. sulphuraria was grown autotrophically or heterotrophically on different substrates, as indicated. RNA was isolated during early exponential growth. Ribosomal RNA was used as loading control for total RNA.
Functional characterization of transporters in the heterologous system
Complementation of yeast cell growth
To determine the functionality and substrate specificity of both, the predicted 6+6 and 3+6 type of transporters, we expressed SPT1, SPT2 and SPT4 in a sugar uptake-deficient yeast strain [24]. We assessed transformands for cell growth on various substrates and determined their uptake capacities for radiolabelled substrates. To evaluate the importance of the first three domains for transport, we deleted the corresponding 148 amino acids from SPT1 to obtain the artificial 3+6 transporter SPT1(Δ1–3).
For complementation of growth, yeast cells were pregrown on maltose before spotting them on to minimal selective plates, supplemented with different carbon sources. We assessed cell growth of transformands on hexoses (glucose, galactose, mannose and fructose), polyols (mannitol, dulcitol and arabitol), pentoses (arabinose and xylose) and deoxysugars (fucose and rhamnose). All carbon sources were used in the D-configuration as yeast is not known to metabolize L-sugars. Heterologous expression of the algal transporters in uptake-deficient yeast did not complement growth on any of the substrates tested (results not shown).
Uptake of radiolabelled substrates
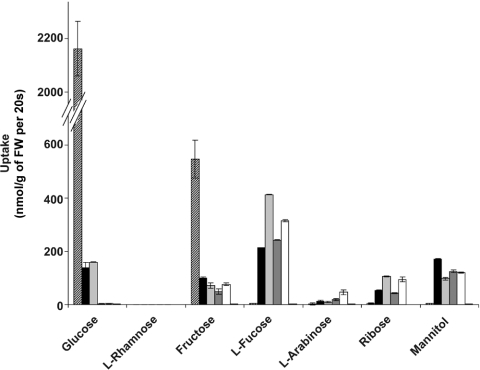
To determine the substrate specificities of transporters independent of cell growth, we followed uptake of radiolabelled substrates in the heterologous expression system. Cells were pregrown on maltose, and subsequently assessed for their uptake capacities. The results are summarized in Figure 2. In contrast with the above results obtained from complementation studies, all three algal transporters, SPT1, SPT2 and SPT4, mediated uptake of hexoses, polyols, pentoses and deoxysugars. Glucose was specifically taken up by SPT1, while fructose transport was also mediated by SPT2 and SPT4. Obviously, the growth defect of yeast transformands on hexoses was not due to a general lack of hexose uptake. We therefore assessed hexose phosphorylation capacities of our yeast strain. We confirmed the presence of glucokinase I, hexokinase I and hexokinase II in the sugar-uptake-deficient host by PCR and by following enzyme activities in crude cell extracts (results not shown). Complementation of yeast growth on glucose and fructose should thus be feasible and the negative results can only be explained if uptake as mediated by the algal transporters was not sufficient to sustain yeast cell growth. For comparison, glucose uptake of wild-type yeast was approx. 15 times higher than in transformands expressing GsSPT1 (Figure 2).
Figure 2. Uptake of radiolabelled substrates by yeast cells expressing transporters from G. sulphuraria.
GsSPT1 (black), GsSPT2 (dark grey), GsSPT4 (white), and the deletion construct GsSPT1(Δ1–3) (light grey) were expressed in the sugar uptake-deficient yeast strain EBY.VW4000. Transformands were assessed for their uptake capacities for radiolabelled substrates. Uptake of cells expressing the empty vector (below resolution, last column position) as well as of wildtype cells (striped) are given as reference. Assays were performed in triplicate.
In contrast with yeast, G. sulphuraria can take up and metabolise a number of substrates in the L-configuration [16,17]. In the heterologous yeast system, uptake of these substrates by SPT1, SPT2 and SPT4 could only be assessed by using radiolabelled compounds. As demonstrated in Figure 2, all three algal transporters mediated high uptake of L-fucose, while another deoxysugar, L-rhamnose, was not taken up. Transport of L-arabinose was very poor and only mediated by SPT4. In general, substrate specificities of SPT1 and its deletion construct SPT1(Δ1–3) were identical, whereas their uptake capacities differed significantly. This effect was most pronounced for L-fucose and D-ribose. Their uptake by SPT1(Δ1–3) was twice that mediated by SPT1. For mannitol, the effect was inverse. Uptake of glucose and fructose was only slightly affected by the deletion (Figure 2). We conclude, that the N-terminus of SPT1 does not determine the transporter's substrate specificity, i.e. it does not take part in substrate recognition, while, on the other hand, it does seem to be involved in determining transport capacities.
Uptake kinetics
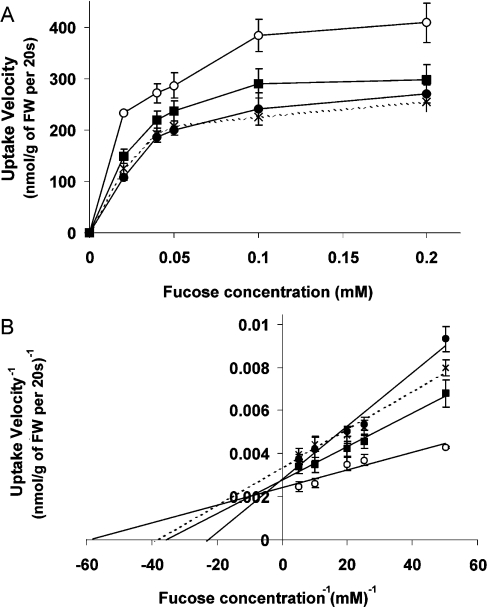
Uptake kinetics were determined for L-fucose, as uptake of this deoxysugar was highest for all transporters (Figure 3). Substrate affinity was highest for SPT1(Δ1–3) (Km=17 μM) and lowest for SPT1 (Km=36 μM), indicating a significant influence of the 148 N-terminal amino acids of SPT1 on substrate binding. SPT2 and SPT4 exhibited similar Km values of 27 and 29 μM respectively. Maximal uptake velocity was highest for SPT1(Δ1–3) (417 nmol/g of FW per 20 s). Apparently, the N-terminal truncation of SPT1 resulted in a reduced sterical hindrance of L-fucose uptake. Vmax values for SPT1 and the predicted 3+6 transporter SPT4 were identical (357 nmol/g of FW per 20 s), that for SPT2 was the lowest (303 nmol/g of FW per 20 s) (Figure 3).
Figure 3. Michaelis–Menten (A) and Lineweaver–Burk (B) plots of uptake kinetics for L-fucose.
The sugar uptake-deficient yeast strain EBY.VW4000, expressing GsSPT1 (closed circles), GsSPT2 (crosses), GsSPT4 (closed squares) and the deletion construct GsSPT1(Δ1–3) (open circles) respectively, was incubated with L-fucose concentrations ranging from 0 to 0.2 mM. Assays were performed in triplicate. Km and Vmax values are given in the text.
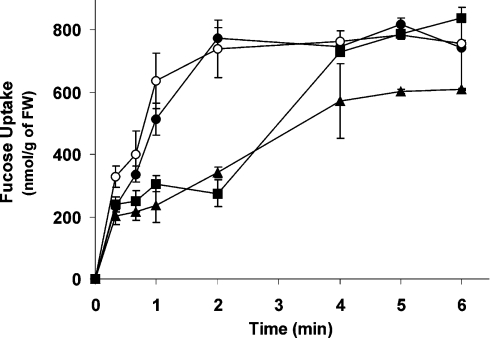
In order to distinguish between active and passive transport, we incubated yeast cells expressing the algal transporters with saturating concentrations of L-fucose (0.1 mM) until a steady state was reached (Figure 4). Intracellular L-fucose concentrations were calculated, estimating an average yeast cell volume of 20 fl and 10×109 cells/g of FW [26,27]. In SPT1 as wells as in SPT1(Δ1–3)-expressing cells, uptake was saturated after 2 min at a level of 750 nmol/g of FW. SPT4-expressing cells reached the same steady state after 4 min, while SPT2-mediated uptake of only 600 nmol/g of FW. Intake of 600–750 nmol of L-fucose/g of FW gives an intracellular concentration of 3.0–3.75 mM, roughly corresponding to a 30–40-fold enrichment as compared with the medium. This intracellular accumulation of sugars has to be based on active transport, and we therefore classify SPT1, SPT2 and SPT4 as active transporters.
Figure 4. Time-dependent uptake of L-fucose.
The sugar uptake-deficient yeast strain EBY.VW4000, expressing GsSPT1 (closed circles), GsSPT2 (closed triangles), GsSPT4 (closed squares) and the deletion construct GsSPT1(Δ1–3) (open circles) respectively, was incubated with saturating L-fucose concentrations (0.1 mM) for up to 6 min. Assays were performed in triplicate.
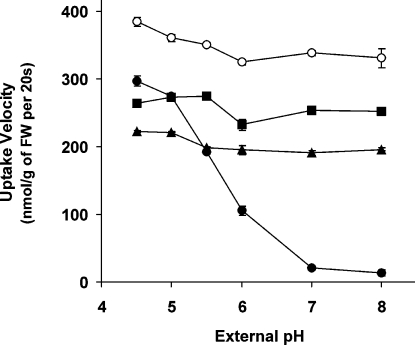
As there are different types of active transporters [e.g. H+ or Na+ co-transporters and ABC transporters (ATP-binding-cassette transporters)], we followed the pH dependency of sugar uptake in yeast (Figure 5). Only SPT1 exhibited a pronounced pH dependency of L-fucose transport with maximal uptake at pH 4.5. The predicted 6+6 transporter is therefore likely to function as an active sugar/H+ symporter. Deletion of the three N-terminal transmembrane-spanning domains resulted in a significant reduction of this pH dependency in SPT1(Δ1–3) expressing cells, while uptake mediated by SPT2 and SPT4 was completely independent of external pH in the range of pH 4.5–8.0. The 3+6 transporters are therefore unlikely to function as H+ symporters and have to rely on alternative energy sources for active transport.
Figure 5. pH-dependent uptake of L-fucose.
The sugar uptake-deficient yeast strain EBY.VW4000, expressing GsSPT1 (closed circles), GsSPT2 (closed triangles), GsSPT4 (closed squares) and the deletion construct GsSPT1(Δ1–3) (open circles) respectively, was incubated with saturating L-fucose concentrations (0.1 mM) at external pH values ranging from pH 4.5 to 8.0. Assays were performed in duplicate.
DISCUSSION
Heterotrophy of G. sulphuraria is mediated by transporters in the plasma membrane, which are specifically encoded in the Galdieria genome, but absent from that of the closely related but obligate autotrophic red alga C. merolae [21]. We have identified two types of transporters in G. sulphuraria – one that follows the conserved structure of 12 transmembrane-spanning domains with a large cytosolic central loop (predicted 6+6 structure), and the other one consisting of nine transmembrane-spanning domains with a cytosolic loop between the third and the fourth hydrophobic domain (predicted 3+6 structure). GsSPT2 and GsSPT4 are two examples of transporters with a putative 3+6 configuration. Their coding regions are significantly shorter than that of GsSPT1, a conserved 6+6 type of transporter, although the cDNA sequences as well as the algal genome still encode the missing N-terminal domains within the putatively non-translated 5′ regions. We suggest that the 3+6 transporters were generated by mutations within the conserved 6+6 type of transporters. The introduction of stop codon(s) has left the resulting proteins with nine instead of 12 transmembrane-spanning domains. Based on molecular clock estimates, Cyanidiaceae are probably more than 1.5 billion years old and thus likely some of the oldest extant eukaryotic organisms [7,8]. This phylogenetic age increases the chance for an accumulation of mutations – as long as they do not impair the cell's viability and evolutionary fitness. From a comparison of SPT1 with its deletion construct SPT1(Δ1–3) (both expressed in the heterologous yeast system), we conclude that loss of the three N-terminal transmembrane-spanning domains does not impair the basic function of active sugar uptake. It has to be kept in mind, though, that uptake specificity as well as kinetics could be affected by heterologous expression. In yeast, both proteins mediated an intracellular accumulation of monosaccharides and polyols and had the same spectrum of substrates. On the other hand, transport kinetics (Km/Vmax) and especially the pH dependency of uptake were significantly altered in SPT1(Δ1–3) expressing cells. H+ symport has been suggested previously as the mechanism for sugar and polyol uptake in Galdieria [17]. Although this still holds true for SPT1, N-terminal truncation released the pH dependency of sugar uptake and therefore appears to significantly impair H+ symport. At the same time, intracellular accumulation of sugars was still possible in SPT1(Δ1–3) expressing yeast cells, and transport therefore had to be active. In line with these results, the predicted 3+6 transporters SPT2 and SPT4 were functional active transporters when expressed in yeast. Their spectrum of substrates did not differ significantly from that of SPT1, yet uptake was pH-independent and therefore unlikely to be mediated by H+ symport. Our attempts to complement the growth defect of an hexose uptake-deficient yeast strain by expressing the algal transporters, however, were not successful. This is quite surprising as we could clearly demonstrate uptake of radiolabelled hexoses by the corresponding yeast transformands. Apparently, hexose uptake rates as mediated by the algal transporters were not sufficient to support yeast cell growth. A second explanation is based on concurrent activity of a sugar exporter in the uptake-deficient yeast strain. EBY.VW4000, deleted in at least 20 transporters [24], still encodes a putative hexose exporter (E. Boles, personal communication). Low hexose uptake by the algal transporters would thus be nullified by this endogenous exporter. We conclude that the inability of the Galdieria transporters, GsSPT1, GsSPT2 and GsSPT4, to complement cell growth of the uptake-deficient yeast strain on hexoses is most probably due to endogenous features of the yeast strain in combination with low uptake rates of the algal transporters in the heterologous system.
With as many as 28 distinct sugar and polyol transporters present in the plasma membrane of Galdieria [21], a highly co-ordinated regulation of their activity has to be applied. Based on in vivo 14C/3H-uptake studies with native cells, we have reported previously a sugar-dependent regulation of transporter activity in the alga [28]. We now corroborate these previous results by Northern blot analyses and conclude that regulation is, at least in part, achieved at the transcriptional level. Transcripts of SPT1 and SPT2 are detectable in autotrophic algae, but are massively up-regulated under heterotrophic conditions. SPT1 was non-specifically induced by all substrates tested in the present study; SPT2 on the other hand was specifically induced by dulcitol. SPT4 transcript was hardly detectable under any of the conditions tested, implying either very high turnover rates or very low transcription of the transporter. For several sugar transporters from higher plants, a stress-related induction has been reported [29,30]. It may well be that SPT4 serves a similar purpose in G. sulphuraria and we did not apply the appropriate conditions for maximal gene induction. Protein phosphorylation as a way of regulating transporter activity [31] was not investigated in the present study, but it should not be excluded as a regulatory level in G. sulphuraria.
The presence of low transcript levels of SPT1 and SPT2 in autotrophic cells is in agreement with previous data indicating marginal sugar uptake already in autotrophic cells [17,28]. Several explanations for this early gene activation come to mind: one is a direct role for SPT1 and SPT2 in sugar sensing, i.e. signalling the presence of extracellular substrates to the cell. Rgt2 and Snf3 are two signalling transporter-homologues known from the yeast system [32,33]. They are no longer active in the uptake of substrates, but are involved in the regulation of the high- and lowaffinity hexose transporters (Hxt1–7) of yeast. Rgt2 and Snf3 exhibit extended cytosolic C-termini which are needed for interaction with the signal transducing proteins Std1 and Mth1 [34,35]. Neither SPT1 nor SPT2 have such putative signalling domains (results not shown). In addition, they proved to be functional transporters when expressed in yeast. A primary role in sugar sensing is therefore unlikely. However, an indirect role of SPT1 and SPT2 in sugar sensing by mediating early uptake of sugars and polyols prior to full activation of the cell's heterotrophic machinery appears possible. In a third model, the presence of SPT1 and SPT2 transcript in autotrophic cells reflects a ‘ready-to-go’ status of the alga. This could speed up the generation of mature transporters upon the arrival of organic carbon.
Until recently, sugar uptake in G. sulphuraria could only be studied in intact cells with an unknown set of transporters obscuring the results. The cloning of SPT1, SPT2 and SPT4 made it possible to assess transporters separately for the first time, to distinguish between active transport and facilitated diffusion, and to determine substrate specificities and uptake kinetics. In the heterologous yeast system, substrate specificities of GsSPT1, GsSPT2 and GsSPT4 were largely redundant, except for glucose, which was only transported by SPT1. These results corroborate previous in vivo studies on intact Galdieria cells, indicating transporters with a broad and partly overlapping spectrum of substrates [17]. However, in contrast with previous models, transporters were not specific for a certain class of substrates, e.g. polyols or hexoses. Furthermore, they mediated transport of substrates in the D-configuration (e.g. fructose) as well as in the L-configuration (e.g. L-fucose). This is in accordance with the spectrum of substrates used for heterotrophic growth of G. sulphuraria [16]. Fucose uptake rates were very high for all three Galdieria transporters, suggesting an important role for this deoxysugar as an in vivo substrate for the alga. Km values ranged from 27 to 36 μM, corresponding to approx. 5 mg of fucose/l. In general, the level of organic compounds is very low at the natural habitat [12,13]. Living in endolithic cell mats, it has been suggested that G. sulphuraria draws substrates from dead cell material of neighbouring Cyanidiaceae and other species. Dissolved organic carbon is highly concentrated by the dense packing of mats and directly available to heterotrophic cells. If fucosylated cell walls should be abundant in these communities, the deoxysugar could be a major source of organic material for Galdieria. N-terminal truncation of conserved 6+6 transporters apparently leads to increased uptake velocity and substrate affinity [as shown for SPT1 and SPT1(Δ1–3)]. Expressing these truncated transporters, cells would thus be able to rapidly deplete organic substrates from the extracellular space.
Comparing the monosaccharide and polyol transporters of G. sulphuraria with those of the green plant line, substrate specificities appear broader in the red alga. The C. kessleri hexose transporters HUP1 and HUP3 are specific for hexoses (glucose, fructose and mannose), while HUP2 takes up glucose, galactose and xylose [2,6]. In higher plants, sugar transporters for glucose, fructose, xylose, arabinose, mannose and galactose [36–40] as well as some polyol-specific transporters [41–45] have been described. To our knowledge, only one transporter from higher plants (Arabidopsis thaliana) exhibits a spectrum of substrates that is comparable with that of G. sulphuraria transporters: AtPLT5 (A. thaliana polyol transporter 5) takes up a large number of polyols in addition to hexoses and pentoses [46]. None of the transporters mentioned has been described as active also after truncation of the three N-terminal transmembrane spans. In this respect, the red algal transporters are unique in the plant kingdom.
Although higher green plants and red algae are only distantly related, our studies on the monosaccharide and polyol transporters of G. sulphuraria will widen the understanding of sugar transport and heterotrophic capacities within the red line of evolution.
Acknowledgments
We thank Professor Dr Andreas Weber for his support, for a critical reading of this paper and for making unpublished sequence data available to us. We are grateful to Professor Dr Eckhard Boles for providing the yeast strain EBY.VW4000, sharing essential strain-related information, and also for commenting on an earlier version of this paper. The present study was funded by the Deutsche Forschungsgemeinschaft (Emmy Noether grant to C. O.).
References
- 1.Sauer N., Tanner W. The hexose carrier from Chlorella cDNA cloning of a eucaryotic H+-cotransporter. FEBS Lett. 1989;259:43–46. doi: 10.1016/0014-5793(89)81489-9. [DOI] [PubMed] [Google Scholar]
- 2.Sauer N., Caspari T., Klebl F., Tanner W. Functional expression of the Chlorella hexose transporter in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. U.S.A. 1990;87:7949–7952. doi: 10.1073/pnas.87.20.7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf K., Tanner W., Sauer N. The Chlorella H+/hexose cotransporter gene. Curr. Genet. 1991;19:215–219. doi: 10.1007/BF00336489. [DOI] [PubMed] [Google Scholar]
- 4.Caspari T., Stadler R., Sauer N., Tanner W. Structure/function relationship of the Chlorella glucose/H+ symporter. J. Biol. Chem. 1994;269:3498–3502. [PubMed] [Google Scholar]
- 5.Caspari T., Will A., Opekarova M., Sauer N., Tanner W. Hexose/H+ symporters in lower and higher plants. J. Exp. Biol. 1994;196:483–491. doi: 10.1242/jeb.196.1.483. [DOI] [PubMed] [Google Scholar]
- 6.Stadler R., Wolf K., Hilgarth C., Tanner W., Sauer N. Subcellular localization of the inducible Chlorella HUP1 monosaccharide-H+ symporter and cloning of a co-induced galactose-H+ symporter. Plant Physiol. 1995;107:33–41. doi: 10.1104/pp.107.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon H., Hackett J., Pinto G., Bhattacharya D. The single, ancient origin of chromist plastids. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15507–15512. doi: 10.1073/pnas.242379899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon H., Hackett J., Ciniglia C., Pinto G., Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya D., Yoon H., Hackett J. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. BioEssays. 2003;26:50–60. doi: 10.1002/bies.10376. [DOI] [PubMed] [Google Scholar]
- 10.Albertano P., Ciniglia C., Pinto G., Pollio A. The taxonomic position of Cyanidium, Cyanidioschyzon and Galdieria: an update. Hydrobiologia. 2000;433:137–143. [Google Scholar]
- 11.Ciniglia C., Yoon H., Pollio A., Pinto G., Bhattacharya D. Hidden biodiversity of the extremophilic Cyanidiales red algae. Mol. Ecol. 2004;13:1827–1838. doi: 10.1111/j.1365-294X.2004.02180.x. [DOI] [PubMed] [Google Scholar]
- 12.Gross W., Küver J., Tischendorf G., Bouchaala N., Büsch W. Cryptoendolithic growth of the red alga Galdieria sulphuraria in volcanic areas. Eur. J. Phycol. 1998;33:25–31. [Google Scholar]
- 13.Gross W., Oesterhelt C. Ecophysiological studies on the red alga Galdieria sulphuraria isolated from South-West Iceland. Plant Biol. 1999;1:694–700. [Google Scholar]
- 14.Rigano C., Fuggi A., DiMartino Rigano V., Aliotta G. Studies on utilization of 2-ketoglutarate, glutamate and other amino acids by the unicellular alga Cyanidium caldarium. Arch. Microbiol. 1976;107:133–138. doi: 10.1007/BF00446832. [DOI] [PubMed] [Google Scholar]
- 15.Rigano C., Aliotta G., Rigano V., Fuggi A., Vona V. Heterotrophic growth patterns in the unicellular alga Cyanidium caldarium. A possible role for threonine dehydrase. Arch. Microbiol. 1977;113:191–196. doi: 10.1007/BF00492024. [DOI] [PubMed] [Google Scholar]
- 16.Gross W., Schnarrenberger C. Heterotrophic growth of two strains of the acido-thermophilic red alga Galdieria sulphuraria. Plant Cell Physiol. 1995;36:633–638. [Google Scholar]
- 17.Oesterhelt C., Schnarrenberger C., Gross W. Characterization of a sugar/polyol-uptake system in the red alga Galdieria sulphuraria. Eur. J. Phycol. 1999;34:271–277. [Google Scholar]
- 18.Archibald J., Keeling P. Recycled plastids: a ‘green movement’ in eukaryotic evolution. Trends Genet. 2002;18:577–584. doi: 10.1016/s0168-9525(02)02777-4. [DOI] [PubMed] [Google Scholar]
- 19.Weber A., Oesterhelt C., Gross W., Braeutigam A., Imboden L., Krassovskaya I., Linka N., Truchina J., Schneidereit J., Voll H., et al. EST-analysis of the thermoacidophilic red microalga Galdieria sulphuraria reveals potential for lipid A biosynthesis and unveils the pathway of carbon export from rhodoplasts. Plant Mol. Biol. 2004;55:17–32. doi: 10.1007/s11103-004-0376-y. [DOI] [PubMed] [Google Scholar]
- 20.Matsuzaki M., Misumi O., Shin-i T., Maruyama S., Takahara M., Miyagishima S., Mori T., Nishida K., Yagisawa F., Nishida K., Kuroiwa T. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 21.Barbier G., Oesterhelt C., Larson M., Halgren R., Wilkerson C., Garavito R., Benning C., Weber A. Comparative genomics of two closely related unicellular thermoacidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of G. sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiol. 2005;137:460–474. doi: 10.1104/pp.104.051169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ford T. W. Ribulose 1,5-bisphosphate carboxylase from the thermophilic, acidophilic alga Cyanidium caldarium Geitler. Purification, characterisation and thermostability of the enzyme. Biochim. Biophys. Acta. 1979;569:239–248. doi: 10.1016/0005-2744(79)90059-7. [DOI] [PubMed] [Google Scholar]
- 23.Sheen J., Bogorad L. Differential expression of the ribulose bisphosphate carboxylase large subunit gene in bundle sheath and mesophyll cells of developing maize leaves is influenced by light. Plant Physiol. 1985;79:1072–1076. doi: 10.1104/pp.79.4.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieczorke R., Krampe S., Weierstall T., Freidel K., Hollenberg C., Boles E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999;464:123–128. doi: 10.1016/s0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- 25.Dohmen R., Strasser A., Höner C., Hollenberg C. An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast. 1991;7:691–692. doi: 10.1002/yea.320070704. [DOI] [PubMed] [Google Scholar]
- 26.Seliber G., Katznelson R. Der Einfluss der Zusammensetzung des Nährbodens auf das Gewicht und den osmotischen Wert der Hefezelle. Protoplasma. 1929;7:204–231. [Google Scholar]
- 27.Tyson C. B., Lord P. G., Wheals A. E. Dependency of size of Saccharomyces cerevisiae cells on growth rate. J. Bacteriol. 1979;138:92–98. doi: 10.1128/jb.138.1.92-98.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oesterhelt C., Gross W. Different sugar kinases are involved in the sugar sensing of Galdieria sulphuraria. Plant Physiol. 2002;128:291–299. [PMC free article] [PubMed] [Google Scholar]
- 29.Truernit E., Schmid J., Epple P., Illig J., Sauer N. The sink-specific and stress-regulated Arabidopsis STP4 gene: enhanced expression of a gene endoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell. 1996;8:2169–2182. doi: 10.1105/tpc.8.12.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer S., Lauterbach C., Niedermeier M., Barth I., Sjolund R. Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiol. 2004;134:684–693. doi: 10.1104/pp.103.033399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roblin G., Sakr S., Bonmort J., Delrot S. Regulation of a plant plasma membrane sucrose transporter by phosphorylation. FEBS Lett. 1998;424:165–168. doi: 10.1016/s0014-5793(98)00165-3. [DOI] [PubMed] [Google Scholar]
- 32.Özcan S., Dover J., Rosenwald A. G., Wölfl S., Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Özcan S., Dover J., Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt M., McCartney R., Zhang X., Tillman T., Solimeo H., Wölfl S., Almonte C., Watkins S. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:4561–4571. doi: 10.1128/mcb.19.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lafuente M., Gancedo C., Jauniaux J., Gancedo J. Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol. Microbiol. 2000;35:161–172. doi: 10.1046/j.1365-2958.2000.01688.x. [DOI] [PubMed] [Google Scholar]
- 36.Reinhold L., Kaplan A. Membrane transport of sugars and amino acids. Annu. Rev. Plant Physiol. 1984;35:45–83. doi: 10.1104/pp.61.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauer N., Baier K., Gahrtz M., Stadler R., Stolz J., Truernit E. Sugar transport across the plasma membranes of higher plants. Plant Mol. Biol. 1994;26:1671–1679. doi: 10.1007/BF00016496. [DOI] [PubMed] [Google Scholar]
- 38.Sherson S., Hemmann G., Wallace G., Forbes S., Germain V., Stadler R., Bechtold N., Sauer N., Smith S. Monosaccharide/proton symporter AtSTP1 plays a major role in uptake and response of Arabidopsis seeds and seedlings to sugars. Plant J. 2000;24:849–857. doi: 10.1046/j.1365-313x.2000.00935.x. [DOI] [PubMed] [Google Scholar]
- 39.Scholz-Starke J., Büttner M., Sauer N. AtSTP6, a new pollen-specific H+ monosaccharide transporter from Arabidopsis. Plant Physiol. 2003;131:70–77. doi: 10.1104/pp.012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneidereit A., Scholz-Starke J., Sauer N., Büttner M. AtSTP11, a pollen tube-specific monosaccharide transporter in Arabidopsis. Planta. 2005;221:48–55. doi: 10.1007/s00425-004-1420-5. [DOI] [PubMed] [Google Scholar]
- 41.Smith S. E., Smith F. A. Uptake of glucose, trehalose and mannitol by leaf slices of the orchid Bletilla hyacinthina. New Phytol. 1973;72:957–964. [Google Scholar]
- 42.Bieleski R. L. Accumulation of sorbitol and glucose by leaf slices of Rosaceae. Aust. J. Plant Physiol. 1977;4:11–24. [Google Scholar]
- 43.Salmon S., Lemoine R., Jamai A., Bouchepillon S., Fromont J. C. Study of sucrose and mannitol transport in plasma-membrane vesicles from phloem and non-phloem tissues of celery (Apium graveolens L.) petioles. Planta. 1995;197:76–83. [Google Scholar]
- 44.Ramsperger-Gleixner M., Geiger D., Hedrich R., Sauer N. Differential expression of sucrose transporter and polyol transporter genes during maturation of common plantain companion cells. Plant Physiol. 2004;124:147–160. doi: 10.1104/pp.103.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider S., Schneidereit A., Konrad K., Hajirezaei M., Gramann M., Hedrich R., Sauer N. Arabidopsis INOSITOL TRANSPORTER4 mediates high-affinity H+ symport of myoinositol across the plasma membrane. Plant Physiol. 2006;141:565–577. doi: 10.1104/pp.106.077123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klepek Y., Geiger D., Stadler R., Klebl F., Landouar-Arsivaud L., Lemoine R., Hedrich R., Sauer N. Arabidopsis POLYOL TRANSPORTER5, a new member of the monosaccharide transporter-like superfamily, mediates H+ symport of numerous substrates, including myo-inositol, glycerol, and ribose. Plant Cell. 2005;17:204–218. doi: 10.1105/tpc.104.026641. [DOI] [PMC free article] [PubMed] [Google Scholar]