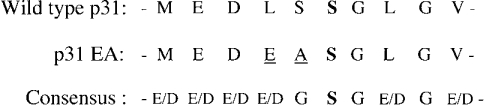Abstract
Targeting of MHCII–invariant chain complexes from the trans-Golgi network to endosomes is mediated by two di-leucine-based signals present in the cytosolic domain of invariant chain. Generation of this endosomal targeting signal is also dependent on multimerization of the invariant chain cytosolic domain sequences, mediated through assembly of invariant chain into homotrimers. A small subset of invariant chain is modified by the addition of chondroitin sulfate and is expressed on the cell surface in association with MHCII. In the present study, we have followed the biosynthetic pathway and route of intracellular transport of this proteoglycan form of invariant chain. We found that the efficiency of chondroitin sulfate modification can be increased by altering the invariant chain amino acid sequence around Ser-201 to the xylosylation consensus sequence. Our results also indicate that, following sulfation, the proteoglycan form is transported rapidly from the trans-Golgi network to the cell surface and is degraded following internalization into an endocytic compartment. Invariant chain–chondroitin sulfate is present in invariant chain trimers that also include conventional non-proteoglycan forms of invariant chain. These data indicate that invariant chain–chondroitin sulfate-containing complexes are transported rapidly from the trans-Golgi network to the cell surface in spite of the presence of an intact endosomal localization signal. Furthermore, these results suggest that invariant chain–chondroitin sulfate may play an important role in the generation of cell-surface pools of invariant chain that can serve as receptors for CD44 and macrophage migration inhibitory factor.
Keywords: chondroitin sulfate, invariant chain (Ii), major histocompatibility complex class II (MHCII), trans-Golgi network (TGN)
Abbreviations: AP, adaptor protein; CS, chondroitin sulfate; DSP, dithiobis(succinimidyl propionate); ER, endoplasmic reticulum; Ii, invariant chain; MIF, macrophage migration inhibitory factor; NF-κB, nuclear factor κB; NP-40, Nonidet P40; TGN, trans-Golgi network
INTRODUCTION
Ii (invariant chain) is a non-polymorphic glycoprotein that participates in a number of immunological functions. The major functions of Ii are mediated through its association with MHCII heterodimers (reviewed in [1]). In the ER (endoplasmic reticulum), newly synthesized Ii self-assembles into a trimer. Three class II heterodimers, each composed of a 32 kDa α- and a 29 kDa β-chain, are sequentially added to the Ii trimer to form a nine-chain complex. In the ER, Ii facilitates MHCII heterodimer assembly and folding and occupies the MHCII-binding groove, preventing premature binding of either peptides or unfolded polypeptides. The nonameric complex transits through the Golgi and is transported to late endosomes/lysosomes, where Ii is degraded, allowing MHCII to be loaded with peptide. Endosomal targeting is mediated by two di-leucine-based signals present in the cytosolic domain of Ii molecules, and mutation of these signals results in expression of MHCII–Ii complexes at the cell surface [2,3]. Thus the best understood function of Ii is to facilitate the assembly, folding and intracellular transport of MHCII molecules.
Ii has also been shown to play a role in B-cell differentiation. Ii-deficient mice exhibit a defect in B-cell development [4], resulting in decreased levels of mature follicular B-cells and increased levels of marginal zone B-cells in the spleen [5,6]. Further studies found that the N-terminal fragment of Ii is liberated into the cytoplasm following sequential endosomal proteolysis of the luminal and intramembrane domains [7]. The resulting N-terminal fragment has been shown to migrate into the nucleus [7], where it stimulates NF-κB (nuclear factor κB) p65/RelA-mediated transcription, resulting in loss of the B-cell maturation block [6,8]. Furthermore, stimulation of surface Ii by MIF (macrophage migration inhibitory factor) binding also appears to induce a signal cascade leading to NF-κB activation, increased expression of Bcl-XL and cell proliferation, suggesting that cell-surface Ii may function as a survival receptor [9].
Finally, Ii has also been shown to function at the cell surface. At steady state, a small pool of Ii can be detected at the cell surface. Some of this cell-surface pool of Ii is modified by the addition of a single glycosaminoglycan side chain [Ii–CS (chondroitin sulfate)]. CS addition is initiated by O-linked xylosylation in the ER/cis-Golgi, and the side chain is elongated and then sulfated during Golgi transport (reviewed in [10]). Ii–CS has been implicated in enhancing T-cell responses through interaction with CD44 [11]. More recently, Ii has been identified as the cell-surface receptor for MIF [12], a cytokine mediator that promotes macrophage activation and is required for expression of pro-inflammatory cytokines [13–15]. MIF can bind directly to the core Ii protein and is thought to utilize CD44 as a co-receptor during signal transduction. The relationship between Ii–CS–CD44 interactions and MIF-mediated signalling through Ii–CD44 has not been examined.
The derivation of the cell-surface pool of Ii is not well understood. It is well established that the majority of Ii is degraded in endosomal/lysosomal compartments shortly after Golgi maturation. Two models have been proposed. The majority of MHCII–Ii may transit briefly through the cell surface on the way from the TGN (trans-Golgi network) to endosomes [16–18]. Alternatively, the bulk of MHCII–Ii complexes may be transported directly from the TGN to endosomes, while a minority of MHCII–Ii may be diverted to the plasma membrane [19]. We have shown previously that multimerization of the cytosolic tail of Ii is required for efficient transport of MHCII–Ii complexes to endosomes [20]. Ii trimers that contained only a single intact cytosolic tail were enriched at the cell surface, suggesting that regulation of Ii trimerization may contribute to the cell-surface pool of Ii. Ii trimerization occurs rapidly after biosynthesis and is mediated through both TM and luminal domain interactions [21,22]. Because Ii–CS is selectively expressed on the cell surface, it raised the possibility that the addition of the CS side chain may disrupt Ii trimerization preventing endosomal localization. Thus it is possible that CS addition may alter MHCII–Ii trafficking, resulting in brief expression of Ii on the surface before internalization.
In the present study, we have examined the biosynthesis and trafficking of Ii–CS. We found that Ii modified by CS addition was rapidly transported from the TGN to the cell surface, then internalized into an endosomal compartment where it was degraded. Ii–CS was present in trimers, indicating that the failure of Ii–CS complexes to trimerize did not account for transport to the cell surface. Interestingly, Ii trimers that contained Ii–CS also contained conventional Ii, suggesting that CS addition on a small subset of Ii may contribute to the cell-surface expression of both conventional and proteoglycan forms of Ii.
MATERIALS AND METHODS
Cell lines and antibodies
Ltk− fibroblast cells were transfected with p31 Ii or p31EA Ii cDNA to generate Lp31 and Lp31EA cell lines. Transfected cells were screened by Western blotting with In-1, specific for the cytosolic domain of Ii [23]. Antisera 657 and 658 were generated by immunizing rabbits with a synthetic peptide corresponding to amino acids 1–29 of the murine Ii cytosolic domain using published protocols [24]. P4H5 is specific for the luminal domain of murine Ii [25], and 16-1-11N is specific for MHCI Kk [26]. Splenocytes were obtained from C3H mice (Jackson).
Site-directed mutagenesis
Amino acids Leu-199 and Ser-200 of p31 Ii were mutated to glutamate and alanine respectively, by overlapping PCR using complementary mutagenic 24-base oligonucleotides, 5′-ATGGAAGACGAAGCTTCTGGCCTG-3′ and 5′-CAGGCCAGAAGCTTCGTCTTCCAT-3′, and flanking oligonucleotides. The PCR product was directionally cloned into a murine p31 cDNA clone to generate pcEXV-p31EA and the constructs were confirmed by dideoxynucleotide sequencing.
Radiolabelling
Cells were metabolically labelled with either [3H]leucine or 35SO4 as described in [11,20]. Cells (2×106 cells/ml) were labelled with [35S]methionine at a concentration of 200 μCi/ml or [3H]leucine at 300 μCi/ml. All biosynthetically labelled cells were pre-labelled for 1–2 h with medium lacking the isotope used, labelled for 10 min to 2 h and chased, where applicable, in medium containing unlabelled amino acids or sulfate [11,27]. To measure surface arrival of Ii–CS, 35SO4-labelled cells were washed three times with cold PBS. Cells were incubated with 0.5 unit/ml chondroitinase ABC (Sigma) in cold PBS for 25 min at 4 °C. The cells were then washed four times with cold PBS to remove chondroitinase, and lysed as described below. In some experiments, all PBS washes and chondroitinase treatments were carried out in the presence of 0.1% NaN3 to prevent internalization of chondroitinase.
Immunoprecipitation
Labelled cells were lysed and pre-cleared, and lysates were immunoprecipitated with the indicated antibody as described previously [20]. Immunoprecipitated protein was eluted, boiled and then separated by SDS/PAGE (10% gels). Gels were then treated with En3Hance™ (New England Nuclear), dried and radiographed as described previously [20]. Immunoprecipitated protein was treated with endoglycosidase H and N-glycosidase (Calbiochem) as described previously [27].
Cross-linking
Metabolically labelled cells were lysed in 0.5% NP-40 (Nonidet P40), 0.13 M NaCl and 0.02 M Bicine (pH 8.2) with or without 200 μg/ml DSP [dithiobis(succinimidyl propionate)] (Pierce) for 30 min at 4 °C. Glycine was added to 0.1 M, and the lysates were incubated at 4 °C for 15 min to quench the reaction. Post-nuclear supernatants were then treated as indicated in each Figure.
Ion exchange
Ii–CS was isolated using DEAE-Sephacel (GE Healthcare) as described previously [28]. Briefly, Ii molecules were isolated by immunoprecipitation with P4H5 as described above. The immunoprecipitates were washed three times with NP-40 wash buffer (0.2% NP-40, 0.15 M NaCl, 0.05 M Tris/HCl and 5 mM EDTA, pH 7.6) and eluted with dissociative buffer (8 M urea, 0.15 M NaCl, 0.05 M sodium acetate, pH 6, and 0.5% NP-40) overnight at room temperature (25 °C). The supernatant was incubated for 15 min at room temperature with DEAE-Sephacel pre-equilibrated in dissociative buffer. The supernatant–DEAE slurry was then loaded on to a 1 ml disposable chromatography column (Bio-Rad) and washed extensively first with dissociative and then with associative (0.15 M NaCl, 0.05 M Tris, pH 6.8, and 0.5% NP-40) buffer. Ii–CS was released from the column with 0.8 M NaCl, 0.05 M Tris/HCl (pH 7.6) and 0.5% NP-40. Column eluate was diluted 10-fold with NP-40 wash buffer and re-precipitated with a combination of In-1, 657 and 658 as described above. These antibodies are conformation-independent.
Gel electrophoresis
Samples were eluted in SDS elution buffer in the absence of the reducing agent, 2-mercaptoethanol, and boiled for 3 min and separated by either SDS/PAGE (10% gels) or SDS/gradient (5–10%) PAGE. The gels were treated with En3Hance, dried and radiographed as described previously [20].
RESULTS
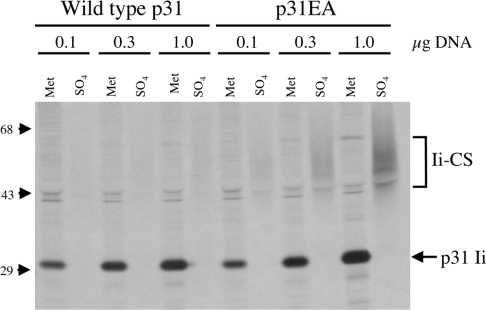
A small amount of total cellular Ii (2–5%) is modified by the addition of the CS glycosaminoglycan side chain at amino acid position 201 [28,29]. When we compared the CS addition site in Ii with the consensus sequence for xylosylation [30–33], we found two significant differences that might account for the paucity of Ii–CS (Figure 1). To determine whether the xylosylation sequence affects the level of Ii–CS expressed, we mutated the Ii cDNA to conform more closely to the consensus sequence. Leu-199 was changed to a glutamate residue to position the two negatively charged amino acids to within a single amino acid of the acceptor serine and the spacer amino acid (Ser-200) was changed to alanine. When Ltk− cells were transiently transfected with increasing concentrations of either wild-type p31 or the mutated Ii construct, p31EA, we found that approx. 3-fold more p31 was modified by CS when compared with wild-type p31 at each concentration of cDNA used (Figure 2). We also found that p31EA increased levels of Ii–CS approx. 3-fold when compared with wild-type p31 in Ltk− transfectants stably expressing either Ii alone or co-expressing MHCII (results not shown). These data suggest that the xylosylation sequence in wild-type Ii is a limiting factor in CS addition to Ii.
Figure 1. Sequence of the xylosylation site in murine Ii.
The wild-type sequence of Ii (amino acids 196–205) is indicated, along with the sequence of the mutated p31EA construct and the consensus xylosylation sequence [43]. The xylose-accepting residue is in bold (Ser-201), and the mutated amino acids are underlined.
Figure 2. Changing the xylosylation site to conform to the consensus sequence results in increased levels of Ii–CS expression.
Ltk− cells were transiently transfected with 0.1, 0.3 or 1.0 μg of cDNA encoding either wildtype p31 or p31EA. The protein expressed by these transfectants was labelled with either [35S]methionine for 10 min or 35SO4 for 30 min. Cell lysate was then immunoprecipitated with P4H5 (anti-Ii) and analysed by SDS/PAGE. The positions of Ii–CS and p31 are indicated on the right, and the positions of the molecular-mass markers are indicated on the left (sizes in kDa).
The increased expression of Ii–CS by the mutated Ii p31EA provides a technical advantage, increasing the ability to detect the otherwise small pool of Ii–CS. We have taken advantage of this increased expression in the biochemical analyses on the trafficking and oligomerization events of Ii–CS included in the present paper. Because this mutation in the xylosyltransferase site could potentially affect Ii structure independently of CS addition, we have verified our results on the assembly and transport of Ii–CS using cells expressing wild-type Ii.
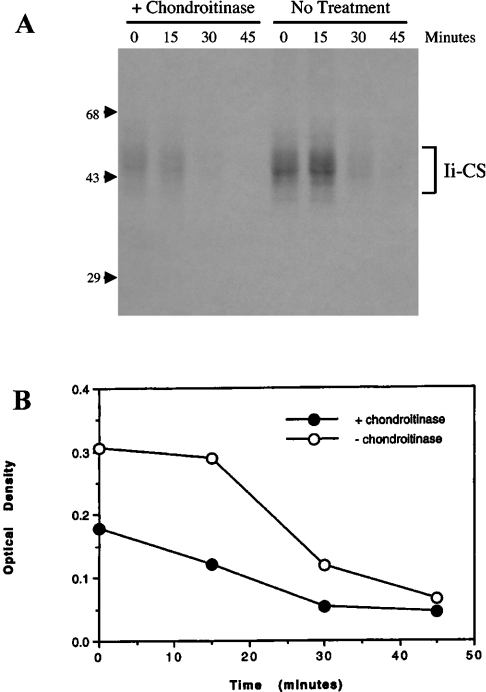
Ii–CS is associated with MHCII at the plasma membrane, suggesting that Ii–CS may be transported directly from the TGN to the cell surface. To test this possibility directly, newly synthesized Ii–CS was pulse-labelled with 35SO4 to label the CS side chain. Sulfation is a late event in proteoglycan maturation and has been shown to occur in the TGN. Arrival of newly sulfated Ii–CS at the cell surface was detected by using a carbohydrate-processing assay, similar to those used in previous studies [16]. In our assay, we treated intact cells with chondroitinase ABC, which selectively cleaves between glucuronic acid and N-acetylglucosamine, digesting the glycosaminoglycan into disaccharides and releasing the 35SO4 label from Ii. Control experiments were performed to assure that all the extracellular chondroitinase was removed before cell lysis. In the absence of chondroitinase treatment, the half-life of sulfate-labelled Ii–CS was found to be approx. 30 min (Figure 3), consistent with earlier reports [28]. Chondroitinase treatment decreased the amount of detectable Ii–CS, indicating that a significant amount of Ii–CS was present on the cell surface (Figure 3). Even after the short pulse of 15 min, chondroitinase treatment reduced levels of detectable Ii–CS, indicating that Ii–CS transits rapidly from the TGN to the cell surface, where it is accessible to chondroitinase treatment.
Figure 3. Ii–CS travels rapidly to the cell surface following sulfation in the TGN.
(A) Splenocytes isolated from C3H mice were labelled with 35SO4 for 15 min and chased for 0, 15, 30 or 45 min at 37 °C. After each chase, cells were treated with or without chondroitinase ABC. After removal of the chondroitinase by washing, the cells were lysed, immunoprecipitated with P4H5 (anti-Ii) and analysed by SDS/PAGE. Position of molecular-mass markers are indicated on the left (sizes in kDa). (B) Optical densitometry was performed on autoradiographs from two independent experiments, and results are means for cells treated with or without chondroitinase ABC. Similar results were obtained with Ltk− transfectants expressing p31EA (not shown).
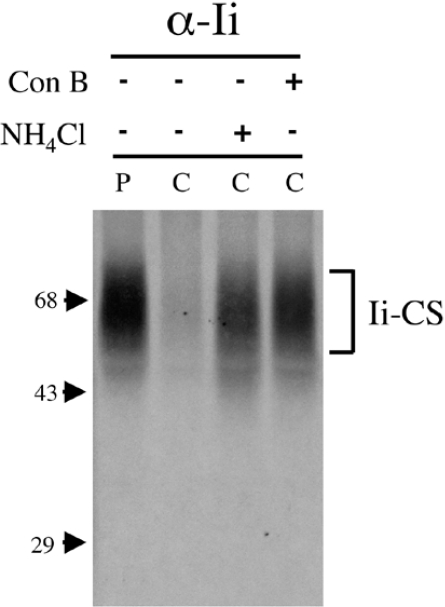
The pathway of Ii–CS degradation resulting in its short half-life is not clear. Some surface-expressed transmembrane proteoglycans are shed from the surface by a proteolytic cleavage of their core protein (reviewed in [34]), resulting in surface degradation. Alternatively, some proteoglycans are degraded in endocytic compartments following internalization [35,36]. The cytosolic domain of Ii functions as an efficient internalization signal [20,37,38]. Therefore rapid endocytosis of Ii–CS from the cell surface and lysosomal degradation could account for the short half-life of Ii–CS. In this case, we should be able to delay Ii–CS degradation by neutralizing the endosomal/lysosomal pathway. NH4Cl is a weak base, which rapidly diffuses through the cell and neutralizes acidic compartments. Concanamycin B is a highly specific inhibitor of the vacuolar H+-ATPases, which acidify the lumen of intracellular compartments, including the Golgi and endosomes/lysosomes [39,40]. By neutralizing acidic compartments, both of these lysosomotropic agents effectively inhibit the activity of endosomal and lysosomal proteases which function optimally at low pH. Cells were pulsed with 35SO4 and chased in unlabelled sulfate in the presence of NH4Cl or concanamycin B. Little Ii–CS remained intact after 1 h of chase in untreated cells, whereas addition of NH4Cl or concanamycin B during the chase inhibits degradation of Ii–CS (Figure 4). Taken together, the rapid transport of Ii–CS to the cell surface and the inhibition of Ii–CS turnover by lysosomotropic agents indicate that degradation of Ii–CS occurs in endocytic or lysosomal compartments following internalization from the cell surface.
Figure 4. Degradation of Ii–CS is inhibited by lysosomotropic agents.
Ltk− cells stably transfected with p31EA were labelled for 1 h with 35SO4 (P) and chased for 1 h (C) without treatment or with 15 mM NH4Cl or 10 μM concanamycin B. Cell lysates were immunoprecipitated with P4H5 (anti-Ii) and analysed by SDS/PAGE. The position of Ii–CS is shown on the right, and molecular-mass markers are indicated on the left (sizes in kDa).
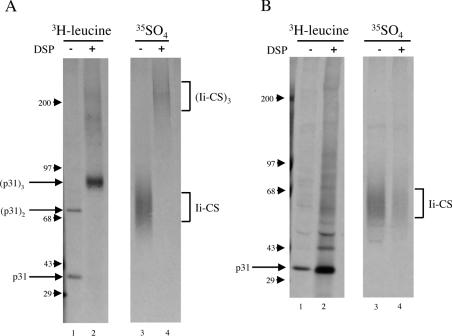
Ii trimerization, which occurs immediately following synthesis in the ER, has been demonstrated to be necessary for efficient endosomal localization [20]. This finding raises the possibility that Ii–CS may escape the TGN to endosome trafficking and travel from the TGN to the cell surface because it is monomeric. To determine whether Ii–CS is present in Ii trimers, we stabilized already formed Ii complexes in cells expressing the mutated p31EA protein using the reducible cross-linker DSP. Cells were biosynthetically labelled with 35SO4, which specifically labels the CS side chain of Ii–CS, or with [3H]leucine, which labels the core protein sequence of Ii. Protein complexes were cross-linked with DSP, and Ii molecules were precipitated and analysed by SDS/PAGE. In the absence of DSP, Ii labelled with [3H]leucine was present in monomers and dimers (Figure 5A, lane 1). Dimers form when cysteine residues in the cytosolic domain of Ii become oxidized following lysis and form disulfide bonds [41]. When cells were treated with DSP during lysis, the Ii trimer was stabilized and migrates at approx. 90 kDa in size (Figure 5A, lane 2). Reduction of the cross-linker destabilizes the trimer, and Ii migrates as a 31 kDa monomer on SDS/PAGE (Figure 5B, lane 2). Because only a small subset of Ii is modified by CS and Ii–CS migrates so diffusely on SDS/PAGE, Ii–CS is not detectable in the [3H]leucine-labelled samples. Rather, Ii–CS is detected by labelling with 35SO4. Ii–CS was also present as a monomer in the absence of DSP (Figure 5A, lane 3) and in a large complex with a mass consistent with trimer formation, in the presence of DSP (Figure 5A, lane 4). Reduction of DSP to remove the cross-linking and thus break apart the protein complexes restored monomeric Ii–CS (Figure 5B, lane 4). These data suggest that Ii–CS may be present in trimers.
Figure 5. Ii–CS cross-links to form a complex whose molecular mass is consistent with Ii trimerization.
Ltk− cells stably transfected with the p31EA construct were labelled with [3H]leucine or 35SO4 for 1.5 h and lysed in the absence (−) or presence (+) of DSP. Lysates were immunoprecipitated with P4H5 (anti-Ii), treated without (A) or with (B) the reducing agent 2-mercaptoethanol to reduce the cross-linker DSP and separated on 5–10% gradient gels. Monomeric, dimeric and trimeric conventional Ii and molecular-mass markers are indicated on the left (sizes in kDa), and Ii–CS complexes are indicated on the right.
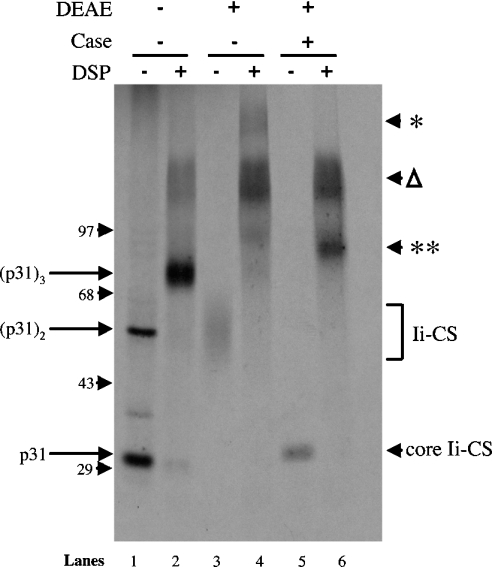
Because the addition of the glycosaminoglycan side chain disproportionately increases the apparent molecular mass of the complex, it is difficult to assess the composition of the putative trimeric complex containing Ii–CS. Therefore, to determine the size of the core protein complex, cells were biosynthetically labelled with [35S]methionine, and complexes containing Ii–CS were enriched by binding to DEAE-Sephacel in 150 mM NaCl. Under these conditions, most proteins do not bind to DEAE, whereas the highly charged side chains on proteoglycans allow for binding. This approach enables us to detect Ii–CS when the Ii–protein backbone is labelled with [35S]methionine (Figure 6, lane 3). In the absence of DEAE-enrichment, primarily conventional Ii is detected, indicative of the relatively small percentage of Ii that is modified by CS addition (Figure 6, lane 1). As seen in Figure 5, DSP cross-linking stabilizes conventional Ii in a 90 kDa trimeric complex (Figure 6, lane 2). When Ii–CS was enriched on DEAE-Sephacel, the Ii–CS monomer was cross-linked into several different high-molecular-mass species (Figure 6, lane 4). To determine which of these contained Ii–CS, the samples were digested with chondroitinase. Chondroitinase treatment effectively removes the CS side chain from Ii–CS, and the diffuse monomeric Ii–CS band migrates at a discrete band of approx. 33 kDa (Figure 6, lane 5). When cross-linked samples were treated with chondroitinase, only the slowest migrating complex (* in Figure 6, lane 4) was eliminated by chondroitinase treatment, and a new species of approx. 96 kDa appeared (** in Figure 6, lane 6), which is consistent with the size of an Ii trimer. Both the monomeric and trimeric core protein complexes released from Ii–CS following chondroitinase treatment migrate at a slightly higher apparent molecular mass than conventional Ii (Figure 6, lane 5 compared with lane 1, and lane 6 compared with lane 2). This increase in molecular mass is not solely due to N-linked sugar maturation of Ii–CS because the difference in apparent molecular mass is still retained after N-glycosidase treatment (results not shown). Rather, at least some of the difference in molecular mass may be due to the hexasaccharide base of Ii–CS, which remains because chondroitinase ABC lyase does not cleave between the first glucuronic acid and the first N-acetylgalactosamine of CS side chains. Taken together, these data indicate that Ii–CS is present within a trimeric complex.
Figure 6. Core Ii–CS is present in complexes whose molecular mass is consistent with trimerization.
Ltk− cells stably transfected with the p31EA construct were labelled with [35S]methionine for 2 h and lysed in the absence (−) or presence (+) of the reducible cross-linker DSP. Lysates were immunoprecipitated with P4H5 (anti-Ii) and eluted. Eluates were then either not treated, or absorbed with DEAE to isolate complexes containing Ii–CS. DEAE-bound material was then eluted, treated without (−) or with (+) chondroitinase ABC (Case) and reprecipitated with a combination of conformation-independent antibodies directed at the cytosolic domain of Ii (In-1 and antisera 657 and 658) and analysed by non-reducing SDS/PAGE. The positions of the monomeric, dimeric and trimeric conventional Ii complexes are labelled on the left. The positions of the core-Ii–CS following chondroitinase treatment (core Ii–CS) and the cross-linked complexes containing Ii–CS both before (*) and after (**) chondroitinase treatment are indicated on the right. The position of the high-molecular-mass Ii aggregates (Δ) is indicated on the right. Molecular-mass markers are indicated on the left (sizes in kDa).
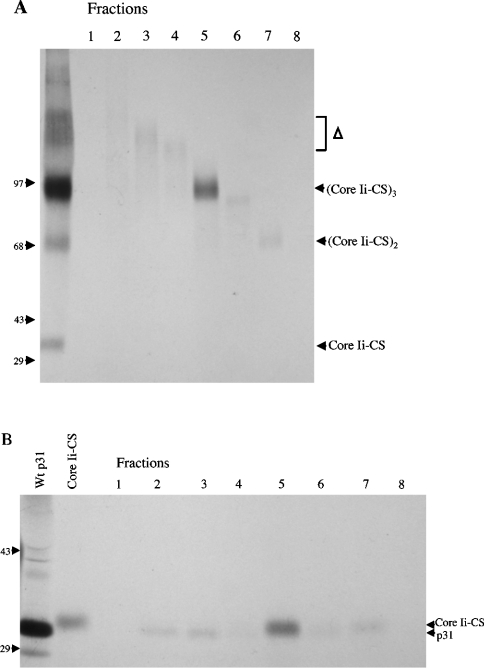
However, the presence of the additional slow-migrating complex that does not contain Ii–CS (Δ in Figure 6) precludes analysis of the components in the putative Ii–CS trimer. This additional complex appears to represent higher-order [42] or aggregated Ii complexes that may be enriched on DEAE due to the increased valency of these protein complexes. When samples containing these complexes are reduced, large amounts of conventional p31 Ii are released (results not shown). To separate these Ii aggregates from Ii–CS-containing trimers, cells were labelled with [35S]methionine and cross-linked with DSP. Ii immunoprecipitates were enriched on DEAE-Sephacel, treated with chondroitinase, and the Ii aggregates were separated from the putative trimeric core protein complex derived from Ii–CS by unreduced SDS/PAGE. The fractions were then re-analysed on reduced and unreduced SDS/PAGE to identify the components present in the cross-linked complexes. Fractions that contain the Ii aggregates released a single discrete band containing p31 Ii when reduced (Figure 7, fractions 2–4). In contrast, the putative trimeric core protein complex derived from Ii–CS contained two closely migrating bands that correspond to conventional p31 and the core protein of Ii–CS (Figure 7, fraction 5). These data indicate that Ii–CS is present in trimeric complexes that contain CS modified and unmodified Ii.
Figure 7. Ii–CS is present in trimers that may contain both CS modified and unmodified Ii.
Ltk− cells stably transfected with p31 were labelled with [35S]methionine for 2 h and lysed in the presence of 200 μg/ml DSP. Lysates were absorbed to DEAE and digested with chondroitinase as described in Figure 6. Anti-Ii immunoprecipitates from DEAE-bound and chondroitinase-treated material was eluted in the absence of 2-mercaptoethanol and run on 7% tube gels. The tube gel was then cut into 1 cm pieces (fractions 1–8), protein was eluted overnight at room temperature, unreduced (A) or reduced (B) to break the cross-linking and separated on 5–10% gradient gels. Lane 1 in (A) contains an aliquot of the material loaded on the tube gel (DSP cross-linked, DEAE-absorbed, chondroitinase-digested and immunoprecipitated with anti-Ii). The positions of the high-molecular-mass Ii aggregates (Δ) and the putative trimeric, dimeric and monomeric p31–CS core complexes are indicated on the right. Lanes 1 and 2 in (B) contain control lanes to indicate the position of [35S]methionine-labelled p31 and p31–CS core proteins. Lane 1 is an anti-Ii precipitate from untreated lysates. The sample in lane 2 was first absorbed to DEAE to enrich for Ii–CS, chondroitinase-treated and then precipitated with anti-Ii. The position of p31 and the slower migrating core Ii–CS are indicated on the right. Molecular-mass markers are indicated on the left of both gels (sizes in kDa).
DISCUSSION
A small percentage of Ii is modified by the addition of CS. This proteoglycan form of Ii is found associated with MHCII at the cell surface [28,29]. In this report, we increased the percentage of Ii modified by CS addition by changing the xylosylation site in Ii to conform more closely to the consensus sequence for xylosyltransferase. We used this modified construct to examine the assembly and trafficking of Ii–CS. We found that Ii–CS is present in Ii trimers, and as such should contain a complete endosomal sorting signal. Nevertheless, Ii–CS is rapidly targeted to the cell surface, where it is internalized into endosomes/lysosomes and degraded. Interestingly, the Ii–CS-containing trimers also include conventional Ii, suggesting that Ii–CS may contribute to the cell-surface expression of both the proteoglycan and conventional forms of Ii.
Glycosaminoglycan chains are added to form proteoglycans through the action of a number of different enzymes in different cellular compartments. The three classes of glycosaminoglycans (chondroitin/dermatan sulfate, heparan sulfate/heparin and keratin sulfate) differ primarily in disaccharide composition, requiring the usage of different enzymes for monosaccharide addition. Chondroitin/dermatan sulfate and heparan sulfate are initiated by xylosylation of a serine residue. Studies have suggested that xylosyltransferase is the rate-limiting enzyme in glycosaminoglycan addition [30,43]. Only a small percentage of Ii is normally modified by CS addition, potentially due to deviation of the Ii sequence from the xylosylation consensus sequence [44]. Modification of the Ii sequence to conform more closely to the consensus sequence (Figure 1) resulted in approx. 3-fold greater levels of Ii–CS. Modification of the amino acids surrounding the xylose-accepting serine residue probably changed the conformation of the protein, potentially affecting the activity of xylosyltransferase, which has been shown to be dependent on protein conformation [44]. However, it is also possible that changing the primary structure around the xylose-accepting serine residue could also affect the activity of other enzymes needed to complete the tetrasaccharide linkage region, thus increasing glycosaminoglycan addition. Discrimination between these possibilities would require the measurement of xylose addition to wild-type Ii and the modified form of Ii.
The intracellular transport pathway of MHCII–Ii complexes from the TGN to endosomes has not been clearly established. Two routes have been suggested: either trafficking directly from the Golgi to endosomes or trafficking indirectly from the Golgi through a surface intermediate followed by internalization into endosomes. The direct endosomal trafficking route is supported by findings that MHCII–Ii complexes can recruit cytosolic AP (adaptor protein) 1 on membranes, an interaction that is strictly dependent on the presence of the Ii cytosolic tail and the di-leucine motifs within the tail [45]. Although the di-leucine-based motifs are able to bind both AP1 and AP2 in vitro, AP1 was found to bind with a higher affinity [46–48]. In human cells, the use of concanamycin B to block early to late endosomal trafficking suggested that most Ii does not pass through a surface intermediate, consistent with direct trafficking of the bulk of MHCII–Ii complexes to endosomes [19]. Our previous studies are also consistent with direct transport from the Golgi to endosomes [20]. Truncation of the di-leucine sequences in the cytosolic tail results in accumulation of trimeric Ii complexes at the cell surface [20,24]. When wild-type and truncated Ii are co-expressed, mixed trimers that contain a single intact cytosolic tail accumulate at the cell surface, suggesting that efficient transport of Ii to endosomes requires multimerization of the cytosolic tail [20]. Interestingly, the single intact cytosolic tail still functioned as an efficient internalization signal, suggesting that efficient internalization alone could not account for the paucity of cell-surface Ii. However, recent data from two groups support the presence of a cell-surface intermediate in Ii transport to endosomes [17,18]. Using RNA interference knockdown, it has been shown that AP2, but not AP1, expression is required for Ii degradation, consistent with trafficking of Ii to endosomes via surface intermediate. However, it is possible that AP2 knockdown prevents recycling of machinery required for direct transport. Furthermore, some data suggest that Ii [49] and transferrin receptor [50] both transit directly from the Golgi to early endosomes, followed by transport out to the surface and rapid re-internalization. This pathway, Golgi to endosomes to plasma membrane, is consistent with the build-up of MHCII–Ii on the cell surface seen in the AP2-knockdown studies. One potential resolution of these data, consistent with [16] and the present paper, is that multiple pathways may be used, and different Ii isoforms may bias trafficking of pools of Ii to endosomes via different routes.
Results of the present study indicate that the majority of Ii–CS is transported to endosomes through a cell-surface intermediate. Furthermore, Ii–CS is assembled into a trimer that also contains conventional Ii. These results raise two implications. First, addition of CS may alter the intracellular sorting of MHCII–Ii complexes. CS modification of Ii may inhibit the direct trafficking mechanism from Golgi to endosomes, may slow internalization from a cell-surface intermediate or may alter the sorting of Ii in early endosomes. How addition of the CS side chain on the lumenal domain of Ii may affect functions that are largely associated with cytosolic domains is not clear. The second implication is that CS addition may contribute to the cell-surface expression of conventional forms of Ii. Both Ii–CS and conventional Ii have been implicated as cell-surface receptors for cell signalling events [9,11,12]. Thus Ii–CS may be essential to both of these functions, either by modulating trafficking of MHCII–Ii complexes or through direct interaction with extracellular ligands.
Acknowledgments
This work was supported in part by NIH (National Institutes of Health) grant GM-42017 to J. M. and NIH training grant GM-07183 to L.S.A.
References
- 1.Hiltbolt E. M., Roche P. A. Trafficking of MHC class II molecules in the late secretory pathway. Curr. Opin. Immunol. 2002;14:30–35. doi: 10.1016/s0952-7915(01)00295-3. [DOI] [PubMed] [Google Scholar]
- 2.Bakke O., Dobberstein B. MHC class II-associated invariant chain contains a sorting signal for endosomal compartments. Cell. 1990;63:707–716. doi: 10.1016/0092-8674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 3.Lotteau V., Teyton L., Peleraux A., Nilsson T., Karlsson L., Schmid S., Quaranta V., Peterson P. Intracellular transport of class II MHC molecules directed by invariant chain. Nature. 1990;348:600–605. doi: 10.1038/348600a0. [DOI] [PubMed] [Google Scholar]
- 4.Shachar I., Flavell R. Requirement for invariant chain in B cell maturation and function. Science. 1996;274:106–108. doi: 10.1126/science.274.5284.106. [DOI] [PubMed] [Google Scholar]
- 5.Benlagha K., Park S.-H., Guinamard R., Forestier C., Karlsson L., Chang C.-H., Bendelac A. Mechanisms governing B cell development defects in invariant chain-deficient mice. J. Immunol. 2004;172:2076–2083. doi: 10.4049/jimmunol.172.4.2076. [DOI] [PubMed] [Google Scholar]
- 6.Matza D., Lantner F., Bogoch Y., Flaishon L., Hershkoviz R., Shachar I. Invariant chain induces B cell maturation in a process that is independent of its chaperonic activity. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3018–3023. doi: 10.1073/pnas.052703299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker-Herman S., Arie G., Medvedovsky H., Kerem A., Shachar I. CD74 is a member of the regulated intramembrane proteolysis-processed protein family. Mol. Biol. Cell. 2005;16:5061–5069. doi: 10.1091/mbc.E05-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matza D., Wolstein O., Dikstein R., Shachar I. Invariant chain induces B cell maturation by activating a TAFII105-NF-κB-dependent transcription program. J. Biol. Chem. 2001;276:27203–27206. doi: 10.1074/jbc.M104684200. [DOI] [PubMed] [Google Scholar]
- 9.Starlets D., Gore Y., Binsky I., Haran M., Harpaz N., Shvidel L., Becher-Herman S., Berrebi A., Schachar I. Cell-surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood. 2006;107:4807–4816. doi: 10.1182/blood-2005-11-4334. [DOI] [PubMed] [Google Scholar]
- 10.Prydz K., Dalen K. T. Synthesis and sorting of proteoglycans. J. Cell Sci. 2000;113:193–205. doi: 10.1242/jcs.113.2.193. [DOI] [PubMed] [Google Scholar]
- 11.Naujokas M., Morin M., Anderson M., Peterson M., Miller J. The chondroitin sulfate form of invariant chain can enhance stimulation of T cell responses through interaction with CD44. Cell. 1993;74:257–268. doi: 10.1016/0092-8674(93)90417-o. [DOI] [PubMed] [Google Scholar]
- 12.Leng L., Metz C. N., Fang Y., Xu J., Donnelly S., Baugh J., Delohery T., Chen Y., Mitchell R. A., Bucala R. MIF signal transduction initiated by binding to CD74. J. Exp. Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calandra T., Bernhagen J., Mitchell R. A., Bucala R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J. Exp. Med. 1994;179:1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bozza M., Satoskare A. R., Lin G., Lu B., Humbles A. A., Gerarad C., David J. R. Targeted disruption of migration inhibitory factor gene reveals its critical role in sepsis. J. Exp. Med. 1999;189:341–346. doi: 10.1084/jem.189.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell R. A., Liao H., Chesney J., Fingerle-Rowson G., Baugh J., David J., Bucala R. Inhibition of macrophage migration inhibitory factor (MIF) tautomerase and biological activities by acetaminophen metabolites. Proc. Natl. Acad. Sci. U.S.A. 2002;99:144–149. doi: 10.1073/pnas.011569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warmerdam P. A., Long E. O., Roche P. A. Isoforms of the invariant chain regulate transport of MHC class II molecules to antigen processing compartments. J. Cell Biol. 1996;133:281–291. doi: 10.1083/jcb.133.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dugast M., Toussaint H., Dousset C., Benaroch P. AP2 clathrin adaptor complex, but not AP1, controls the access of the major histocompatibility complex (MHC) class II to endosomes. J. Biol. Chem. 2005;280:19656–19664. doi: 10.1074/jbc.M501357200. [DOI] [PubMed] [Google Scholar]
- 18.McCormick P. J., Martina J. A., Bonifacino J. S. Involvement of clathrin and AP-2 in the trafficking of MHC class II molecules to antigen-processing compartments. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7910–7915. doi: 10.1073/pnas.0502206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benaroch P., Yilla M., Raposo G., Ito K., Miwa K., Geuze H. J., Ploegh H. L. How MHC class II molecules reach the endocytic pathway. EMBO J. 1995;14:37–49. doi: 10.1002/j.1460-2075.1995.tb06973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arneson L. S., Miller J. Efficient endosomal localization of MHC class II-invariant chain complexes requires multimerization of the invariant chain targeting sequence. J. Cell Biol. 1995;129:1217–1228. doi: 10.1083/jcb.129.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashman J. B., Miller J. A role for the transmembrane domain of the trimerization of the MHC class II-associated invariant chain. J. Immunol. 1999;163:2704–2712. [PubMed] [Google Scholar]
- 22.Biljmakers M. J., Benaroch P., Ploegh H. L. Mapping functional regions in the luminal domain of the class II-associated invariant chain. J. Exp. Med. 1994;180:623–629. doi: 10.1084/jem.180.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koch N., Hammerling G., Tada H., Kimura S., Hammerling U. Cross-blocking studies with monoclonal antibodies against I-A molecules of haplotypes b, d and k. Eur. J. Immunol. 1982;12:909–914. doi: 10.1002/eji.1830121103. [DOI] [PubMed] [Google Scholar]
- 24.Anderson M., Swier K., Arneson L., Miller J. Enhanced antigen presentation in the absence of the invariant chain endosomal localization signal. J. Exp. Med. 1993;178:1959–1969. doi: 10.1084/jem.178.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehringer J., Harris M., Kindle C., McCourt D., Cullen S. Characterization of fragments of the murine Ia-associated invariant chain. J. Immunol. 1991;146:920–927. [PubMed] [Google Scholar]
- 26.Ozato K., Mayer N., Sachs D. Hybridoma cell lines secreting monoclonal antibodies to mouse H-2 and Ia antigens. J. Immunol. 1980;124:533–540. [PubMed] [Google Scholar]
- 27.Anderson M., Miller J. Invariant chain can function as a chaperone protein for class II major histocompatibility complex molecules. Proc. Natl. Acad. Sci. U.S.A. 1992;89:2282–2286. doi: 10.1073/pnas.89.6.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sant A., Cullen S., Giacoletto K., Schwartz B. Invariant chain is the core protein of the Ia-associated chrondroitin sulfate proteoglycan. J. Exp. Med. 1985;162:1916–1934. doi: 10.1084/jem.162.6.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller J., Hatch J. A., Simonis S., Cullen S. E. Identification of the glycosaminoglycan-attachment site of mouse invariant-chain proteoglycan core protein by site-directed mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 1988;85:1359–1363. doi: 10.1073/pnas.85.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kearns A. E., Vertel B. M., Schwartz N. B. Topography of glycosylation and UDP-xylose production. J. Biol. Chem. 1993;268:11097–11104. [PubMed] [Google Scholar]
- 31.Bourdon M. A., Krusius T., Campbell S., Schwartz N. B., Ruoslahti E. Identification and synthesis of a recognition signal for the attachment of glycosaminoglycans to proteins. Proc. Natl. Acad. Sci. U.S.A. 1987;84:3194–3198. doi: 10.1073/pnas.84.10.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber S., Winterhalter K. H., Vaughan L. Isolation and sequence analysis of the glycosaminoglycan attachment site of type IX collagen. J. Biol. Chem. 1988;263:752–756. [PubMed] [Google Scholar]
- 33.Zhang L., Esko J. D. Amino acid determinants that drive heparan sulfate assembly in a proteoglycan. J. Biol. Chem. 1994;269:19295–19299. [PubMed] [Google Scholar]
- 34.Bernfield M., Gotte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 35.Egeberg M., Kjeken R., Kolset S. O., Berg T., Prydz K. Internalization and stepwise degradation of heparan sulfate proteoglycans in rat hepatocytes. Biochim. Biophys. Acta. 2001;1541:135–149. doi: 10.1016/s0167-4889(01)00132-x. [DOI] [PubMed] [Google Scholar]
- 36.Kasevayuth K., Yanagishita M. Catabolism of heparan sulfate proteoglycans in Drosophila cell lines. Biochem. Biophys. Res. Commun. 2004;324:205–211. doi: 10.1016/j.bbrc.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 37.Odorizzi G., Trowbridge I. S., Xue L., Hopkins C. R., Davis C. D., Collawn J. F. Sorting signals in the MHC class II invariant chain cytoplasmic tail and transmembrane region determine trafficking to an endocytic processing compartment. J. Cell Biol. 1994;126:317–330. doi: 10.1083/jcb.126.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roche P. A., Teletski C. L., Stang E., Bakke O., Long E. O. Cell surface HLA-DR-invariant chain complexes are targeted to endosomes by rapid internalization. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8581–8585. doi: 10.1073/pnas.90.18.8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinashi H., Someno K., Sakaguchi K. Isolation and characterization of concanamycins A, B and C. J. Antibiot. 1984;37:1333–1343. doi: 10.7164/antibiotics.37.1333. [DOI] [PubMed] [Google Scholar]
- 40.Bowman E. J., Siebers A., Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells and plant cells. Proc. Natl. Acad. Sci. U.S.A. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biljmakers M. J., Benaroche P., Ploegh H. L. Assembly of HLA DR1 molecules translated in vitro: binding of peptide in the endoplasmic reticulum precludes association with invariant chain. EMBO J. 1994;13:2699–2707. doi: 10.1002/j.1460-2075.1994.tb06560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marks M., Blum J., Cresswell P. Invariant chain trimers are sequestered in the rough endoplasmic reticulum in the absence of association with HLA class II antigens. J. Cell Biol. 1990;111:839–855. doi: 10.1083/jcb.111.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kresse H., Seidler D. G., Muller M., Breuer E., Hausser H., Roughley P. J., Schonherr E. Different usage of the glycosaminoglycan attachment sites of biglycan. J. Biol. Chem. 2001;276:13411–12316. doi: 10.1074/jbc.M009321200. [DOI] [PubMed] [Google Scholar]
- 44.Brinkmann T., Weilke C., Kleesiek K. Recognition of acceptor proteins by UDP-D-xylose proteoglycan core protein β-D-xylosyltransferase. J. Biol. Chem. 1997;272:11171–11175. doi: 10.1074/jbc.272.17.11171. [DOI] [PubMed] [Google Scholar]
- 45.Salamero J., Le Borgne R., Saudrais C., Goud B., Hoflack B. Expression of major histocompatibility complex class II molecules in HeLa cells promotes the recruitment of AP-1 Golgi-specific assembly proteins on Golgi membranes. J. Biol. Chem. 1996;271:30318–30321. doi: 10.1074/jbc.271.48.30318. [DOI] [PubMed] [Google Scholar]
- 46.Rodionov D. G., Bakke O. Medium chains of adaptor complexes AP-1 and AP-2 recognize leucine-based sorting signals from the invariant chain. J. Biol. Chem. 1998;273:6005–6008. doi: 10.1074/jbc.273.11.6005. [DOI] [PubMed] [Google Scholar]
- 47.Hofmann M. W., Honing S., Rodionov D., Dobberstein B., von Figura K., Bakke O. The leucine-based sorting motifs in the cytoplasmic domain of the invariant chain are recognized by the clathrin adaptors AP1 and AP2 and their medium chains. J. Biol. Chem. 1999;274:36153–36158. doi: 10.1074/jbc.274.51.36153. [DOI] [PubMed] [Google Scholar]
- 48.Kongsvik T. L., Honing S., Bakke O., Rodionov D. G. Mechanism of interaction between leucine-based sorting signals from the invariant chain and clathrin-associated adaptor protein complexes AP1 and AP2. J. Biol. Chem. 2002;277:16484–16488. doi: 10.1074/jbc.M201583200. [DOI] [PubMed] [Google Scholar]
- 49.Lindner R. Transient surface delivery of invariant chain–MHC II complexes via endosomes: a quantitative study. Traffic. 2002;3:133–146. doi: 10.1034/j.1600-0854.2002.030206.x. [DOI] [PubMed] [Google Scholar]
- 50.Futter C. E., Connolly C. N., Cutler D. F., Hopkins C. R. Newly synthesized transferrin receptors can be detected in the endosome before they appear on the cell surface. J. Biol. Chem. 1995;270:10999–11003. doi: 10.1074/jbc.270.18.10999. [DOI] [PubMed] [Google Scholar]