Abstract
In a search for genes that modify iron homoeostasis, a gene (1300017J02Rik) was located immediately upstream of the murine TF (transferrin) gene. However, expression of the 1300017J02Rik gene product was not responsive to a number of modulators of iron metabolism. Specifically, expression was not altered in mouse models of iron disorders including mice with deficiencies in the haemochromatosis protein Hfe, the recombination-activating protein, Rag, β2-microglobulin, TF, ceruloplasmin or Hb, or in mice with microcytic anaemia. Additionally, neither lipopolysaccharide nor hypoxia treatment resulted in any significant changes in the 1300017J02Rik expression level. The genomic DNA sequence suggested that the 1300017J02Rik gene product might be a protein equivalent to the pICA {porcine ICA [inhibitor of CA (carbonic anhydrase)]}. The coding region for the murine 1300017J02Rik gene was placed into the pNUT expression vector. Transformed BHK cells (baby-hamster kidney cells) were transfected with this plasmid, resulting in secretion of recombinant mICA (murine ICA) into the tissue culture medium. Following purification to homogeneity, the yield of mICA from the BHK cells was found to be considerably greater (at least 4-fold) than the yield of pICA from a previously reported Pichia pastoris (yeast) expression system. MS showed that the recombinant mICA was a glycoprotein that associated with CA in a 1:1 stoichiometry. Despite its high sequence similarity to TF, titration experiments showed that mICA was unable to bind iron specifically. Although enzymatic assays revealed that mICA was able to inhibit CA, it is unclear if this is its sole or even its major function since, to date, humans and other primates appear to lack functional ICA. Lastly, we note that this member of the TF superfamily is a relatively recent addition resulting from a tandem duplication event.
Keywords: baby-hamster kidney cell (BHK cell), evolution, inhibitor of carbonic anhydrase, iron homoeostasis, transferrin superfamily, melanotransferrin
Abbreviations: BHK cell, baby-hamster kidney cell; CA, carbonic anhydrase; DMEM-F12, Dulbecco's modified Eagle's medium/Ham's F-12; ESI, electrospray ionization; FBS, fetal bovine serum; HRP, horseradish peroxidase; TF, transferrin; hTF, human TF; hTF-NG, non-glycosylated recombinant iron-containing human serum TF; ICA, inhibitor of CA; KLH, keyhole-limpet (Diodora aspera) haemocyanin; LTF, lactotransferrin; mICA, murine ICA; mTF, melanotransferrin; NTA, nitrilotriacetate; Ni-NTA, Ni2+-NTA; oTF, ovotransferrin; ORF, open reading frame; pICA, porcine ICA; PNGase F, peptide N-glycosidase F; QTL, quantitative trait loci; RBC, red blood cell; TOPBP1, topoisomerase (DNA) II-binding protein 1; TFR, TF receptor
INTRODUCTION
CAs (carbonic anhydrases) constitute a ubiquitous family of enzymes found at high concentrations in various tissues, with a particularly high abundance in RBCs (red blood cells) [S. Dutta and D. S. Goodsell (2004) PDB Molecule of the Month: carbonic anhydrase; http://www.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb49_1.html]. CA carries out the biologically critical function of reversible hydration of CO2 to produce bicarbonate ions. Metabolic CO2 is generated as a result of the breakdown of sugars and fats and must be excreted from the body. Although 20% of CO2 is bound to Hb and ∼10% is dissolved directly in plasma, most of the CO2 (∼70%) is converted by CA into bicarbonate within RBCs. Upon reaching the lungs, CA catalyses the conversion of bicarbonate back into CO2, so that the CO2 can be expelled. There are at least three classes of CA that are distinguished by their species of origin: α (mammals), β (plants) and γ (some bacteria). Interestingly, in spite of their identical function and requirement for a zinc atom, CA proteins from different species share little sequence or structural homology, implying that they evolved in a convergent manner. The α class comprises at least 11 isoforms [1]. Common to all of these isoforms is a zinc atom that co-ordinates a water molecule in the active site and breaks it down into a hydroxy group and a proton. The hydroxy group is transferred to CO2 to form bicarbonate. When the reaction runs in reverse, CO2 is regenerated. A specific histidine residue swings towards and away from the zinc in a cyclic manner [1]. In carrying out these reversible reactions, CA has roles in the regulation of pH and in fluid balance in a tissue-specific manner. Other examples of CA activity include participation in acidification of the gastric juice, alkylation of the pancreatic juice, and neutralization of the saliva. Renal failure and glaucoma can be caused by CA malfunctions in the kidney and eye respectively, leading to fluid build up. An autosomal recessive disorder that causes a deficiency in one of the CAII isoforms results in osteopetrosis, kidney acidosis, mental and growth retardation as well as calcification within the brain [2,3]. Clearly, the activity of such an important and multifaceted enzyme as CA must be carefully regulated.
Nearly 10 years ago, the Fierke laboratory identified an ICA (inhibitor of CA) in the serum of pigs [pICA (porcine ICA)] [4]. Fierke and co-workers [5] subsequently suggested that this ICA binds to CA (isoforms I, II and III) that escapes into the plasma (from, for example, RBC lysis) and transports the CA to the liver for degradation (and recovery of the zinc atom) in a manner analogous to the haptoglobin–Hb system. Both ICA and haptoglobin are glycoproteins present at high concentrations (∼1 μM) in the plasma of some species. Furthermore, and perhaps not coincidentally, Hb and CA are the two most abundant proteins in RBCs. Curiously, although CA inhibitory activity has been found in the sera of rats, cats, sheep, dogs and rabbits [4], it has never been detected in the sera of humans or monkeys or any avian species [4–6].
Since pICA has 63% sequence similarity to porcine serum TF (transferrin), it was proposed that it is a member of the growing TF superfamily of proteins. The founding member of the superfamily, serum TF, is synthesized in the liver and secreted into blood plasma where it circulates at a concentration of ∼35 μM. TF comprises two homologous lobes (designated the N-lobe and C-lobe) that function in the transport of iron. Each lobe of TF binds a single Fe(III) ion deep in a cleft formed by two subdomains; the iron is co-ordinated to two tyrosine residues, a histidine residue and an aspartic acid residue. Additionally, an arginine and a threonine residue stabilize a synergistic carbonate anion that is essential to high-affinity iron binding. Iron is acquired from the diet after transport across the gut and is delivered to cells following the binding of diferric TF to specific TFRs (TF receptors) on the cell surface [7]. The complex is then taken up by receptor-mediated endocytosis. Within the endosome (aided by a decrease in pH and the TFR), iron is released and the TF–TFR complex returns to the cell surface where the TF (lacking iron) dissociates to continue the cycle [8]. In spite of identical ligands, the two lobes of hTF (human TF) differ in their affinity for iron and in the effect of the TFR on the rate of iron release [9–13]. The binding sites of family members, oTF (ovotransferrin) and LTF (lactotransferrin), are also the same, although again the affinity for iron differs between individual lobes and between family members [14]. LTF serves as a powerful antimicrobial agent, tightly sequestering iron and withholding it from microbes which require it in order to replicate [15]. The family member oTF is present in avian serum to deliver iron and in hen's-egg white to sequester iron, thus carrying out two distinct roles in the same species [16]. In the past couple of decades, new family members have been identified with functions that are definitely unrelated to iron metabolism. For example, saxiphilin from the bullfrog tightly binds to and transports a neurotoxin called saxitonin [4,16,17]. Unfortunately, in spite of much research effort, the precise function(s) of family members ICA and mTF (melanotransferrin) has not been firmly established. As first identified on melanoma cells, mTF retains a single iron-binding site in the N-lobe [18], is attached to the cell membrane by a glycosylphosphatidylinositol anchor and does not appear to play a significant role in iron transport [19]. Recent studies using mTF−/− mice, in combination with gene array analysis and post-transcriptional gene silencing, have allowed a more quantitative and thorough evaluation of both the distribution and role of mTF in mice [20–22] (see the Discussion section).
In the present study, a search for genes that modify iron homoeostasis led to the identification of a gene immediately telomeric to the gene encoding TF on mouse chromosome 9. The translated sequence was consistent with its putative identification as the mouse equivalent of the pICA. As a result, the 1300017J02Rik gene product was tentatively named mICA (murine ICA). A polyclonal antibody to a 15-amino-acid peptide derived from the mICA sequence was generated and used to determine the mICA tissue distribution and sensitivity to modifiers of iron metabolism, including the mICA gene expression level in various transgenic mice. A hexahistidine-tagged version of the putative mICA was expressed in BHK cells (baby-hamster kidney cells) to allow direct assessment of its iron-binding properties and to evaluate its CA inhibitory activity.
EXPERIMENTAL
Materials
DMEM-F12 (Dulbecco's modified Eagle's medium/Ham's F-12) nutrient mixture and antibiotic/antimycotic solution (100×) were from Gibco BRL-Life Technologies. FBS (fetal bovine serum) was obtained from Atlanta Biologicals. Ultroser G is a serum replacement from BioSepra. Ferrous ammonium sulfate, Protein A–Sepharose and CelLytic-MT lysis buffer were purchased from Sigma. CA derived from bovine RBCs was obtained from MP Biomedicals. PNGase F (peptide N-glycosidase F) was from New England Biolabs. A protease inhibitor cocktail and O-glycosidase were from Roche. KLH [keyhole-limpet (Diodora aspera) haemocyanin] was from Cambridge Research Biochemicals. Corning expanded surface roller bottles were obtained from Fisher Scientific. The EndoFree Plasmid Maxi kit and the Ni-NTA [Ni2+-NTA (nitrilotriacetate)] resin were from Qiagen. HisProbe–HRP (horseradish peroxidase) was from Pierce. The HiPrep Sephacryl S-200HR (26/60) column was from Amersham Biosciences. Methotrexate from Cetus was purchased at a local hospital pharmacy. A Kaleidoscope molecular-mass standard and SDS/4–15% PAGE gels were from Bio-Rad. Anti-mouse TF antibody was a gift from Dr Mark Fleming (Department of Pathology, Children's Hospital and Harvard Medical School, Boston, MA, U.S.A.) [23]. The anti-hTF antibody was obtained from Santa Cruz Biotechnology. Chemiluminescent reagent was from PerkinElmer or Amersham Biosciences. Centricon 30 microconcentrators, YM-30 ultrafiltration membranes and a spiral cartridge concentrator fitted with an S1Y10 cartridge were from Millipore/Amicon. All other chemicals and reagents were of analytical grade.
Mouse models
C57BL/10J mice were originally purchased from the Jackson ImmunoResearch Laboratory and were housed and bred at the animal facility at the Children's Hospital Boston. All mice were maintained under institutionally approved protocols for humane treatment of animals. Hfe−/−, Rag1−/−, β2m−/− (β2-microglobulin-null), Trfhpx/hpx (hypotransferrinaemic), mk (microcytic anaemia) and hbd (Hb deficit) mice used in the present study have been described in detail previously [23–28]. Experimental and control animals of the same age and same sex were used for each comparison.
Preparation of a polyclonal antibody and Western blotting
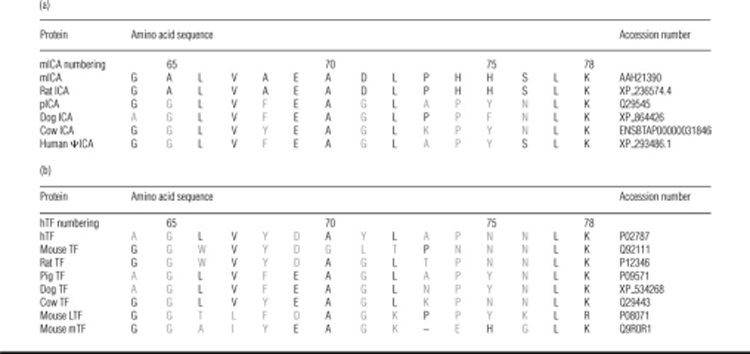
Rabbit antiserum was produced by Covance Research Products against a peptide (GALVAEADLPHHSLK) derived from the predicted mICA sequence (Table 1) and conjugated to KLH. Mouse tissues were harvested from 8-week-old male mice and immediately homogenized in CelLytic-MT mammalian tissue lysis buffer containing protease inhibitors. Loading buffer containing 5 mM dithiothreitol was added to the protein extracts and incubated at 37 °C for 30 min. Samples (50 μg of protein per well) were separated by SDS/PAGE. Similarly, serum was isolated from blood collected from 8-week-old male or female transgenic or wild-type mice and treated as described above for the tissue samples. Volumes equivalent to 0.2 μl of original serum were run on SDS/4–15% PAGE, transferred on to nitrocellulose membranes and probed using either rabbit antiserum (diluted 1:2000) to the mouse peptide sequence or sheep antiserum to mouse TF (1:2000). The blots were then treated with HRP conjugates of a secondary goat antibody to rabbit IgG (1:5000) or a donkey antibody to sheep IgG (1:10000) and visualized by chemiluminescent detection. Enzymatic removal of glycosyl groups was performed by PNGase F (500 units/μl) or O-glycosidase (0.5 m-unit/μl) treatment of mouse serum according to the manufacturer's instructions. Briefly, 2 μl of glycosidase was added to mouse serum (1 μl in 100 μl of 50 mM NaPO4 buffer, pH 7.5, containing 1% Nonidet P40) and incubated overnight at 37 °C. A final sample equivalent to 0.25 μl of the original serum was loaded on to an SDS/PAGE gel prior to immunodetection as described above.
Table 1. Sequence alignment of the peptide from mICA to which the antibody was produced.
Conserved residues are shown in black and non-conserved residues in grey.
 |
Immunoprecipitation
For analysis by MS, blood was obtained from C57BL/10J mice (6–9 weeks old). The serum was diluted (1:100, v/v) into ice-cold lysis buffer [50 mM Tris/HCl, pH 8.0, containing 150 mM NaCl and 1% (v/v) Triton X-100] supplemented with protease inhibitors. The samples were incubated with Protein A–Sepharose (20 μl/ml) at 4 °C for 1 h. After centrifugation at 5000 g for 5 min, the supernatants were removed and incubated with anti-mICA serum (1:1000) at 4 °C overnight. Serum immune complexes were then precipitated by addition of Protein A–Sepharose at 4 °C for 1 h. The immunoprecipitates were washed four times with lysis buffer, taken up in SDS/PAGE sample buffer containing reductants, boiled for 5 min, and run on SDS/8% PAGE. After staining with Bio-Rad Bio-Safe Coomassie Blue stain, bands were cut out of the gel and submitted to the Taplin Biological Mass Spectrometry Facility (Harvard Medical School).
Northern-blot analysis
Mouse liver total RNA was isolated from 8-week-old male Trfhpx/+ and Trfhpx/hpx mice using RNA STAT-60 (ISO-TEX Diagnostics) in accordance with the manufacturer's instructions. Tissue blots were probed using either a 585 bp 32P-labelled fragment containing the mouse mICA ORF (open reading frame; 1–585 bp) or a 587 bp fragment containing the mouse TF ORF (1–87 bp) with mouse β-actin as the loading control.
Preparation of plasmid
The murine homologue of ICA, identified as hypothetical protein LOC71775 [NCBI (National Center for Biotechnology Information) accession number NP_082194] was cloned into the pCMV vector (Stratagene, La Jolla, CA, U.S.A.). For expression in BHK cells, the mICA cDNA was modified to be cloned into the pNUT expression vector [29,30] as follows: (i) the coding region for the native mICA signal peptide was removed and replaced with the signal peptide and the first four residues of hTF to ensure efficient removal of the signal peptide by the signal peptidase and subsequent secretion from the BHK cells; (ii) a hexahistidine tag followed by the Factor Xa recognition sequence, IEGR, was incorporated 5′ of the first codon for Leu20 of mature mICA; and (iii) NotI sites were incorporated into the 5′- and 3′-ends of the construct. The 5′-end of the mICA construct is shown in Table 2.
Table 2. Oligonucleotide sequence of the 5′-end of the mICA construct.
The 5′-end of the mICA construct is shown (mICA residues are denoted with superscript numbers, the reading frame is indicated by spaces, and the NotI site is underlined).
| M | R | L | A | V | G | A | L | L | V | C | A | V | L | G | L | |||
| 5′-GCGGCCGCACC | ATG | AGG | CTC | GCC | GTG | GGA | GCC | CTG | CTG | GTC | TGC | GCC | GTC | CTA | GGG | CTG | ||
| 3′-CGCCGGCGTGG | TAC | TCC | GAG | CGG | CAC | CCT | CGG | GAC | GAC | CAG | ACG | CGG | CAG | GAT | CCC | GAC | ||
| C | L | A | V | P | D | K | H | H | H | H | H | H | I | E | G | R | L20 | P21 |
| TGT | CTG | GCT | GTC | CCT | GAT | AAA | CAT | CAT | CAT | CAT | CAT | CAT | ATC | GAG | GGA | AGG | CTT | CCT-3′ (sense) |
| ACA | GAC | CGA | CAG | GGA | CTA | TTT | GTA | GTA | GTA | GTA | GTA | GTA | TAG | CTC | CCT | TCC | GAA | GGA-5′ (antisense) |
The mICA construct was excised from the pCMV vector via the 5′- and 3′-NotI restriction sites, ligated into the NotI-digested pNUT vector, and propagated in Escherichia coli strain DH5α. The nucleotide sequences of both strands of all constructs were verified by automated DNA sequence analysis using the BigDye Terminator kit and an ABI 3700 DNA sequencer (Applied Biosystems). For transfection into BHK cells, plasmid DNA was purified from DH5α E. coli using an EndoFree Plasmid Maxi kit.
Production and purification of mICA
The transfection, selection and expansion of BHK cells have been described previously [30–32]. Briefly, transfected BHK cells containing the pNUT vector coding for N-terminal hexahistidine-tagged mICA were grown in DMEM-F12 supplemented with 10% (v/v) FBS. At ∼80% confluence, the cells were sequentially transferred to T-150 flasks, and then expanded in surface roller bottles. The cell culture medium was collected (200 ml/roller bottle) at 2–3 day intervals. The first three batches contained DMEM-F12 with 10% FBS to stimulate cell growth and adhesion to the surface and were discarded. Subsequent batches contained the same medium but with the serum substitute, Ultroser G (1%), in place of FBS. Additionally, butyric acid (1 mM) was included in the medium as previous work showed as much as a 65% increase in production of recombinant protein in its presence [33]. The batches were pooled and the His-tagged mICA was purified as described in detail for His-tagged hTF and TFR [33,34]. Briefly, after reduction and exchange using a Millipore spiral wound membrane concentrator fitted with an S1Y10 cartridge and dilution with 5× Qiagen start buffer, the sample in 1× buffer (50 mM Tris/HCl, pH 7.5, containing 300 mM NaCl, 20 mM imidazole, 10% glycerol and 0.05% NaN3) was pumped over a Ni-NTA column (1 cm×10 cm) to specifically isolate the His-tagged mICA. The sample from this column was concentrated to <5 ml and loaded on to a Sephacryl S200HR column (2.6 cm×60 cm) for final purification and complete exchange into 100 mM NH4HCO3. The fractions from the peak were pooled and reduced to the appropriate volume to prepare a 15 mg/ml stock solution. Purity was verified by SDS/10% PAGE.
Western-blot analysis of recombinant mICA
To monitor production of recombinant mICA, an aliquot from each collection (5 μl) was loaded on to an SDS/PAGE gel. Following electrophoretic transfer on to a nitrocellulose membrane, the membrane was washed, blocked with 1% BSA, incubated in a solution containing HisProbe–HRP (a nickel activated derivative of HRP) at 25 °C for 1 h and visualized by chemiluminescent detection.
Determination of molar absorption coefficient (ϵ)
Molar absorption coefficients for CA and recombinant mICA were calculated by the method of Pace et al. [35], which is based on the amino acid composition. For CA, the calculated ϵ280 of a 0.1% solution is 1.74 compared with 1.9 reported previously (i.e. within 8%) [36]; a 0.1% solution of recombinant mICA has an ϵ280 of 1.1. In both cases the calculated estimates were used since they provided a reasonable estimate of the concentrations.
Analysis by MS
Samples of CA, recombinant mICA, recombinant mICA post-iron titration, and the CA–mICA complex were evaluated by ESI (electrospray ionization)–MS under both native and denaturing conditions on a JMS-700 MStation (JEOL) two-sector mass spectrometer equipped with a standard ESI source. Typically, a 10 μM protein solution in 100 mM NH4HCO3 was continuously injected into the source at a flow rate of 3 μl/min. To avoid in-source oxidation of the protein ions, the spray needle potential was kept below 1.9 kVa. The acceleration voltage was maintained at 2.8 kV and the nominal resolution was set at 1000. All spectra were recorded by scanning the magnet at a rate of 5 s/decade. Typically, 80–180 scans were averaged for each spectrum to ensure an adequate signal-to-noise ratio. Protein denaturation was carried out with 50% (v/v) methanol and 3% (v/v) acetic acid.
Iron removal and iron titration
Solutions of hTF-NG (non-glycosylated recombinant iron-containing human serum TF) and recombinant mICA (both with N-terminal hexahistidine tags) were placed in Centricon 30 microconcentrators with 2 ml of iron removal buffer (0.5 M sodium acetate, pH 4.9, containing 1 mM EDTA/1 mM NTA) and reduced to <0.1 ml. Addition of 2 ml of 100 mM sodium perchlorate removed any residual chelator. The samples were washed (four times) with 100 mM KCl and then exchanged (five times) into 10 mM NaHCO3. Apoprotein (∼10 μM) was placed into a stirred cuvette and steady-state tryptophan fluorescence spectra were obtained using a Quantamaster spectrofluorimeter (Photon Technology International, South Brunswick, NJ, U.S.A.) equipped with a 75 W xenon arc lamp excitation source, excitation/emission monochromators, and a WG-320 nm cut-off emission filter. Samples were excited at 280 nm and emission scans were collected (305–400 nm) using slit widths of 0.5 nm (excitation) and 1 nm (emission) respectively. All spectra were corrected for Raman scattering and background fluorescence by subtraction of the appropriate buffer blank. A solution of 5 mM Fe(III)-NTA2 was freshly prepared from stocks of 25 mM ferrous ammonium sulfate in 10 mM HCl and 100 mM disodium NTA. Aliquots of 2 μl of Fe(III)-NTA2 were added to the cuvette and scans were collected 10 min after each addition. Titration continued until there was no further change in the fluorescent signal.
Enzymatic assay of CA and mICA
A protocol from Sigma's Technical Service Department and posted at http://www.sigmaaldrich.com/img/assets/18220/Carbonic_Anhydrase_Titrimetric.pdf was used to measure the activity of the CA. It is an adaptation of one described by Wilbur and Anderson [37]. All solutions were stored at 4 °C until use. Tris sulfate buffer (20 mM) was prepared by addition of Trizma base to 200 ml of water, and adjustment to pH 8.3 with 100 mM sulfuric acid. Solutions of both CA (34.5 μM) and recombinant mICA (12.5 μM) were prepared in water. Prior to beginning the assay, CO2 was bubbled through 100 ml of water at 4 °C in a 150 ml bottle for 1 h (CA substrate). The bottle was capped and left at 4 °C for 20 min prior to use. To conduct the assay, 3 ml of buffer and 2 ml of substrate solution were added to a 6 ml scintillation vial at 4 °C containing a micro stir bar with a hole bored in the cap of the vial large enough to hold a pH meter microelectrode. Immediately after the buffer and substrate were mixed, a pH electrode was inserted into the vial, and the experiment began. The time required for the pH of the mixture to decrease from ∼8.3 to 6.3 was recorded until three trials were within 15 s of each other. Although the protocol indicated that the blank should be in the 70–100 s range, we typically recorded values of 60–70 s. For the enzyme assay, an aliquot of CA solution was added to the buffer/substrate mixture in place of the water (final concentration 38 nM). To test for inhibition, premixed CA–mICA in a 1:1 stoichiometry was added. Additionally, the effect of adding mICA alone was measured.
RESULTS AND DISCUSSION
Identification of putative mICA
Grant et al. [38] undertook a QTL (quantitative trait loci) analysis of two mouse strains, C57BL/6J (low iron) and SWR mice (high iron), due to significant differences in the level of non-haem iron that exist between them. They reported a locus on chromosome 9 that was highly significant (P<0.01) for spleen iron loading. Although gene array and real-time PCR analyses failed to detect any difference in the expression of TF in liver or spleen and there were no measurable differences in the serum level of TF, both iron levels and TF saturation differed in the two mouse strains. Unlike mouse TF, which shows no differences in the coding region in different mouse strains, this upstream gene has a cDNA polymorphism between the strains (107 C>T) corresponding to a change in amino acid 36 from threonine (C57BL/6J, C57BL/10J) to isoleucine (SWR/J). Additionally, there is an increase in the level of mRNA in the spleen of SWR mice compared with C57BL/6J mice [38]. In a search for genes that could play a role in iron homoeostasis in mice, we identified a gene (1300017J02Rik) just upstream of the gene coding for murine TF that had been previously identified, but never characterized. Since the translated protein is 72% similar to the amino acid sequence of the pICA, this gene was proposed to encode the mouse equivalent of pICA [38]. This gene was thus considered as a strong candidate for a mouse spleen iron loading QTL.
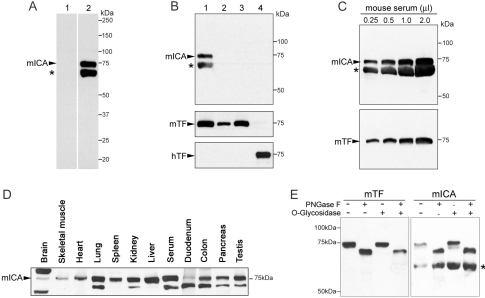
To study the expression of the murine gene, a polyclonal antibody was raised to a peptide from the N-terminal portion of this protein (residues 64–78, hTF numbering) (Table 1). Western blotting of serum from control mice revealed the presence of two bands, one corresponding to the predicted mass of mICA, and another running just below it (Figure 1A). Importantly, this antibody does not cross-react with either mouse or hTF (Figure 1B) as might be predicted from the sequence differences in the peptide to which the antibody was raised (Table 1). Western blots of 50 μg of tissue from 8-week-old mice (Figure 1D) showed substantial amounts of the immunoreactive upper band in lung, spleen, kidney, liver, colon, pancreas and testes with smaller amounts in brain, skeletal muscle, heart and duodenum. Interestingly, immunoreactive material is observed at a higher mass in brain tissue although the significance of this observation is unknown. The apparent wide distribution of this protein is very interesting. In immunohistological studies of pig tissues, pICA was confined to the liver sinusoids and kidney glomeruli, with no evidence of its presence in lung, heart, muscle or eye [5]. The upper band was sensitive to treatment with an N-glycosidase, but not an O-glycosidase, showing that it contains one or more N-linked glycosylation sites (Figure 1E). Following immunoprecipitation and gel electrophoresis, the lower band was identified by MS as albumin (an apparent contaminant probably due to its abundance in plasma). This finding is consistent with early work on oTF, which was originally named conalbumin because it stayed bound to albumin during isolation from hen's-egg white [39,40].
Figure 1. Western-blot analysis of mICA from different mouse tissues.
(A) Lanes 1 and 2 were loaded with 0.5 μl of mouse serum and probed with 1:2000 pre-bleed serum or mICA antiserum respectively. Note that the mICA-positive band is at 75 kDa. (B) Lane 1 contains 0.5 μl of mouse serum, lanes 2 and 3 contain 0.1 or 0.5 μg of mouse TF, and lane 4 contains 0.5 μg of hTF. The upper blot was probed with mICA antiserum; the middle blot was probed with an antibody to mouse TF after stripping the membrane. The bottom blot was reprobed with an antibody to hTF after stripping. (C) Increasing amounts of mouse serum were electrophoresed and probed with mICA antiserum. (D) Tissues (50 μg) and a serum control (0.2 μl) from 8-week-old C57BL/10J male mice were electrophoresed, transferred and probed with mICA antiserum. (E) Mouse sera were treated with PNGase F or O-glycosidase as described in the Experimental section and probed with anti-mouse TF or anti-mICA serum respectively. As confirmed by MS analysis, the lower band (indicated by an asterisk) is albumin.
Level of mICA in mouse models
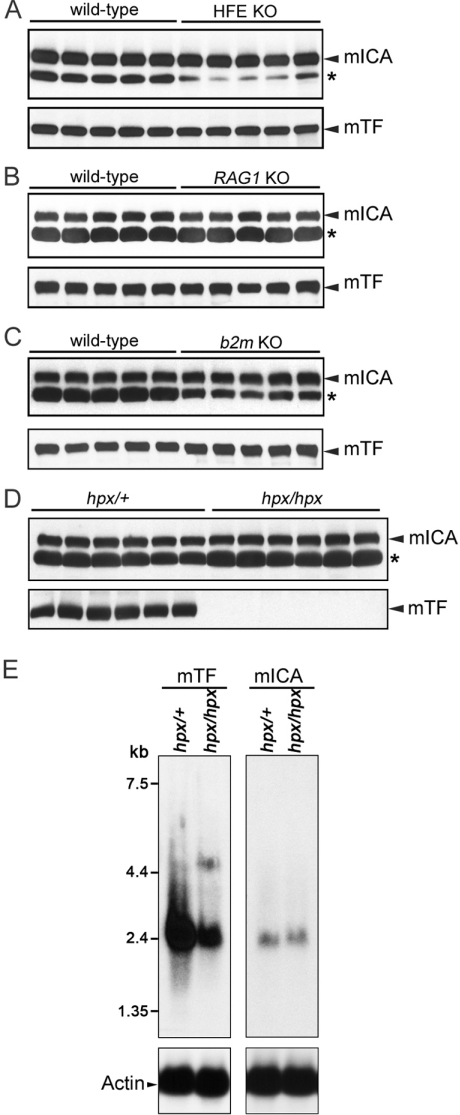
Because of the homology to TF and the proximity of the two genes, the possibility that the putative mICA might play a role in iron homoeostasis was evaluated. A number of transgenic mice including Rag1−/−, β2m−/− and Hfe−/−, accumulate substantial amounts of iron in various tissues. Serum samples were collected from experimental mice carrying these mutations and from age- and sex-matched control mice. Western blotting showed no change in either mICA or mTF from these mice (Figures 2A–2C). Additionally, experimentally induced iron overload (iron dextran for 2 weeks) did not lead to any alteration in the serum level of mICA (results not shown). Likewise, sera from iron-deficient mouse models, including Dmt1mk/mk, Sec1511hbd/hbd and ceruloplasmin-deficient mice revealed no change in mICA (results not shown). Although serum iron concentrations decreased significantly 24 h after injection of 8-week-old female 129S6/SvEvTac wild-type mice with 5 μg of lipopolysaccharide, levels of serum mICA and TF remained unaltered (results not shown).
Figure 2. mICA protein and mRNA expression levels in various mouse strains with mutations in genes involved in iron metabolism.
(A–D) Western blots of serum from control and transgenic mice as indicated. (E) Northern blots of 10 μg of total RNA from Trfhpx/+ or Trfhpx/hpx mice. The blots were probed using either a 587 bp 32P-labelled fragment containing the mouse TF ORF (1–587 bp) (left) or a 585 bp 32P-labelled fragment containing the mouse mICA ORF (1–585 bp) (right). The molecular masses are indicated on the left. After stripping, the blots shown on the bottom were reprobed with the loading control mouse actin.
Trfhpx/hpx mice have a single point mutation in the mTF gene [23] resulting in a circulating level that is ∼1% of normal. The mice are born alive but die from severe anaemia if they are not treated with exogenous TF or transfused with RBCs during the first week after birth. Therefore these mice provide a good model for determining whether mICA has any role in iron regulation. No differences were found in the level of mICA in the serum of the Trfhpx/hpx animals (Figure 2D). Additionally, Northern blots clearly show that, in contrast with the mouse TF mRNA, the level of mRNA for mICA from the liver did not change (Figure 2E). Cumulatively, these studies appear to rule out a role for the mICA gene product in iron regulation. However, since the level of mouse TF varies relatively little in the various mouse models (except in Trfhpx/hpx), a small possibility remained that mICA could influence iron metabolism. To allow further investigation, we used BHK cells to express substantial amounts of recombinant mICA and purified the secreted protein to homogeneity.
Production of mICA
Culture medium (5 μl) from BHK cells secreting the recombinant hexahistidine-tagged putative mICA was probed for the presence of the tag by Western-blot analysis. As expected, each successive collection contained an increasing amount of recombinant mICA (results not shown). Importantly, the recombinant mICA is recognized by the antiserum raised to the 15-amino-acid peptide (Table 1). As little as 30 ng of recombinant protein elicited a strong signal on a Western blot (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/406/bj4060085add.htm). The antibody also gives a robust signal to rat serum, but no specific signal was observed in pig or human serum (as might be predicted from the lack of sequence conservation in the peptide shown in Table 1). Following final purification, the total amount of N-His mICA originally produced was estimated to be ∼60 mg/l in one run and 21 mg/l in a second run. This approximation is based on a final recovery of 60% of the original recombinant mICA [33]. The amount of recombinant mICA produced in the BHK cell expression system is considerably more than the 5 mg/l of pICA achieved using a yeast (Pichia pastoris) expression system [4].
CA–mICA complex formation
Application of a mixture of bovine CA (excess) and mICA to an S200HR column resulted in a chromatogram with two peaks. The profile, and evaluation of the pooled fractions by SDS/PAGE, was consistent with identification of a complex of CA–mICA (in an apparent 1:1 stoichiometry) in the first peak and free CA in the second peak (shown in Supplementary Figure 2 at http://www.BiochemJ.org/bj/406/bj4060085add.htm).
MS analyses
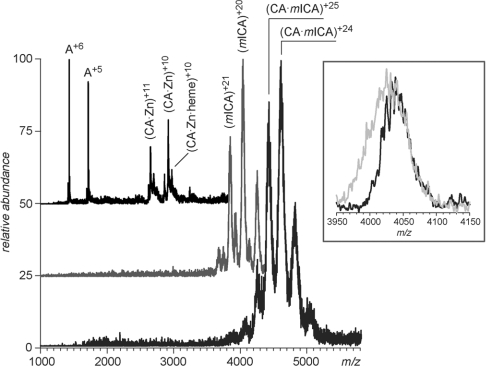
As shown in Table 2, MS analyses of the CA and mICA samples were carried out under both denaturing and native conditions. The experimental masses obtained for CA differed by less than 0.02% from the calculated masses. Three major species were found in the CA samples under near-native conditions. A low-molecular-mass species (8563 Da) indicated as ‘A’ in Table 4 was a contaminant of unknown identity, while two other species were identified as CA bound to a Zn atom, as well as a ternary complex formed by CA, a Zn atom and a haem group (Figure 3, upper trace). We note that the solution of CA had a reddish tinge that was not eliminated by passage over a gel-filtration column. A Soret band at A410 (absorbance; consistent with the presence of haem) was observed in a scan of a solution of CA (results not shown). Since the CA was isolated from bovine RBCs, the presence of a small amount of residual haem is not surprising. Analysis of the recombinant mICA revealed that under denaturing conditions a difference of 4000 Da was found between the experimental and calculated masses (Table 4). This is attributed to the presence of two N-linked carbohydrate moieties. There are two potential linkage sites in the mICA sequence: NQT at position 470–472 and NKT at position 645–647. The ESI–MS analysis of pICA isolated from plasma gave a value of 79036 Da ±13 of which ∼3000 Da is attributed to one N-linked carbohydrate (NQT at position 472–474) [41]. Attempts to detect the presence of Fe-NTA bound to mICA after titration were not conclusive due to extensive adduct formation, which led to noticeable peak broadening and did not allow ionic masses to be measured with the desired degree of accuracy (see inset in Figure 3). Analysis of the CA–mICA complex indicated the presence of both species at 1:1 stoichiometry. The CA–mICA complex remained intact in the gas phase even under conditions that normally result in dissociation of non-covalent complexes (elevated energy of collisions between the complex ions and residual solvent molecules in the ESI interface). This suggests very strong electrostatically driven binding of the two proteins, consistent with the work of Wuebbens et al. [4] in which it was reported that pICA binds to human CA(II) with very high (nanomolar) affinity.
Table 4. Determination of masses by ESI–MS.
| Sample | Solution | Species designation | Measured mass (Da) | Calculated average mass* |
|---|---|---|---|---|
| CA | Denaturing | Species ‘A’† | 8563±5 | |
| CA | 29017±10 | 29024 | ||
| Near-native | Species ‘A’ | 8563±5 | ||
| CA+Zn | 29088±15 | 29089 | ||
| CA+Zn+haem | 29700±15 | 29705 | ||
| Recombinant mICA | Denaturing | mICA | 80450±800 | 76512* |
| Near-native | mICA | 80525±800 | 76512* | |
| Recombinant mICA+CA | Denaturing | CA | 29019±10 | 29024 |
| mICA | 80588±800 | 76512* | ||
| Near-native | ZnCA/mICA | 109988±1500 | 105601* |
*The calculated mass of mICA (residues 1–681) is 74793 Da, to which is added 440 Da for the VPDK sequence, 823 Da for the hexahistidine tag and 456 Da for the Factor Xa cleavage sequence for a total mass of 76512 Da. The difference is attributed to the carbohydrate (see the Results and discussion section).
†Unidentified species found in the sample.
Figure 3. ESI–MS of CA, mICA and mICA–CA.
The spectra represent CA (top trace), mICA (middle trace) and mICA–CA complex (bottom trace) acquired under near-native conditions. The inset shows peak shapes of protein ions desorbed from an Fe-NTA saturated mICA solution in ESI mass spectra acquired under mild (grey) and strong (black) desolvation in the ESI interface.
Iron titrations
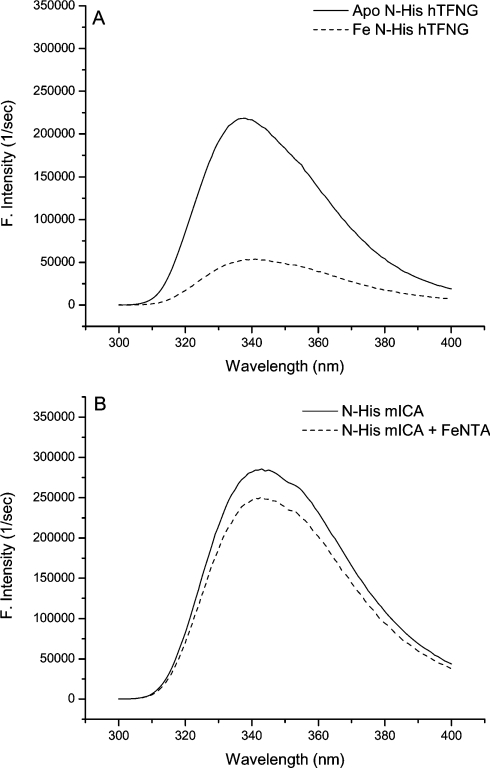
As shown in Figure 4(A), titration of apo-N-His–hTF-NG with Fe(III)-NTA2 leads to significant quenching of the intrinsic tryptophan fluorescent signal (75% reduction). This is ascribed to energy transfer to an absorption band created by metal tyrosine interaction [42,43]. Monitoring by fluorescence provides a sensitive and definitive signature of specific iron binding. As shown in Figure 4(B), addition of an equivalent saturating amount of Fe(III)-NTA2 produced a minimal change in the intrinsic signal (∼12% decrease). These results indicate that although mICA has 58% similarity to murine TF, it does not specifically bind iron. As shown in Table 3, mICA is missing one of the two tyrosine ligands, and also the arginine residue that stabilizes the synergistic carbonate anion is replaced by a tryptophan residue in its N-terminal half. In the C-terminal half of mICA, the equivalent tyrosine ligand is absent in addition to the histidine ligand and the arginine residue. Our previous studies with recombinant hTF clearly demonstrated that the particular tyrosine ligand at this position in each lobe (replaced by serine in mICA) is absolutely essential to iron binding [13,44]. Interestingly, mICA has a greater fluorescent intensity (∼26% higher with a maximum that is 7 nm red-shifted compared with hTF) (Figures 4A and 4B). This is attributed to the extra tryptophan residue at position 124 which is probably relatively solvent-exposed. Assuming that mICA folds in a manner similar to other members of the TF superfamily it would be predicted that the cleft in each lobe would be open since there is no metal binding to keep it closed.
Figure 4. Titration of apo-N-His–hTF-NG and mICA with Fe-NTA monitored by fluorescence.
Apoprotein was excited at 280 nm and scanned (300 and 400 nm). Aliquots of Fe(III)-NTA2 were added until no further change in the signal was observed, i.e. in the case of the hTF sample until the protein was completely iron-saturated. An equivalent amount was added to the ICA sample.
Table 3. Sequence comparison of iron-liganding residues.
The amino acids shown for hTF are conserved in all serum TFs, oTFs and LTFs [17]. Conserved residues are shown in black and non-conserved residues in grey.
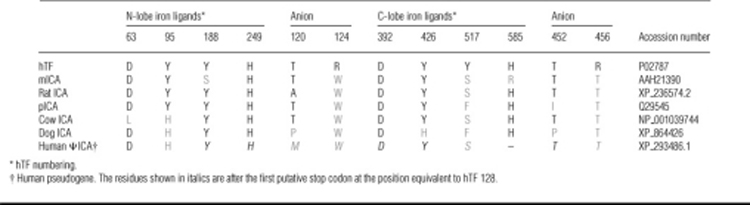 |
Given the extensive sequence differences in the iron- and anion-binding residues of the N- and C-lobes of the other putative ICAs (including the rat, cow and dog sequences), it appears very likely that the ICAs from these species, like mICA, would be unable to bind iron specifically (Table 3 and Supplementary Figure 4 at http://www.BiochemJ.org/bj/406/bj4060085add.htm). A similar conclusion was reached in the case of pICA [4] although it has the most remaining conserved residues (five of six in the N-lobe). Note that the sequence for the human pseudogene (indicated as human ΨICA) has a predicted stop codon following Trp128 (see below).
Enzymatic activity of CA and inhibition by mICA
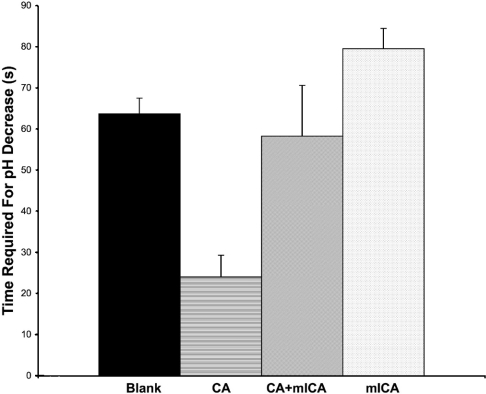
A simple protocol to analyse the activity of CA was modified to measure the ability of the recombinant mICA to inhibit CA. The assay of CA resulted in a significant decrease in the time required for the decrease in pH demonstrating the presence of functional CA (Figure 5). When CA and recombinant mICA were premixed, the time required for the decrease in pH was restored to the value of the blank, indicating that recombinant mICA inhibited the activity of CA. Furthermore, the time for the pH decrease was similar to the blank when mICA was assayed alone, indicating that it had no significant effect on the rate of the pH decrease.
Figure 5. CA inhibition assay.
The time required for the pH of the reaction mixture to decrease from 8.3 to 6.3 is shown. Results are means±S.D. for three trials within 15 s of each other. Addition of CA drastically reduced the time required for the pH decrease compared with the assay blank. Mixing of CA and mICA in a 1:1 stoichiometry before addition to the reaction mixture restored the time to the baseline level. As indicated, mICA alone had no effect.
Evolution of ICA
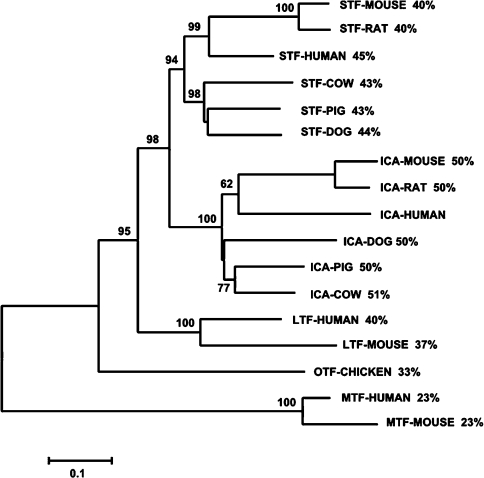
One very intriguing aspect of ICA biology is the apparent absence of a functional transcript in primates (specifically, humans and chimpanzees) or in avian species [4–6]. This is especially interesting considering the abundance of ICA (∼1 μM) in the sera of the species in which it is found. Previously, GenBank® listed a transcript and partial protein sequence at the ICA locus in humans (annotated as XP_293486.1, similar to RIKEN cDNA 1300017J02, now removed). Using GeneWise2 [45] at the European Bioinformatics Institute website, analysis of DNA from the region between TF and TOPBP1 [topoisomerase (DNA) II-binding protein 1] revealed that both the human and chimpanzee genomes contain remnant ICA sequences with multiple stop codons, the first occurring at a position equivalent to Trp128 in the hTF sequence. The partial human sequence is aligned against other TF, ICA and mTF sequences (Supplementary Figure 4). Like the chimpanzee (which is not included), the partial human sequence appears to be more closely related to the ICA sequences than to the other TF family members. As shown in Figure 6, the human sequence clearly segregates with the other ICA sequences. In addition, certain individual amino acids or triads of amino acids are found in the ICA sequences that are not found in other family members. A list of these residues is provided in Supplementary Figure 3 (http://www.BiochemJ.org/bj/406/bj4060085add.htm); a Figure showing placement of these residues on the apo hTF scaffold is also included (Supplementary Figure 3). In the N-lobe there are only two residues that are highly conserved in and unique to ICA. Of interest, both are important in the function of hTF. The first at position 124 is a tryptophan residue instead of the arginine residue which stabilizes the binding of the synergistic anion critical for specific iron binding (see below). The second is a TXA motif in place of KSX found in TF, mTF and LTF. The lysine at position 296 makes up half of a dilysine pair which is essential to efficient iron release from hTF N-lobe [46,47]. In the C-lobe there are ten residues or motifs unique to ICA. Again the arginine residue at position 456 (equivalent to Arg124) binds the synergistic anion; in the C-lobe of ICA it is mutated to a threonine residue. The conservation and location of the YXM and PAH motifs at the lips of the cleft could have functional significance although this is, of course, completely speculative.
Figure 6. Phylogenetic tree of selected TF family member protein sequences.
The consensus tree was constructed with MEGA3.1 using the NJ (neighbour-joining) method, JTT model, complete deletion options [53]. Numbers at the nodes represent the percentage of 2000 bootstrap samples supporting that branch. The numbers to the side of each sequence label represent percentage identity with the predicted human ICA pseudogene sequence. The scale bar indicates an evolutionary distance of 0.1 amino acid substitutions per position.
We believe that serum TF and ICA are the result of a tandem duplication event and that genes for both will probably be found in all eutherian mammals. A map of the relevant area of human chromosome 3 and mouse chromosome 9 is presented in Supplementary Figure 5 (http://www.BiochemJ.org/bj/406/bj4060085add.htm). A search of the newly available genomic databases for dogs and cows shows that they contain genes that encode both serum TF and ICA (Supplementary Figure 4). In each case, the location of the putative ICA gene is found in a similar region to the other ICA orthologues (pig, mouse and rat), lying between the serum TF and the TOPB1 genes. Although the equivalent region is lacking detail in the cow, it is clearly seen in the dog. Unfortunately, Ensembl annotates dog TF and another gene, the SRPRB (signal recognition particle receptor B subunit), as fused, which is clearly not correct.
Function of ICA
On an evolutionary time scale, the ICA sequences appear to be a relatively recent addition to the TF superfamily [16,48]; the ICA gene is not present in marsupials, monotremes, avians or fish [48]. For reasons that are unknown, ICA has become inactive in primates, but more sequences are needed to confirm that this observation is true in all primate species. The loss of a ‘functional’ ICA gene may be an example of the phenomenon termed ‘pseudogenization’, in which loss of function confers some kind of selective advantage [49,50]. The question is whether the ability of ICA to inhibit CA in some species is ‘coincidental’ and/or the only function of this protein [5]. As suggested earlier, ICA could have a transport function assuring efficient recovery of zinc from CA [5]. We note that the search for the function of mTF has been long and difficult. Over the years, many suggestions have been made and refuted (described in detail in [22]). As also seen for the LTF−/− mouse, mTF−/− mice showed no phenotype [21,51]. Only after microarray analysis of the mTF−/− mice, in conjunction with gene silencing in a melanoma cell line, was it possible to suggest that mTF plays a role in proliferation and tumorigenesis (although the mechanism remains obscure) [22]. As with mTF it may be more of a challenge to determine what ICA does than what it does not do. The wide distribution of ICA in mouse tissues (Figure 1D) is of interest. Clearly more studies are needed to clarify its precise role and explain its apparent inactivation in some species including human.
A final curious aspect of the TF/ICA story is that both of these proteins directly or indirectly involve carbonate. In TF, carbonate serves as the synergistic anion that is essential for high-affinity iron binding; in its absence iron becomes hydrolysed and is unable to bind [52]. The carbonate anion in each lobe of TF is stabilized by highly conserved arginine and threonine residues (Table 4). As mentioned above in ICA, the conserved arginine residue is replaced by a tryptophan residue in the N-lobe and threonine in the C-lobe. The tryptophan substitution, in particular, seems unlikely to allow interaction with a carbonate anion further preventing the specific binding of iron in this lobe.
In summary, the focus of the present study was to determine whether the mouse equivalent of pICA has any role in iron homoeostasis either as a modifier of genes involved in iron regulation or by direct binding of iron. Our results show that neither appears to be the case; mICA has no ability to specifically bind iron and protein expression levels do not appear to change as a result of iron status.
Online data
Acknowledgments
We thank Dr Cindy N. Roy and Dr Hiromi Gunshin for providing the transgenic mouse sera and tissues. This work was supported by USPHS (United States Public Health Service) grants R01 DK 21739 (A. B. M.), R01 DK066373 (N. C. A.) and R01 GM061666 and NSF grant CHE-0406302 (I. A. K.) and a grant from the CIHR (Canadian Institutes of Health Research) – Canadian Blood Services Blood Utilization and Conservation Program (to R. T. A. M.). T. A. M. G. was supported in part by a Graduate Fellowship from the Strategic Training Program in Transfusion Science funded by the CIHR and the Heart and Stroke Foundation of Canada.
References
- 1.Tripp B. C., Smith K., Ferry J. G. Carbonic anhydrase: new insights for an ancient enzyme. J. Biol. Chem. 2001;276:48615–48618. doi: 10.1074/jbc.R100045200. [DOI] [PubMed] [Google Scholar]
- 2.Shah G. N., Bonapace G., Hu P. Y., Strisciuglio P., Sly W. S. Carbonic anhydrase II deficiency syndrome (osteopetrosis with renal tubular acidosis and brain calcification): novel mutations in CA2 identified by direct sequencing expand the opportunity for genotype–phenotype correlation. Hum. Mutat. 2004;24:272. doi: 10.1002/humu.9266. [DOI] [PubMed] [Google Scholar]
- 3.McMahon C., Will A., Hu P., Shah G. N., Sly W. S., Smith O. P. Bone marrow transplantation corrects osteopetrosis in the carbonic anhydrase II deficiency syndrome. Blood. 2001;97:1947–1950. doi: 10.1182/blood.v97.7.1947. [DOI] [PubMed] [Google Scholar]
- 4.Wuebbens M. W., Roush E. D., Decastro C. M., Fierke C. A. Cloning, sequencing, and recombinant expression of the porcine inhibitor of carbonic anhydrase: a novel member of the transferrin family. Biochemistry. 1997;36:4327–4336. doi: 10.1021/bi9627424. [DOI] [PubMed] [Google Scholar]
- 5.Ridderstrale Y., Fierke C. A., Roush E. D., Wistrand P. J. Localization of a protein inhibitor of carbonic anhydrase in pig tissues. Acta Physiol. Scand. 2002;176:27–31. doi: 10.1046/j.1365-201X.2002.01010.x. [DOI] [PubMed] [Google Scholar]
- 6.Booth V. H. The carbonic anhydrase inhibitor in serum. J. Physiol. 1938;91:474–489. doi: 10.1113/jphysiol.1938.sp003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung J. Y., Wessling-Resnick M. Molecular mechanisms and regulation of iron transport. Crit. Rev. Clin. Lab. Sci. 2003;40:151–182. doi: 10.1080/713609332. [DOI] [PubMed] [Google Scholar]
- 8.Klausner R. D., Ashwell G., van Renswoude J., Harford J. B., Bridges K. R. Binding of apotransferrin to K562 cells: explanation of the transferrin cycle. Proc. Natl. Acad. Sci. U.S.A. 1983;80:2263–2266. doi: 10.1073/pnas.80.8.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aisen P., Leibman A., Zweier J. Stoichiometric and site characteristics of the binding of iron to human transferrin. J. Biol. Chem. 1978;253:1930–1937. [PubMed] [Google Scholar]
- 10.Bali P. K., Aisen P. Receptor-modulated iron release from transferrin: differential effects on N- and C-terminal sites. Biochemistry. 1991;30:9947–9952. doi: 10.1021/bi00105a019. [DOI] [PubMed] [Google Scholar]
- 11.Zak O., Aisen P. Iron release from transferrin, its C-lobe, and their complexes with transferrin receptor: presence of N-lobe accelerates release from C-lobe at endosomal pH. Biochemistry. 2003;42:12330–12334. doi: 10.1021/bi034991f. [DOI] [PubMed] [Google Scholar]
- 12.Halbrooks P. J., He Q. Y., Briggs S. K., Everse S. J., Smith V. C., MacGillivray R. T. A., Mason A. B. Investigation of the mechanism of iron release from the C-lobe of human serum transferrin: mutational analysis of the role of a pH sensitive triad. Biochemistry. 2003;42:3701–3707. doi: 10.1021/bi027071q. [DOI] [PubMed] [Google Scholar]
- 13.Mason A. B., Halbrooks P. J., James N. G., Connolly S. A., Larouche J. R., Smith V. C., MacGillivray R. T. A., Chasteen N. D. Mutational analysis of C-lobe ligands of human serum transferrin: insights into the mechanism of iron release. Biochemistry. 2005;44:8013–8021. doi: 10.1021/bi050015f. [DOI] [PubMed] [Google Scholar]
- 14.Halbrooks P. J., Giannetti A. M., Klein J. S., Bjorkman P. J., Larouche J. R., Smith V. C., MacGillivray R. T. A., Everse S. J., Mason A. B. Composition of pH sensitive triad in C-lobe of human serum transferrin: comparison to sequences of ovotransferrin and lactoferrin provides insight into functional differences in iron release. Biochemistry. 2005;44:15451–15460. doi: 10.1021/bi0518693. [DOI] [PubMed] [Google Scholar]
- 15.Baker E. N., Baker H. M., Kidd R. D. Lactoferrin and transferrin: functional variations on a common structural framework. Biochem. Cell Biol. 2002;80:27–34. doi: 10.1139/o01-153. [DOI] [PubMed] [Google Scholar]
- 16.Lambert L. A., Perri H., Halbrooks P. J., Mason A. B. Evolution of the transferrin family: conservation of residues associated with iron and anion binding. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2005;142:129–141. doi: 10.1016/j.cbpb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Li Y., Llewellyn L., Moczydlowski E. Biochemical and immunochemical comparison of saxiphilin and transferrin, two structurally related plasma proteins from Rana catesbeiana. Mol. Pharmacol. 1993;44:742–748. [PubMed] [Google Scholar]
- 18.Baker E. N., Baker H. M., Smith C. A., Stebbins M. R., Kahn M., Hellström K. E., Hellström I. Human melanotransferrin (p97) has only one functional iron-binding site. FEBS Lett. 1992;298:215–218. doi: 10.1016/0014-5793(92)80060-t. [DOI] [PubMed] [Google Scholar]
- 19.Food M. R., Sekyere E. O., Richardson D. R. The soluble form of the membrane-bound transferrin homologue, melanotransferrin, inefficiently donates iron to cells via nonspecific internalization and degradation of the protein. Eur. J. Biochem. 2002;269:4435–4445. doi: 10.1046/j.1432-1033.2002.03140.x. [DOI] [PubMed] [Google Scholar]
- 20.Sekyere E. O., Dunn L. L., Richardson D. R. Examination of the distribution of the transferrin homologue, melanotransferrin (tumour antigen p97), in mouse and human. Biochim. Biophys. Acta. 2005;1722:131–142. doi: 10.1016/j.bbagen.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Sekyere E. O., Dunn L. L., Rahmanto Y. S., Richardson D. R. Role of melanotransferrin in iron metabolism: studies using targeted gene disruption in vivo. Blood. 2006;107:2599–2601. doi: 10.1182/blood-2005-10-4174. [DOI] [PubMed] [Google Scholar]
- 22.Dunn L. L., Sekyere E. O., Rahmanto Y. S., Richardson D. R. The function of melanotransferrin: a role in melanoma cell proliferation and tumorigenesis. Carcinogenesis. 2006;27:2157–2169. doi: 10.1093/carcin/bgl045. [DOI] [PubMed] [Google Scholar]
- 23.Trenor C. C., III, Campagna D. R., Sellers V. M., Andrews N. C., Fleming M. D. The molecular defect in hypotransferrinemic mice. Blood. 2000;96:1113–1118. [PubMed] [Google Scholar]
- 24.Levy J. E., Montross L. K., Andrews N. C. Genes that modify the hemochromatosis phenotype in mice. J. Clin. Invest. 2000;105:1209–1216. doi: 10.1172/JCI9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muckenthaler M., Roy C. N., Custodio A. O., Miñana B., DeGraaf J., Montross L. K., Andrews N. C., Hentze M. W. Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat. Genet. 2003;34:102–107. doi: 10.1038/ng1152. [DOI] [PubMed] [Google Scholar]
- 26.Roy C. N., Custodio A. O., de Graaf J., Schneider S., Akpan I., Montross L. K., Sanchez M., Gaudino A., Hentze M. W., Andrews N. C., Muckenthaler M. U. An Hfe-dependent pathway mediates hyposideremia in response to lipopolysaccharide-induced inflammation in mice. Nat. Genet. 2004;36:481–485. doi: 10.1038/ng1350. [DOI] [PubMed] [Google Scholar]
- 27.Fleming M. D., Trenor C. C., Su M. A., Foernzler D., Beier D. R., Dietrich C. F., Andrews N. C. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat. Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 28.Lim J. E., Jin O., Bennett C., Morgan K., Wang F., Trenor C. C., III, Fleming M. D., Andrews N. C. A mutation in Sec15l1 causes anemia in hemoglobin deficit (hbd) mice. Nat. Genet. 2005;37:1270–1273. doi: 10.1038/ng1659. [DOI] [PubMed] [Google Scholar]
- 29.Palmiter R. D., Behringer R. R., Quaife C. J., Maxwell F., Maxwell I. H., Brinster R. L. Cell lineage ablation in transgenic mice by cell-specific expression of a toxic gene. Cell. 1987;50:435–443. doi: 10.1016/0092-8674(87)90497-1. [DOI] [PubMed] [Google Scholar]
- 30.Funk W. D., MacGillivray R. T. A., Mason A. B., Brown S. A., Woodworth R. C. Expression of the amino-terminal half-molecule of human serum transferrin in cultured cells and characterization of the recombinant protein. Biochemistry. 1990;29:1654–1660. doi: 10.1021/bi00458a043. [DOI] [PubMed] [Google Scholar]
- 31.Mason A. B., Miller M. K., Funk W. D., Banfield D. K., Savage K. J., Oliver R. W. A., Green B. N., MacGillivray R. T. A., Woodworth R. C. Expression of glycosylated and nonglycosylated human transferrin in mammalian cells: characterization of the recombinant proteins with comparison to three commercially available transferrins. Biochemistry. 1993;32:5472–5479. doi: 10.1021/bi00071a025. [DOI] [PubMed] [Google Scholar]
- 32.Mason A. B., He Q. Y., Halbrooks P. J., Everse S. J., Gumerov D. R., Kaltashov I. A., Smith V. C., Hewitt J., MacGillivray R. T. A. Differential effect of a His tag at the N- and C-termini: functional studies with recombinant human serum transferrin. Biochemistry. 2002;41:9448–9454. doi: 10.1021/bi025927l. [DOI] [PubMed] [Google Scholar]
- 33.Mason A. B., Halbrooks P. J., Larouche J. R., Briggs S. K., Moffett M. L., Ramsey J. E., Connolly S. A., Smith V. C., MacGillivray R. T. A. Expression, purification, and characterization of authentic monoferric and apo-human serum transferrins. Protein Expression Purif. 2004;36:318–326. doi: 10.1016/j.pep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 34.Byrne S. L., Leverence R., Klein J. S., Giannetti A. M., Smith V. C., MacGillivray R. T. A., Kaltashov I. A., Mason A. B. Effect of glycosylation on the function of a soluble, recombinant form of the transferrin receptor. Biochemistry. 2006;45:6663–6673. doi: 10.1021/bi0600695. [DOI] [PubMed] [Google Scholar]
- 35.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nyman P., Lindskog S. Amino acid composition of various forms of bovine and human erythrocyte carbonic anhydrase. Biochim. Biophys. Acta. 1964;85:141–151. doi: 10.1016/0926-6569(64)90174-9. [DOI] [PubMed] [Google Scholar]
- 37.Wilbur K. M., Anderson N. G. Electrometric and colorimetric determination of carbonic anhydrase. J. Biol. Chem. 1948;176:147–154. [PubMed] [Google Scholar]
- 38.Grant G. R., Robinson S. W., Edwards R. E., Clothier B., Davies R., Judah D. J., Broman K. W., Smith A. G. Multiple polymorphic loci determine basal hepatic and splenic iron status in mice. Hepatology. 2006;44:174–185. doi: 10.1002/hep.21233. [DOI] [PubMed] [Google Scholar]
- 39.Alderton J., Ward W. H., Fevold H. L. Identification of the bacteria inhibiting, iron binding protein of egg white as conalbumin. Arch. Biochem. Biophys. 1946;11:9–13. [PubMed] [Google Scholar]
- 40.Woodworth R. C., Schade A. L. Conalbumin: a rapid, high-yield preparation from egg white. Arch. Biochem. Biophys. 1959;82:78–82. doi: 10.1016/0003-9861(59)90091-8. [DOI] [PubMed] [Google Scholar]
- 41.Roush E. D., Fierke C. A. Purification and characterization of a carbonic anhydrase II inhibitor from porcine plasma. Biochemistry. 1992;31:12536–12542. doi: 10.1021/bi00164a034. [DOI] [PubMed] [Google Scholar]
- 42.Lehrer S. S. Fluorescence and absorption studies of the binding of copper and iron to transferrin. J. Biol. Chem. 1969;244:3613–3617. [PubMed] [Google Scholar]
- 43.Patch M. G., Carrano C. J. The origin of the visible absorption in metal transferrins. Inorg. Chim. Acta. 1981;56:L71–L73. [Google Scholar]
- 44.He Q.-Y., Mason A. B., Woodworth R. C., Tam B. M., MacGillivray R. T. A., Grady J. K., Chasteen N. D. Inequivalence of the two tyrosine ligands in the N-lobe of human serum transferrin. Biochemistry. 1997;36:14853–14860. doi: 10.1021/bi9719556. [DOI] [PubMed] [Google Scholar]
- 45.Birney E., Clamp M., Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dewan J. C., Mikami B., Hirose M., Sacchettini J. C. Structural evidence for a pH-sensitive dilysine trigger in the hen ovotransferrin N-lobe: implications for transferrin iron release. Biochemistry. 1993;32:11963–11968. doi: 10.1021/bi00096a004. [DOI] [PubMed] [Google Scholar]
- 47.He Q.-Y., Mason A. B., Tam B. M., MacGillivray R. T. A., Woodworth R. C. Dual role of Lys206–Lys296 interaction in human transferrin N-lobe: iron-release trigger and anion-binding site. Biochemistry. 1999;38:9704–9711. doi: 10.1021/bi990134t. [DOI] [PubMed] [Google Scholar]
- 48.Lambert L. A., Perri H., Meehan T. J. Evolution of the duplications in the transferrin family of proteins. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2005;140:11–25. doi: 10.1016/j.cbpc.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 49.Olson M. V. When less is more: gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gross L. When less is more: losing genes on the path to becoming human. PLoS Biol. 2006;4:e76. doi: 10.1371/journal.pbio.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward P. P., Mendoza-Meneses M., Cunningham G. A., Conneely O. M. Iron status in mice carrying a targeted disruption of lactoferrin. Mol. Cell. Biol. 2003;23:178–185. doi: 10.1128/MCB.23.1.178-185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlabach M. R., Bates G. W. The synergistic binding of anions and Fe(III) by transferrin. J. Biol. Chem. 1975;250:2182–2188. [PubMed] [Google Scholar]
- 53.Kumar S., Tamura K., Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.