Abstract
Molecular engineering of ligand-binding proteins is commonly used for identification of variants that display novel specificities. Using this approach to introduce novel specificities into CBMs (carbohydrate-binding modules) has not been extensively explored. Here, we report the engineering of a CBM, CBM4-2 from the Rhodothermus marinus xylanase Xyn10A, and the identification of the X-2 variant. As compared with the wild-type protein, this engineered module displays higher specificity for the polysaccharide xylan, and a lower preference for binding xylo-oligomers rather than binding the natural decorated polysaccharide. The mode of binding of X-2 differs from other xylan-specific CBMs in that it only has one aromatic residue in the binding site that can make hydrophobic interactions with the sugar rings of the ligand. The evolution of CBM4-2 has thus generated a xylan-binding module with different binding properties to those displayed by CBMs available in Nature.
Keywords: aromatic residue, binding specificity, carbohydrate-binding module, molecular engineering, thermodynamics, xylan
Abbreviations: AE, affinity electrophoresis; CBM, carbohydrate-binding module; HSQC, heteronuclear single quantum coherence spectroscopy; ITC, isothermal titration calorimetry
INTRODUCTION
CBMs (carbohydrate-binding modules) comprise a diverse group of non-catalytic protein modules that are parts of a variety of glycoside-hydrolysing or -modifying enzymes. Their primary function is to increase the enzyme concentration on the substrate and thereby enhance the efficiency of degradation [1,2]. Recent analysis of CBMs from different species has led to the identification and characterization of many new modules and today there are several hundred known sequences classified into 49 sequence-based families that display differences in structure and specificity (http://afmb.cnrs-mrs.fr/CAZY/). Structural studies on CBMs have led to a greater understanding of how these modules recognize their ligands as reviewed recently by Boraston et al. [3]. Based on the topology of their binding site, CBMs are divided into three groups: type A, which has a flat binding surface, type B, which displays a binding cleft, and type C that possesses a solvent-exposed binding pocket or blind canyon [4].
Type B CBMs exhibit a wide range of specificities, recognizing single glycan chains comprising hemicellulose (xylans, mannans, galactans and glucans of mixed linkages) and/or non-crystalline cellulose. Recognition is primarily through stacking/hydrophobic interactions between the sugar rings and aromatic residues in the CBMs [5,6] and by the conformational fitting of the glycan chains in the binding cleft [3]. Xylan-binding properties have been reported for several type B CBMs in families 4 [7], 6 [8], 15 [9] and 22 [10]. One of the family 4 modules, CBM4-2 from Rhodothermus marinus xylanase Xyn10A, binds strongly to both xylan and β-glucan (barley β-glucan) and has significant affinity for laminarin, lichenan [7] and xyloglucan [11]. The solution structure of this module has been resolved [12], providing insight into the unusually broad specificity displayed by CBM4-2. An important feature of CBM4-2 is the location of two aromatic residues that stack with the sugar rings of the ligand, which are at the top of two flexible loops, presumably allowing conformational changes to occur upon binding.
We have previously reported on the molecular engineering of CBM4-2, where a molecular library was created and variants specific for different carbohydrates, as well as a glycoprotein, were successfully identified using a phage-display selection system [11,13]. Characterization studies of the modules selected for binding to unfucosylated xyloglucan or an IgG4 molecule confirmed the isolation of highly specific binders based on the molecular scaffold of CBM4-2 [11,14]. In vitro molecular evolution of this kind, although attractive, has only rarely been employed to introduce novel specificities into CBMs. It appears that the CBM4-2 scaffold is amenable to such engineering strategies which, in addition, has the capacity to unlock important information about the function of specificity-determining residues in both the wild-type and engineered proteins.
In the present study, we have investigated the properties of a module, X-2, selected from the CBM4-2 molecular library for binding to birchwood xylan. This xylan-specific CBM has lost the wild-type affinity for glucan-containing carbohydrates and displays greatly reduced affinity for xylo-oligosaccharides. Intriguingly, one of the aromatic residues that sandwich a xylose residue in the ligand has been replaced by an aliphatic residue. This finding illustrates for the first time that a CBM can bind strongly and specifically to xylan in a mode likely to involve only one aromatic residue that can form the stacking interactions with the ligand.
EXPERIMENTAL
Genes, strains and vectors
Genes encoding the wild-type CBM4-2, a CBM variant selected on xylan, X-2, and a protein-variant, G-4, selected on a human IgG4 molecule (GenBank® accession numbers AY347870, AY534546 and AY534558 respectively), as well as variants of the wild-type protein with single mutations were cloned into the expression vector pET-22b(+) (Novagen, Madison, WI, U.S.A.). Production of soluble recombinant protein fused to a hexahistidine tag at the C-terminus was achieved using Escherichia coli strain BL21(DE3) (Novagen).
Construction of CBM4-2 variants with single mutations
Site-directed mutagenesis was used to construct mutants of the wild-type CBM4-2 as previously described [14]. Point mutations were introduced at position 69 or 110 by replacing the tryptophan and phenylalanine residues with alanine, leucine, histidine, tyrosine and, in the case of residue 69, also phenylalanine residues. PCRs were performed using a set of primers, the AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA, U.S.A.) or DNA polymerase platinum Pfx (Invitrogen, Paisley, Renfrewshire, Scotland, U.K.), and a vector encoding the wild-type CBM4-2 as the template DNA. The PCR products or intermediary cloned products were digested with NdeI and XhoI and inserted into suitably restricted pET-22b(+) vector and transformed into competent E. coli BL21(DE3). All constructs were validated by DNA sequencing.
Protein production and purification
Production of CBM containing a C-terminal hexahistidine tag in E. coli BL21(DE3) and purification of the soluble protein using metal-ion-affinity chromatography have been described earlier [13]. The concentration of each purified protein was determined spectrophotometrically at 280 nm using extinction coefficients individually calculated for each CBM variant [15].
Modelling the structure of X-2
A modelled three-dimensional structure of the X-2 module was obtained using CPHmodels 2.0, which is a homology modelling server (http://www.cbs.dtu.dk/services/CPHmodels/). Databases were searched through and once the appropriate template, the CBM4-2, was found, neural networks and probability density functions were used to predict protein distance constraints.
AE (affinity electrophoresis)
The AE method was performed in the Bio-Rad (Hercules, CA, U.S.A.) mini-gel apparatus as described earlier for the wild-type CBM4-2 [7]. Purified CBM variants (3 μg per gel) were separated on native gels, containing no carbohydrate or different concentrations of oat-spelt xylan, lichenan, laminarin, glucomannan or xyloglucan (all purchased from Sigma–Aldrich, St. Louis, MO, U.S.A.), β-glucan (Megazyme, Bray, Ireland) or birchwood xylan (Roth, Karlsruhe, Germany). The gels were run at room temperature (25 °C) and 90 V. A kaleidoscope prestained standard (Bio-Rad) was included as a control in each gel and the proteins were detected by staining with Coomassie Brilliant Blue or Simply Blue Safe stain (Invitrogen).
Qualitative binding assay
The binding of the X-2 module to insoluble birchwood xylan was assessed as follows: 30 μg of protein in PBS containing 0.05% (v/v) Tween 20 (incubation buffer) was mixed with 6 mg of slurry containing the insoluble part of birchwood xylan (Sigma) after it had been washed in PBS overnight. The total reaction volume was 200 μl and the mixture was incubated on a head-to-tail shaker for 3 h at room temperature. After centrifugation at 15100 g for 1 min, the supernatant containing the unbound protein was removed and the xylan was washed in 200 μl of incubation buffer. The bound protein was then eluted by boiling the xylan in 200 μl of 10% (w/v) SDS containing 10% (v/v) 2-mercaptoethanol for 15 min. The bound, unbound and washed off protein fractions were analysed by SDS/PAGE using 10% (w/v) precast acrylamide gels (NuPAGE; Invitrogen). An IgG4-specific module (G-4) selected from the same library as X-2 was used as a negative control protein in this experiment. Controls containing only protein with no xylan were assayed in parallel to ensure that no precipitation of the proteins occurred during the incubations.
ITC (isothermal titration calorimetry)
ITC measurements were made at 25 °C following standard procedures [16] using a Microcal Omega titration calorimeter (Microcal, Northampton, MA, U.S.A.). Proteins were dialysed, extensively, against 50 mM Hepes/HCl buffer (pH 7.5) containing 1 mM CaCl2, and the ligands [wheat arabinoxylan, barley β-glucan, konjac glucomannan, carob galactomannan, amyloid xyloglucan, Icelandic moss lichenan and xylopentaose (Megazyme), and oat-spelt xylan, birchwood xylan and laminarin (Sigma)] were dissolved in the same buffer to minimize the heat of dilution. During a titration experiment, the protein sample (generally 50 μM), stirred at 300 rev./min in a 1.4331 ml reaction cell, was injected with a single 1 μl aliquot, followed by 24 successive 10 μl aliquots of ligand comprising 0.7 mM xylopentaose or 0.15% xylan at 300 s intervals. Integrated heat effects, after correction for heats of dilution, were analysed by nonlinear regression using a single site binding model (Microcal Origin, version 7.0). The fitted data yield the association constant (KA) and the enthalpy of binding (ΔH). Other thermodynamic parameters were calculated using the standard thermodynamic equation (eqn 1),
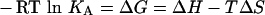 |
(1) |
NMR titrations
All spectra were acquired on a Bruker DRX-500 spectrometer. A sample of uniformly 15N, 13C double-labelled CBM4-2 was prepared at a concentration of 1.2 mM as detailed by Simpson et al. [12]. Titration data for cellohexaose were obtained by a series of eight 15N-HSQC (heteronuclear single quantum coherence spectroscopy) spectra, in which the sugar concentration was successively increased until the protein was saturated. Spectral processing and analysis was carried out using Felix (Accelrys, San Diego, CA, U.S.A.) and in-house macros. A dissociation constant was calculated for the cellohexaose interaction by fitting to the shift changes [17].
RESULTS
Engineering of a type B CBM
In a previous study, we created a molecular library of CBM4-2 by introducing restricted variation in 12 residues within the carbohydrate-binding site of the protein [13]. The amino acids targeted were predicted to play a key role in ligand recognition as NMR studies had shown that these residues undergo large chemical shift changes upon titration of wild-type CBM4-2 with xylopentaose [12] or were located in close proximity to such residues. This module, typical of many other xylan-binding CBMs, displays promiscuous ligand recognition, interacting with both xylo- and gluco-configured saccharides [7]. In order to characterize the residues responsible for interactions with yet another carbohydrate, an NMR study was carried out defining the interaction of wild-type CBM4-2 with cellohexaose.
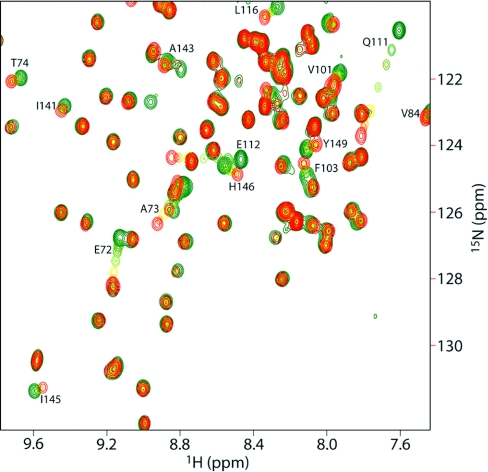
As shown in Figure 1, a number of amino acids have specific chemical shift changes on addition of the oligosaccharide. These residues make up a well-defined cleft on the surface of the protein. Indeed, the binding site is very similar to that previously found for xylopentaose [12]. The largest chemical shift changes (Table 1) demonstrate that the same amino acids are affected by both ligands, with the largest shifts being to the same residues in both cases. In general, cellohexaose affects a smaller set of residues than does xylopentaose, as one might expect from its lower affinity (NMR titrations were well fitted to a cellohexaose affinity of 1.3 mM, compared with 5.1 μM for xylopentaose). The residue showing the clearest difference between xylopentaose and cellohexaose is Glu72, which shows a significantly larger maximum shift for cellohexaose (Table 1).
Figure 1. CBM4-2 binding to cellohexaose.
HSQC titration of wild-type CBM4-2 with cellohexaose. Residues are coloured from green (free) to red (bound), and residues with significant chemical shift changes are labelled.
Table 1. Backbone amide chemical shift changes on addition of ligands to CBM4-2.
| Xylopentaose | Cellohexaose | |||
|---|---|---|---|---|
| Residue | H | N | H | N |
| Gln111*† | 0.29 | 1.22 | 0.21 | 3.17 |
| Arg115* | 0.09 | 1.36 | 0.15 | 0.68 |
| Arg142* | 0.25 | 0.46 | 0.25 | 0.94 |
| Phe110*† | 0.06 | 0.80 | 0.06 | 0.61 |
| Glu112* | 0.10 | 0.48 | 0.38 | 0.07 |
| Glu118* | 0.08 | 0.38 | NS‡ | NS |
| Trp69*† | 0.03 | 0.44 | 0.19 | 0.63 |
| Glu72*† | 0.08 | 0.29 | 0.04 | 1.39 |
| His146* | NS | NS | 0.07 | 0.27 |
| Asp70* | 0.11 | 0.26 | NS | NS |
* Residues targeted by the library design. These residues are listed in order of largest weighted shift change with xylopentaose.
† Residues modified in the X-2 module.
‡ NS, no shift or a value below the cutoff set for a significant shift change.
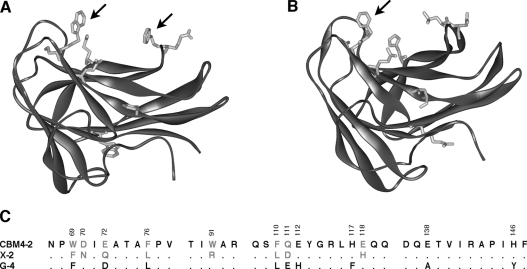
The objective of the herein described molecular engineering work on this scaffold has focused on the construction of a molecular variant that displays substantially increased binding specificity for a single ligand. Despite the rather small size of the library (1.6×106 clones), CBM variants with altered binding specificities were successfully isolated using a phage-display system. One of the selected CBMs was X-2, which was isolated using the insoluble part of birchwood xylan as the selection ligand [13]. Compared with wild-type CBM4-2, this module had eight mutated residues (Figure 2) but since the library mainly included substitutions restricted to related, surface-exposed residues, the overall structure of the CBM is very likely to be similar to native CBM4-2. This view is supported by its CD spectrum that is compatible with a β-sheet-type fold [13] and through the modelling of the structure of X-2 to a reasonable solution (Figure 2B). In common with wild-type CBM4-2, X-2 appears to display a cleft that can easily accommodate single carbohydrate chains.
Figure 2. Differences between CBM4-2 and X-2.
NMR structure of CBM4-2 (1K45.pdb) (A) and a model of X-2 (B) displaying the amino acids mutated in the X-2 module as compared with the wild-type CBM4-2. The arrows show the aromatic residues Trp69 and Phe110 of the wild-type protein and Phe69 of X-2. Differences in the primary sequence between CBM4-2, X-2 and the negative control module G-4 used in some experiments are shown in (C).
The residues mutated in X-2 are widely spread over the binding site (Figure 2) and in addition to six mutations introduced by library design, the X-2 module had two spontaneous mutations, D70N and W91R that were probably introduced into the protein-encoding gene during the PCR-amplification process. Of the eight mutations, all of the affected residues except F76L and W91R shifted during NMR titrations with oligosaccharide ligands (Table 1). Interestingly, the residues affected include the two aromatics that are key to binding saccharide ligands, Trp69 and Phe110, as well as Glu72, which shows the biggest difference in shift between xylopentaose and cellohexaose (Table 1). Thus the evolutionary path taken to generate X-2 involved mutation of residues that are involved in the interaction of CBM4-2 with different carbohydrates.
Binding properties of the engineered X-2 module
Initial tests using phage-displayed X-2 protein [13] showed an increased selectivity for the insoluble xylan as compared with other targets, and consequently we initiated a more complete investigation of the binding properties of this module. Soluble X-2, similar to phage-displayed X-2, displayed binding to insoluble birchwood xylan (Figure 3), which was the selection target for this variant [13]. Screening for binding to a range of ligands, using both AE (results not shown) and ITC (Table 2) techniques, and a comparison with the wild-type CBM4-2 demonstrated that the X-2 module exhibited increased specificity for xylan, binding only to the different xylans among the tested carbohydrates and showing no recognition of gluco-configured polysaccharides. In addition, ITC-based studies revealed that although X-2 had a slight decrease in affinity for xylans (2–9-fold), it had a much larger, 100-fold, decrease in affinity for xylopentaose as compared with the wild-type CBM4-2 (Table 2) and showed no measurable binding to cellopentaose (results not shown). Thus the selection process had been successful in identifying a variant that displayed higher specificity for its natural ligand, xylan, irrespective of whether the backbone of the polysaccharide was relatively unsubstituted (oat-spelt xylan) or highly decorated with arabinofuranose side chains (arabinoxylan).
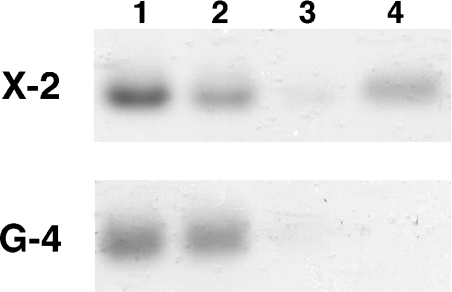
Figure 3. Binding to insoluble xylan.
Qualitative binding of the X-2 and G-4 (a CBM variant specific for human IgG4) modules to insoluble birchwood xylan. The proteins (lane 1) were mixed with the xylan and, after removing the unbound fractions (lane 2), the xylan was washed once with incubation buffer (lane 3) before the bound proteins were eluted by boiling in SDS (lane 4). As expected, the X-2 module had a clear fraction of protein eluted from the xylan, whereas in G-4, which has no affinity for xylan and served as a negative control in this assay, all the protein was present in the incubation supernatant.
Table 2. Affinities and thermodynamic properties of CBM–carbohydrate interactions as determined by ITC.
| Ligand | Protein | KA (M−1) | ΔH (kcal/mol) | TΔS (kcal/mol) | n* |
|---|---|---|---|---|---|
| Xylopentaose | CBM4-2 | 5.13×106 | −19.08±0.06 | −9.92 | 0.95±0.002 |
| X2 | 4.80×104 | −12.65±0.89 | −6.26 | 1.08±0.050 | |
| Birchwood xylan | CBM4-2 | 1.71×105 | −15.21±0.29 | −8.07 | 1.06±0.015 |
| X2 | 8.39×104 | −14.94±1.19 | −8.22 | 1.09±0.066 | |
| Oat-spelt xylan | CBM4-2 | 6.67×105 | −17.18±0.39 | −9.23 | 1.07±0.002 |
| X2 | 1.15×105 | −10.53±1.01 | −3.62 | 0.91±0.060 | |
| Wheat arabinoxylan | CBM4-2 | 1.92×105 | −15.69±0.40 | −8.46 | 1.00±0.019 |
| X2 | 2.19×104 | −18.95±0.71 | −13.02 | 1.01±0.025 | |
| β-Glucan | CBM4-2 | 5.63×104 | −12.69±0.40 | −6.20 | 1.03±0.025 |
| X2 | Weak† | ||||
| Glucomannan | CBM4-2 | 1.65×103 | −16.75±32.4 | −12.34 | 1.02±1.700 |
| X2 | NB‡ | ||||
| Galactomannan | CBM4-2 | NB | |||
| X2 | NB | ||||
| Lichenan | CBM4-2 | Weak | |||
| X2 | NB | ||||
| Laminarin | CBM4-2 | 7.82×103 | −27.14±9.04 | −21.82 | 0.995±0.260 |
| X2 | NB | ||||
| Xyloglucan | CBM4-2 | Weak | |||
| X-2 | NB |
* n is the number of binding sites on the protein.
† Extremely weak binding was detected but too low to quantify.
‡ NB, no binding.
Binding of a CBM to its target is usually driven by enthalpic forces, while entropic effects counteract this interaction [3]. This also holds true for the interaction of CBM4-2 and X-2 with both soluble xylan and xylo-oligosaccharides (Table 2). The thermodynamics revealed that the mutations introduced into the X-2 module had caused relative alternations in enthalpic and entropic contributions to binding most of the targets as compared with the wild-type CBM4-2. The driving forces for binding to the target used in selection of the X-2 variant, birchwood xylan, though, were very similar for both wild-type CBM4-2 and X-2. In conclusion, the evolutionary pathway used to select for X-2 substantially tightened the ligand specificity of the selected CBM and affected the thermodynamics of binding to some xylan variants as compared with wild-type CBM4-2.
Single point mutants of stacking residues in wild-type CBM4-2
The X-2 module is characterized by mutations in both surface-exposed aromatic residues, which in the wild-type protein are believed to make important stacking interactions with the bound ligand. In order to investigate the importance of these residues in carbohydrate recognition, Trp69 and Phe110 in wild-type CBM4-2 were replaced with an alanine, leucine, histidine or tyrosine residue and in position 69 also with a phenylalanine residue. Except for alanine, all these amino acids were allowed by our library design in both positions, and Phe69 and Leu110 were found in the X-2 variant.
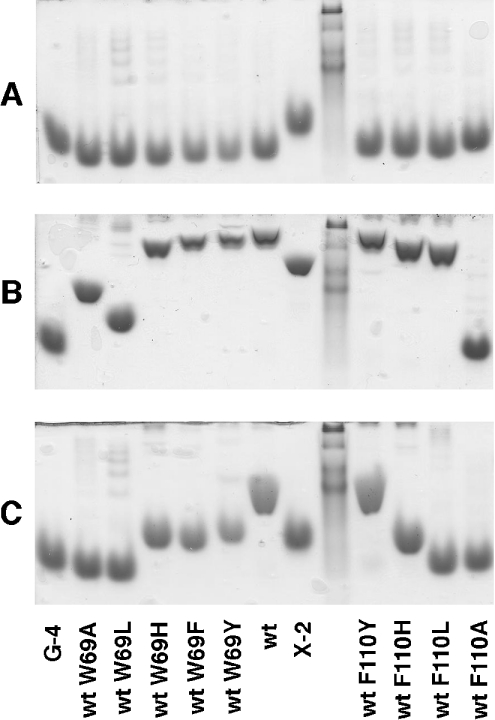
Binding of the mutated CBM variants to both xylan and β-glucan was studied using AE (Figure 4). Mutants that had the tryptophan in position 69 replaced with other aromatic residues or histidine showed some decrease in affinity for both xylan and β-glucan. Alanine and leucine in that same position caused a more substantial decrease in the affinity for xylan and a complete loss of binding to β-glucan, indicating that the character of the residue 69 of CBM4-2 is very important for ligand binding, but the presence of the tryptophan is not an absolute requirement for carbohydrate recognition.
Figure 4. Xylan and β-glucan binding assessed by AE.
Native AE was performed in the absence of ligand (A) or in the presence of oat-spelt xylan (B) or barley β-glucan (C) by using wild-type (wt) CBM4-2 or soluble variants thereof. A kaleidoscope prestained standard was used as a non-interacting migration control.
Replacing the phenylalanine in position 110 had almost the same effects on ligand binding as mutations in Trp69, except for the single mutant F110L, which interestingly still had affinity for xylan but displayed no binding to β-glucan. Since leucine is the amino acid found in position 110 of the X-2 module, these findings strongly suggest that this mutation is one of the factors responsible for the loss of binding to β-glucan.
DISCUSSION
The X-2 module that was evolved in the present study has lost the affinity for glucan-containing polysaccharides displayed by the wild-type CBM4-2 and thus exhibits higher specificity for xylan. Using insoluble xylan as a target during the selection of this module imposed a higher pressure on isolation of a CBM with a higher binding preference for the decorated polysaccharide over the oligosaccharide. This is clearly indicated by the 100-fold decrease in affinity for xylopentaose as compared with the wild-type CBM4-2, whereas the affinity for xylan was only 2–9-fold lower.
Binding of CBM4-2, X-2 and other type B CBMs to xylan is enthalpically driven [5,6,10], indicating that the hydrogen bonds between the residues buried in the binding cleft and the ligand are very important for overall affinity. The lower ΔH of binding suggests the presence of less overall enthalpic forces in the X-2 ligand binding compared with CBM4-2. Two residues that were believed to be involved in hydrogen-bonding of the wild-type protein to xylan, Glu72 and Glu118, [12] were mutated in the X-2 module to glutamine and histidine respectively, and it is possible that these mutations influenced the hydrogen-bonding network between the CBM and ligand.
In addition to hydrogen bonds, hydrophobic interactions between aromatic residues in the binding sites of CBMs and the sugar rings of the ligand play a pivotal role in carbohydrate recognition [5,18–20]. Two such residues, Trp69 and Phe110, are situated on loops edging the binding cleft of CBM4-2 and they are most likely to be involved in ligand binding through stacking interactions with the sugar rings of the target carbohydrate [12]. These residues were both mutated in the X-2 module; the tryptophan was replaced by the aromatic amino acid phenylalanine, while at position 110 the phenylalanine was replaced by leucine, a hydrophobic but non-aromatic amino acid. These mutations are most likely involved in the change in binding specificity of the CBM scaffold and the consequently improved xylan specificity displayed by the X-2 module. Replacing phenylalanine in position 110 with leucine led to the complete loss of binding of wild-type CBM4-2 to β-glucan and had presumably the same effect in X-2. The residue at position 69 was proved to be important for ligand binding of CBM4-2 and although tryptophan is replaced by phenylalanine in the X-2 module, the aromatic character of the substituted residue and thus its capacity to stack against the sugar moieties of the carbohydrate polymer are retained. Phenylalanine in position 69 is thus suggested to be one of the key residues in the interaction of the X-2 module with the xylan chain.
The exact same combination of mutations in positions 69 and 110 (W69F and F110L) were also found in two clones selected from the molecular library of CBM4-2 for binding to an IgG4 molecule [13,14]. One of these modules, G-4, had completely lost its capacity to recognize xylan and all other carbohydrates that the wild-type CBM4-2 was shown to have an affinity for [14]. The affinity for xylan exhibited by X-2 but not by G-4 is thus a result of differences in the sequence between these modules other than at positions 69 and 110 (Figure 2C). Structural studies of the X-2 module alone and in complex with the ligand are needed to identify the residues in this particular module that confer high specificity on xylan.
Similarities in the location of the extended binding cleft and the overall protein fold of members belonging to the families CBM4, CBM6, CBM15 and CBM22 have led to the proposal that these proteins should be grouped into a superfamily [21]. The CBM22 module of Clostridium thermocellum xylanase Xyn10B [20] is a xylan-specific CBM that, similar to wild-type CBM4-2 in the present study and family 6 CBMs, sandwiches a single xylose molecule between two aromatic residues, and this interaction makes a significant contribution to xylan recognition. The mechanism of ligand recognition in this superfamily is not, however, completely conserved. Thus the tryptophan residues in the xylan-binding CBM15 derived from Pseudomonas cellulosa xylanase Xyn10C are located on one side of the binding cleft, leading to formation of stacking interactions to only one face of the ligand chain [9]. This type of binding is also found in the X-2 module engineered in the present study since it has lost the ability of wild-type CBM4-2 to sandwich the xylan chain between two aromatic residues. In X-2 it is only phenylalanine in position 69 that can form the stacking interaction between itself and the ligand. The comparably high affinity (KA=105 M−1 for oat-spelt xylan) of the X-2 module for xylan demonstrates that only one residue, instead of the usual two or three amino acids that make the normally critical stacking interactions with the ligand, is required for binding in this protein. Thus molecular evolution strategies targeting binding sites of this type can successfully explore variants that lack the multiple aromatic residues in the binding site of wild-type CBMs. This substantially extends the possible variation and it may even contribute to the improved specificity profile of X-2, which can be exploited, for example, in analytical applications (L. Filonova, L. Cicortas Gunnarsson, G. Daniel and M. Ohlin, unpublished work).
In summary, we demonstrate the engineering process of a CBM that had broad ligand specificity into a xylan-specific CBM. The evolution of the xylan specificity has not only led to the loss of glucan recognition but has also changed the binding characteristics of the protein compared with other xylan-binding CBMs.
Acknowledgments
We thank Professor Olle Holst and Dr Mats Andersson for fruitful discussions and Linda Dexlin and Kristina Lundberg for their contribution to the site-directed mutagenesis study. The Swedish Research Council (VR) is gratefully acknowledged for financial support.
References
- 1.Bolam D. N., Ciruela A., McQueen-Mason S., Simpson P., Williamson M. P., Rixon J. E., Boraston A., Hazlewood G. P., Gilbert H. J. Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem. J. 1998;331:775–781. doi: 10.1042/bj3310775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilkes N. R., Warren R. A., Miller R. C., Jr, Kilburn D. G. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J. Biol. Chem. 1988;263:10401–10407. [PubMed] [Google Scholar]
- 3.Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boraston A. B., McLean B. W., Kormos J. M., Alam M., Gilkes N. R., Haynes C. A., Tomme P., Kilburn D. G., Warren R. A. J. Carbohydrate binding modules: diversity of structure and function. In: Gilbert H. J., Davies G. J., Svensson B., Henrissat B., editors. Recent Advances in Carbohydrate Engineering. Cambridge, U.K.: Royal Society of Chemistry; 1999. pp. 202–211. [Google Scholar]
- 5.Pell G., Williamson M. P., Walters C., Du H., Gilbert H. J., Bolam D. N. Importance of hydrophobic and polar residues in ligand binding in the family 15 carbohydrate-binding module from Cellovibrio japonicus Xyn10C. Biochemistry. 2003;42:9316–9323. doi: 10.1021/bi0347510. [DOI] [PubMed] [Google Scholar]
- 6.Xie H., Bolam D. N., Nagy T., Szabo L., Cooper A., Simpson P. J., Lakey J. H., Williamson M. P., Gilbert H. J. Role of hydrogen bonding in the interaction between a xylan binding module and xylan. Biochemistry. 2001;40:5700–5707. doi: 10.1021/bi010034z. [DOI] [PubMed] [Google Scholar]
- 7.Abou Hachem M., Nordberg Karlsson E., Bartonek-Roxa E., Raghothama S., Simpson P. J., Gilbert H. J., Williamson M. P., Holst O. Carbohydrate-binding modules from a thermostable Rhodothermus marinus xylanase: cloning, expression and binding studies. Biochem. J. 2000;345:53–60. [PMC free article] [PubMed] [Google Scholar]
- 8.Czjzek M., Bolam D. N., Mosbah A., Allouch J., Fontes C. M., Ferreira L. M., Bornet O., Zamboni V., Darbon H., Smith N. L., et al. The location of the ligand-binding site of carbohydrate-binding modules that have evolved from a common sequence is not conserved. J. Biol. Chem. 2001;276:48580–48587. doi: 10.1074/jbc.M109142200. [DOI] [PubMed] [Google Scholar]
- 9.Szabo L., Jamal S., Xie H., Charnock S. J., Bolam D. N., Gilbert H. J., Davies G. J. Structure of a family 15 carbohydrate-binding module in complex with xylopentaose. Evidence that xylan binds in an approximate 3-fold helical conformation. J. Biol. Chem. 2001;276:49061–49065. doi: 10.1074/jbc.M109558200. [DOI] [PubMed] [Google Scholar]
- 10.Charnock S. J., Bolam D. N., Turkenburg J. P., Gilbert H. J., Ferreira L. M., Davies G. J., Fontes C. M. The X6 ‘thermostabilizing’ domains of xylanases are carbohydrate-binding modules: structure and biochemistry of the Clostridium thermocellum X6b domain. Biochemistry. 2000;39:5013–5021. doi: 10.1021/bi992821q. [DOI] [PubMed] [Google Scholar]
- 11.Cicortas Gunnarsson L., Zhou Q., Montanier C., Nordberg Karlsson E., Brumer H., III, Ohlin M. Engineered xyloglucan specificity in a carbohydrate-binding module. Glycobiology. 2006;16:1171–1180. doi: 10.1093/glycob/cwl038. [DOI] [PubMed] [Google Scholar]
- 12.Simpson P. J., Jamieson S. J., Abou-Hachem M., Nordberg Karlsson E., Gilbert H. J., Holst O., Williamson M. P. The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase. Biochemistry. 2002;41:5712–5719. doi: 10.1021/bi012093i. [DOI] [PubMed] [Google Scholar]
- 13.Cicortas Gunnarsson L., Nordberg Karlsson E., Albrekt A. S., Andersson M., Holst O., Ohlin M. A carbohydrate binding module as a diversity-carrying scaffold. Protein Eng. Des. Sel. 2004;17:213–221. doi: 10.1093/protein/gzh026. [DOI] [PubMed] [Google Scholar]
- 14.Cicortas Gunnarsson L., Dexlin L., Nordberg Karlsson E., Holst O., Ohlin M. Evolution of a carbohydrate binding module into a protein-specific binder. Biomol. Eng. 2006;23:111–117. doi: 10.1016/j.bioeng.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 16.Flint J., Nurizzo D., Harding S. E., Longman E., Davies G. J., Gilbert H. J., Bolam D. N. Ligand-mediated dimerization of a carbohydrate-binding molecule reveals a novel mechanism for protein–carbohydrate recognition. J. Mol. Biol. 2004;337:417–426. doi: 10.1016/j.jmb.2003.12.081. [DOI] [PubMed] [Google Scholar]
- 17.Charlton A. J., Baxter N. J., Khan M. L., Moir A. J., Haslam E., Davies A. P., Williamson M. P. Polyphenol/peptide binding and precipitation. J. Agric. Food Chem. 2002;50:1593–1601. doi: 10.1021/jf010897z. [DOI] [PubMed] [Google Scholar]
- 18.Nagy T., Simpson P., Williamson M. P., Hazlewood G. P., Gilbert H. J., Orosz L. All three surface tryptophans in Type IIa cellulose binding domains play a pivotal role in binding both soluble and insoluble ligands. FEBS Lett. 1998;429:312–316. doi: 10.1016/s0014-5793(98)00625-5. [DOI] [PubMed] [Google Scholar]
- 19.Ponyi T., Szabo L., Nagy T., Orosz L., Simpson P. J., Williamson M. P., Gilbert H. J. Trp22, Trp24, and Tyr8 play a pivotal role in the binding of the family 10 cellulose-binding module from Pseudomonas xylanase A to insoluble ligands. Biochemistry. 2000;39:985–991. doi: 10.1021/bi9921642. [DOI] [PubMed] [Google Scholar]
- 20.Xie H., Gilbert H. J., Charnock S. J., Davies G. J., Williamson M. P., Simpson P. J., Raghothama S., Fontes C. M., Dias F. M., Ferreira L. M., Bolam D. N. Clostridium thermocellum Xyn10B carbohydrate-binding module 22-2: the role of conserved amino acids in ligand binding. Biochemistry. 2001;40:9167–9176. doi: 10.1021/bi0106742. [DOI] [PubMed] [Google Scholar]
- 21.Sunna A., Gibbs M. D., Bergquist P. L. Identification of novel β-mannan- and β-glucan-binding modules: evidence for a superfamily of carbohydrate-binding modules. Biochem. J. 2001;356:791–798. doi: 10.1042/0264-6021:3560791. [DOI] [PMC free article] [PubMed] [Google Scholar]