Abstract
CgNa (Condylactis gigantea neurotoxin) is a 47-amino-acid- residue toxin from the giant Caribbean sea anemone Condylactis gigantea. The structure of CgNa, which was solved by 1H-NMR spectroscopy, is somewhat atypical and displays significant homology with both type I and II anemone toxins. CgNa also displays a considerable number of exceptions to the canonical structural elements that are thought to be essential for the activity of this group of toxins. Furthermore, unique residues in CgNa define a characteristic structure with strong negatively charged surface patches. These patches disrupt a surface-exposed cluster of hydrophobic residues present in all anemone-derived toxins described to date. A thorough characterization by patch–clamp analysis using rat DRG (dorsal root ganglion) neurons indicated that CgNa preferentially binds to TTX-S (tetrodotoxin-sensitive) voltage-gated sodium channels in the resting state. This association increased the inactivation time constant and the rate of recovery from inactivation, inducing a significant shift in the steady state of inactivation curve to the left. The specific structural features of CgNa may explain its weaker inhibitory capacity when compared with the other type I and II anemone toxins.
Keywords: Condylactis gigantea (giant Caribbean sea anemone), nuclear magnetic resonance (NMR), sea anemone toxin, site 3 toxin, sodium channel inactivation
Abbreviations: ApA, anthopleurin A from Anthopleura xanthogrammica (giant green anemone); ApB, anthopleurin B from Anthopleura xanthogrammica; ApC, anthopleurin C from Anthopleura elegantissima (aggregating anemone); CgNa, Condylactis gigantea neurotoxin; DRG, dorsal root ganglion; Hyp, 4-hydroxyproline; NOE, nuclear Overhauser effect; rmsd, root mean square deviation, Sh I, neurotoxin I from Stichodactyla helianthus (sun anemone); TTX, tetrodotoxin; TTX-R, tetrodotoxin (resistant); TTX-S, TTX-sensitive
INTRODUCTION
Sea anemones are sessile cnidarians that protect themselves against predators by releasing different types of polypeptide toxins. These toxins mainly affect ion-channel activity, although some of them exhibit cytolytic activity [1]. The first polypeptide toxins purified from Anemonia sulcata (Mediterranean snakelocks anemone) and Condylactis gigantea (giant Caribbean sea anemone) were reported to affect voltage-gated sodium channels [2,3]. These voltage-gated sodium channels represent an important target for different natural toxins and at least six receptor sites have been identified on sodium channels for these kinds of neurotoxins.
Sodium-channel toxins from the sea anemone can be classified into types I and II, according to their sequence homology [4], and these types of toxins exhibit comparable electrophysiological properties. Type I toxins elicit important neurotoxic repetitive (firing) and cardiotoxic (arrhythmia) effects, delaying the inactivation of mammalian voltage-gated sodium channels. Moreover, the type I toxins target receptor-site 3 located at the extracellular face of the sodium channel [5]. There are many toxins that target this site on mammalian voltage-gated sodium channels, they are a structurally diverse group of peptides which have been isolated from scorpions [6,7], sea anemones [4,8], spiders [9,10] and wasps [11,12]. These toxins are important and are useful tools with which to study the mechanism of sodium-channel inactivation and to emulate certain important periodic paralysis pathologies such as paramyotonia congenita and hyperkalaemic periodic paralysis. Indeed, the effects of such toxins are similar to those caused by the mutations responsible for these diseases [13–16].
We recently isolated and performed the preliminary characterization of CgNa (Condylactis gigantea neurotoxin), the main peptide toxin from Condylactis gigantea [17]. On the basis of sequence homology, CgNa is a type I toxin group member along with the following anemone toxins: ATXI, ATXII and ATXV from Anemonia sulcata [18–20]; AftI and AftII from the sea anemone Anthopleura fuscoviridis [21]; ApA and ApB from Anthopleura xanthogrammica (giant green anemone) [22–24]; ApC from Anthopleura elegantissima (aggregating anemone) [25]; and BgII and BgIII from the Caribbean sea anemone Bunodosoma granulifera [26,27]. The three-dimensional structure of CgNa in solution has been resolved using 1H-NMR spectroscopy, and the electrophysiological effects of CgNa in mammalian DRG (dorsal root ganglion) neurons have been described. Moreover, all these data have correlated with those available for other closely related type I and II anemone toxins. These results may assist to define further the structural basis underlying the electrophysiological properties of this family of sea-anemone sodium-channel toxins.
MATERIALS AND METHODS
Biological materials
Animal care and all experimental procedures were carried out in accordance with the Helsinki Declaration. The number of animals used in the present study was kept to the minimum necessary for meaningful interpretation of the data. CgNa was isolated and purified from the sea anemone Condylactis gigantea [17]. Aliquots of a stock solution in deionized water were prepared and stored at −20 °C. Prior to each experiment, the aliquot of CgNa was diluted in the corresponding perfusion solution.
DRG neurons were isolated and cultured from Wistar rats (P5–9) of either sex, according to the procedure described previously [27].
Electrophysiological measurements
Whole-cell recording was carried out using an Axopatch-1D amplifier (Axon Instruments, Union City, CA, U.S.A.). The generation pulse and the data sampling was controlled by Pclamp 8.0 software (Axon Instruments) using a 16-bit data-acquisition system (Digidata 1320A, Axon Instruments). All experiments were performed at room temperature (23–25 °C).
DRG neurons are known to express both TTX-R [TTX (tetrodotoxin)-resistant] and TTX-S (TTX-sensitive) sodium channels [28,29]. Thus the types of sodium currents present in the cell under study were determined before each experiment. Only cells in which the TTX-R sodium current was less than 10% of the total current, as defined from a steady-state inactivation profile, were used to determine the effects of the toxin on TTX-S sodium currents [30].
Leakage and capacity currents were digitally subtracted using the P-P/n method [30a], and the capacitance and series resistance (80%) were electronically compensated for. Experiments were rejected when the voltage error exceeded 5 mV after compensation for series resistance at the maximum peak current, but no corrections were made for smaller values.
CgNa toxin was ejected under pressure using a microinjector (Baby Bee; Bass, West Lafayette, IN, U.S.A.) from a micropipette positioned in the vicinity of the cell being recorded. The solutions used in the experiments are detailed in Table 1. Unless otherwise stated, all numerical data are presented as the means±S.E.M. of at least four measurements. A more detailed description of the methods and data analysis has been reported previously [27].
Table 1. Concentrations of solutions employed in electrophysiological experiments (in mM; except for TTX, which is expressed in nM).
4-AP, 4-aminopiridine; cc, current clamp; Chol-Cl, choline chloride; NMDG, N-methyl-D-glucamine; TEA-Cl, tetraethylamonium chloride.
| Concentration | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KCl | NaCl | CsCl | CdCl | CsF | MgCl2 | CaCl2 | NMDG | Chol-Cl | Hepes | EGTA | TEA-Cl | 4-AP | TTX | |
| Extracellular cc | 5.4 | 140 | − | − | − | 1.2 | 3.6 | − | − | 10 | − | − | − | − |
| Intracellular cc | 145 | 10 | − | − | − | − | 0.1 | − | − | 5 | 10 | − | − | − |
| Extracellular INa | − | 20 | − | − | − | 1 | 1.8 | − | 70 | 10 | − | 45 | 10 | 300 |
| Intracellular INa | − | 10 | 30 | − | 100 | − | − | − | − | 5 | 8 | 10 | − | − |
| Extracellular IK | 5.4 | − | − | 100 | − | 1.2 | 1.8 | − | 140 | 10 | − | − | − | − |
| Intracellular IK | 72.5 | − | − | − | − | − | 0.1 | 72.5 | − | 5 | 10 | − | − | − |
| Extracellular ICa | − | − | 5 | − | − | 1.2 | 10 | − | − | 10 | − | 125 | 10 | − |
| Intracellular ICa | − | − | − | − | − | − | 0.134 | − | − | 5 | 10 | 10 | − | − |
NMR spectroscopy
The concentration of CgNa in 90% 1H2O/10% 2H2O and in 100% 2H2O was 1.4 mM in order to acquire NMR spectra.
1H-NMR spectra were recorded at 500 MHz on a Bruker AVANCE-500 spectrometer (Bruker BioSpin), using an inverse multinuclear probehead fitted with gradients along the z-axes. Spectra were referenced to external TSP {3-(trimethylsilyl)propionic acid-[2H4], sodium salt} and were recorded using the States-TPPI (time proportional phase incrementation) method, with 3–9–19 watergate pulse sequence water suppression. In general, 256 equally spaced evolution time period t1 values were acquired, averaging 16 transients of 2048 points, with a spectral width of 7002.8 Hz (14.00 p.p.m.). Time-domain data matrices were all zero-filled to 4K complex points in both dimensions, thereby yielding a digital resolution of 3.41 Hz/pt (point). Prior to Fourier transformation, a Lorentz–Gauss window with different parameters was applied to both the t1 and t2 dimensions for all of the experiments. NOESY spectra were obtained using different mixing times (0.15 and 0.25 s), and TOCSY experiments were recorded using 0.025 and 0.07 s DIPSI (decoupling in the presence of scalar interactions) mixing pulses. Both NOESY and TOCSY experiments were performed at 298 K.
NOESY cross-peak assignment and structure calculation
Peak lists for the NOESY spectra recorded with a 0.25 s mixing time were generated by interactive peak picking using the XEASY software [31]. NOESY cross-peak volumes were determined by the automated peak integration routine peakint [32] implemented in XEASY. The three-dimensional structure of CgNa was determined using the standard protocol of combined automated NOE (nuclear Overhauser effect) assignment and the structure calculation of the CYANA program (version 2.1) [33]. Seven cycles of combined automated NOESY assignment and structure calculations were followed by a final structure calculation. The structure calculation started in each cycle from 100 randomized conformers, and the standard simulated annealing schedule [33a] was used. The 20 conformers with the lowest final CYANA target function values were retained for analysis and passed to the next cycle. Constraint combination spanning at least three residues was applied in the first two cycles to all NOE distance restraints, in order to minimize structural distortion by erroneous distance restraints. The covalent parameters of Engh and Huber were used [34]. Restraints that involved degenerate groups of protons were expanded into ambiguous distance restraints between all corresponding pairs of hydrogen atoms, whereas non-degenerate diastereotopic pairs were swapped periodically for minimal target function values during simulated annealing in cycles 1–7. Three standard upper and lower limit distance constraints were also used to enforce disulfide bonds between the pairs of cysteine residues at positions 4–44, 6–34 and 27–45. Weak restraints on ψ/ψ torsion-angle pairs and on side-chain torsion angles between tetrahedral carbon atoms were applied temporarily during the high-temperature and cooling phases of the simulated annealing schedule in order to favour the permitted regions of the Ramachandran plot and staggered rotamer positions respectively. The list of upper-distance bonds for the final structural calculation consists of unambiguously assigned upper-distance bonds and does not require the possible swapping of diastereotopic pairs.
The 20 conformers with the lowest final CYANA target function values were subjected to restrained energy-minimization in a water shell using the AMBER 8.0 program [35]. The ω dihedral angles were restrained in the range 175–185°. The protein was immersed in a shell of water molecules created using the TIP3P model, with a thickness of 10 Å (1 Å=0.1 nm). The number of additional water molecules oscillated between 2979 and 4000 over the 20 conformers. The restrained energy minimization was performed in three stages. In the first stage, only the water molecules were optimized. Subsequently, the protein alone was relaxed, maintaining the water molecules fixed, and, finally, the whole system was minimized. In the last stage, a maximum of 1500 steps of restrained energy minimization and a combination of the steepest descent and conjugate gradient algorithms were applied using, in addition to the force field of Cornell et al. [37], a parabolic or linear penalty function for the NOE upper-distance bonds and torsion-angle restraints. The force constants were chosen such that restraint violations of 0.1 Å and torsion violations of 1° contributed by 0.84 kJ/mol (0.2 kcal/mol) to the potential energy. The resulting 20 energy-minimized CYANA conformers represent the solution structure of CgNa.
The MOLMOL program [38] was used to build the co-ordinates for the non-standard amino acid residue 4-hydroxyproline (Hyp). The topological information for this residue has been reported previously [39] and was used here without any further modification. No additional force field parameters for 4-hydroxyproline were needed.
The MOLMOL program was used to visualize the three-dimensional structures. CYANA was used to obtain statistics on target function values, restraint violations and Ramachandran plots according to PROCHECK-NMR conventions etc. [40]. Rmsd (root mean square deviation) values were calculated using CYANA for superpositions of the backbone N, Cα and C′ atoms; the heavy atoms over the whole protein, or over its structured regions. To obtain the rmsd of a structure represented by a bundle of conformers, all conformers were superimposed upon the first one and the average of the rmsd values between the individual conformers and their average co-ordinates was calculated. Conformational energies were calculated using AMBER 8.0 [35] and applying the force field of Cornell et al. [37].
RESULTS
NMR structural analysis of CgNa
Protein-resonance assignments were obtained using standard strategies based on two-dimensional NMR experiments. In total, 98.7% of the amide and 1H shifts of the side-chain moieties, CHn, were unambiguously identified and assigned. 1H assignments were also obtained for the indole NH group of Trp31 and for the side-chain NH2 resonances of all asparagine and glutamine residues in CgNa.
Structure determination
The statistics regarding the quality and precision of the 20 energy-minimized conformers that represent the solution structure of CgNa are summarized in Table 2. The structures were well defined and are in perfect agreement with the experimental data. Of the 1618 cross-peaks identified in the two-dimensional NOESY spectrum, 1554 (96%) were unambiguously assigned by the CYANA program. Aditionally, standard upper- and lower-limit distance constraints were applied to enforce the disulfide bridges. The disulfide arrangement (CysI–CysV, CysII–CysIV, CysIII–CysVI) was predicted on the basis of sequence homology and the conservation of the spacing of cysteine residues with another related type I sea-anemone toxin. Test calculations with alternative disulfide bond pairings were also assessed, and these generated structures that possessed less favourable conformation energies and deviated from the experimental NMR data to a greater extent from the disulfide bond pairings described above. Furthermore, 292 long-range distance restraints between protons that were separated in the sequence by five or more residues were used in the final calculations with CYANA and in the subsequent energy refinement with the AMBER program. No violations of NOE distance restraints greater than 0.2 Å were found in the final bundle of structures. The low rmsd values of 0.31 Å for the mean co-ordinates of the backbone and of 0.58 Å for all heavy atoms except the flexible loop of residues 8–16, reflected the high convergence of the ensembled structures. The quality of the structures was also reflected by the presence of 67% of the (ψ/ψ) backbone torsion-angle pairs in the most favoured regions, and 31% of these were found within the regions additionally permitted by the Ramachandran plot, according to the PROCHECK-NMR program.
Table 2. Statistics for the NMR solution structure of CgNa in 90% 1H2O/10% 2H2O at 298 K.
Average values over the 20 energy-minimized CYANA conformers. The final CYANA target function value was calculated for the structure before energy minimization with AMBER 8.0.
| Parameter | Value |
|---|---|
| NOE cross-peaks | 1618 |
| Assigned (%) | 96 |
| NOE upper distance limits | 829 |
| Short-range, |i–j|≤1 | 435 |
| Medium-range, 1<|i–j|<5 | 99 |
| Long-range, |i–j|≥5 | 295 |
| Maximal violation (Å) | 0.19 |
| Violations > 0.2 Å | 0 |
| CYANA target function (Å2) | 0.84 |
| AMBER energy [kJ/mol (kcal/mol)] | −1.826×105 (−4.38×104) |
| Rmsd from idealized geometry | |
| Bond lengths (Å) | 3.26 |
| Bond angles (°) | 0.05 |
| Rmsd to mean co-ordinates (Å) | |
| Backbone N,Cα,C′ of residues 1–47 | 1.20 |
| All heavy atoms of residues 1–47 | 1.45 |
| Backbone N,Cα,C′ of residues 1–7,17–47 | 0.31 |
| All heavy atoms of residues 1–7, 17–47 | 0.58 |
| Ramachandran plot statistics (%) | |
| Residues in the most favoured region | 67 |
| Residues in the additionally allowed regions | 31 |
| Residues in the generously allowed regions | 2 |
| Residues in the disallowed regions | 0 |
Solution structure
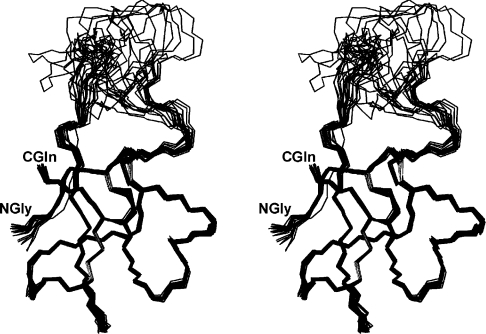
The final 20 structures calculated for CgNa have been superimposed on the backbone heavy atoms (Figure 1). The structure consists of four β-strands from antiparallel β-sheets (involving residues 1–3, 21–23, 32–33 and 43–46), and three β-turns at residues 13–16, 27–30 and 31–34. The first β-strand is connected to the second one by a long poorly defined loop exposed to the solvent (Asp7–Ser19). The peptide bonds at Hyp3 and Pro11 were predominantly determined to be in trans orientation due to the presence of αi–δi+i NOE cross-peaks. There was also evidence of isomers, with a cis peptide bond at Hyp3 identified by one NOE between the Hα protons of Val2 and Hyp3. The relative slow isomerization between cis and trans forms of the prolyl-peptide bond usually induces additional cross-peaks for nearby residues. In the CgNa spectra, two sets of Val2 cross-peaks were observed in the TOCSY fingerprint region.
Figure 1. Stereo view of the 20 representative structures of CgNa.
The structures are superimposed over the backbone atoms N, Cα and C′ of residues 1–47.
Comparison with other long sea-anemone polypeptides
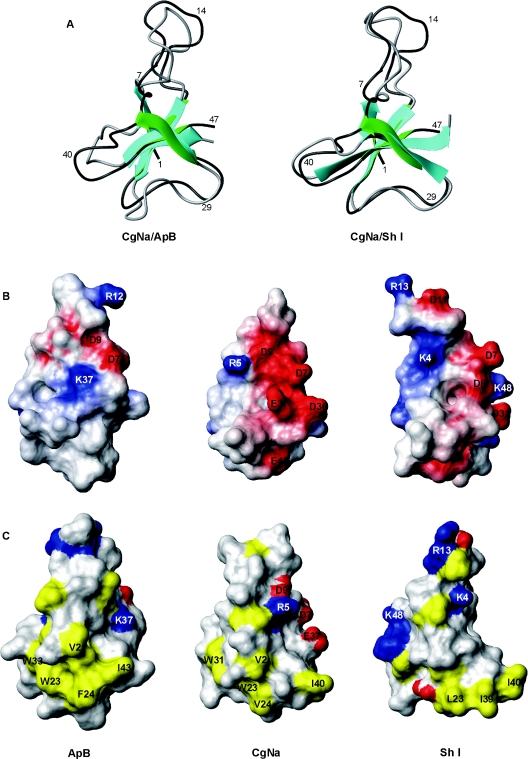
Other polypeptides which have a three-dimensional structure similar to that of CgNa were identified by a search of the DALI three-dimensional protein structure database [41] using CgNa as the query. Only sea-anemone toxin peptides classified as sodium-channel blockers were retrieved, and a pairwise comparison was made between the averaged three-dimensional structures of the toxins singled out by DALI and CgNa (Table 3). The highest DALI Z-scores for structure comparison corresponded to the anthopleurins (ApA and ApB) and the ATX Ia neurotoxin. These are proteins that displayed more than 50% sequence identity with CgNa in structure-based sequence alignments (Figure 2). Although CgNa should be classified as a group I sodium toxin, Sh I (neurotoxin I from the sun anemone, Stichodactyla helianthus), a group II sodium toxin, was also retrieved in the DALI search. Although Sh I only demonstrated 44% identity with CgNa, it was structurally similar to CgNa, with a significant DALI Z-score (Figure 2 and Table 3). Although the similarity of folding between CgNa, ApB and Sh I is evident (Figure 3A), important differences in the primary structure between CgNa, ApB and Sh I reflected the distinct distributions of electrostatic charge at functionally important regions on the surface of the proteins (Figure 3B). Moreover, these differences extended to the geometry of a surface-exposed hydrophobic patch (Figure 3C) that is thought to be involved in the recognition of the toxin by the sodium channel [42].
Table 3. Statistical comparison of the CgNa structure with those of type I and type II toxins.
| Aligned residues† | Rmsd to CgNa (Å)‡ | |
|---|---|---|
| Type I* | ||
| ApA | 1–25, 28–49; 1–25, 26–47 | 3.00/1.30 |
| ApB | 1–25, 28–49; 1–25, 26–47 | 2.09/1.62 |
| ATX Ia | 1–34, 42–46; 1–34, 42–46 | 3.22/2.02 |
| Type II* | ||
| Sh I | 1–46; 2–47 | 2.35/1.25 |
*PDB ID codes: 1AHL, ApA; 1APF, ApB; 1ATX, ATX Ia; 1SHI, Sh I.
†PDB residue numbers of equivalent residues. On the left, the numbers corresponding to the different toxins already reported in the literature and on the right, the equivalent residue in CgNa.
‡Rmsd between mean structures of the superimposed backbone atoms N, Cα and C′ of equivalent fragments including (left-hand value) and excluding (right-hand value) the unstructured flexible loop 8–16 respectively.
Figure 2. Structure-based sequence alignment of the CgNa toxin with type I (above CgNa) and type II (below CgNa) sea-anemone polypeptides of known structure.
Following the notation from Norton [4], identical residues in each class are boxed. Those residues that are conservatively substituted are shaded in grey. *, Residues that are identical in all type I and type II sequences shown, †, Residues which are conservatively substituted. The residue numbering scheme is based on the CgNa sequence. ApA and ApB have a Pro-Ser insertion following Gly25, causing the numbering of the residues after this position to be shifted by two units in relation to CgNa.
Figure 3. Ribbon structure representation and space-filling models of CgNa, ApB and Sh I toxins.
(A) Superimposed ribbon representation of CgNa and ApB (left) and CgNa and Sh I (right) structures. CgNa is coloured green and dark grey. For CgNa and ApB, the structure that is closest to the average is shown, while for Sh I the closest to the average refined minimized structures of Wilcox et al. [56] is shown. (B) Solvent-accessibility surface representations of ApB (left), CgNa (middle) and Sh I (right), which represents an approx. 90° counterclockwise rotation around the vertical axis of the structures shown in (A). Electrostatic potential at the surface is represented by graded colours (blue, highly positive; red, highly negative). (C) Solvent-accessibility of the surface of ApB (left), CgNa (middle) and Sh I (right). Hydrophobic residues are coloured yellow, and positively and negatively charged amino acids are blue and red respectively. The views are rotated approx. 180° counterclockwise around the vertical axis compared with those in (A).
Electrophysiology in mammalian DRG neurons
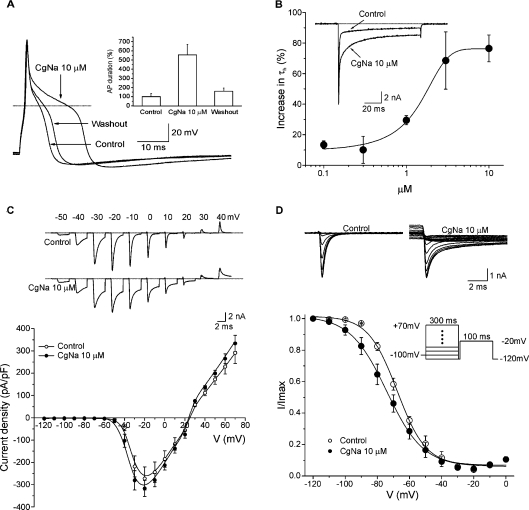
The mean capacitance of the neurons examined in the present study was 47±1.9 pF (n=83), which corresponds to a cell diameter of approx. 38 μm. In current-clamp experiments, the neurons had a mean resting membrane potential of −53.7±2.4 mV (n=3). Action potentials were evoked by application of a depolarizing current pulse of 5 ms and 100 pA with an 8 s interpulse interval (Figure 4A). The duration of the action potential at −60 mV was 3.3±1.1 ms when measured at 50% of the peak amplitude. Perfusion with CgNa (10 μM) for 1 min increased the action potential duration to 18.6±3.8 ms, producing a 4.6-fold increase with respect to control values. The resting membrane potential was not significantly affected by the toxin (paired Student's t test, P>0.05). After a 5 min washout, the duration of the action potential returned to 3.7±1.2 ms, which represents a recovery of approx. 80% with respect to the maximum effect of treatment with CgNa.
Figure 4. Current- and voltage-clamp experiments on TTX-S voltage-gated sodium channels in the presence of CgNa.
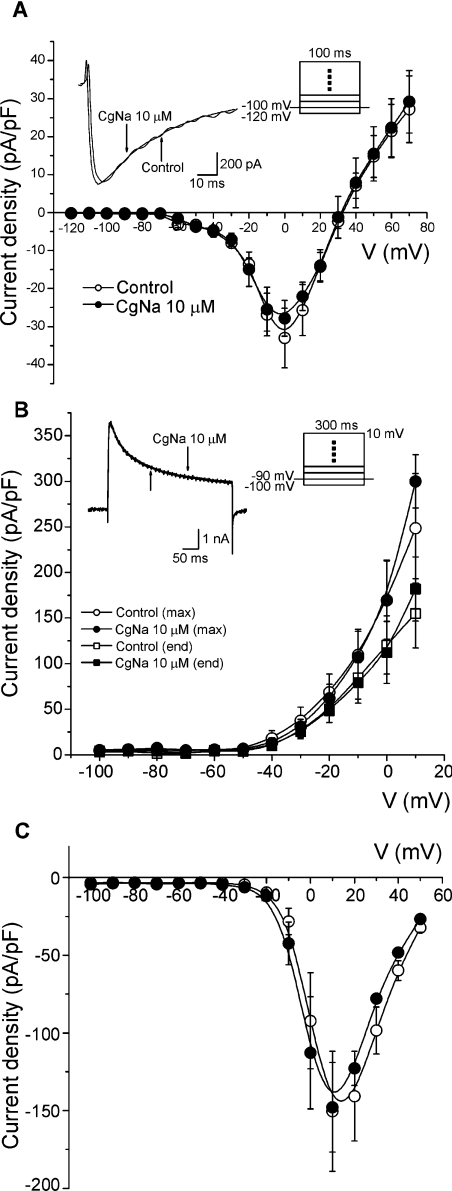
(A) CgNa increases the length of the action potential. Action potentials of a DRG neuron recorded under control conditions, in the presence of CgNa (10 μM) and after washout (5 min). In this particular cell, a small depolarization of approx. 2 mV was observed. The dotted line in all panels indicates the zero voltage level. Inset: data for the entire experimental group in which current-clamp experiments were carried out. Exposure to the toxin increased the duration of action potentials measured at 50% of its amplitude by 463% (n=3). (B) CgNa increased the inactivation time course of TTX-S INa. Inset: representative experiment showing the effect of CgNa (10 μM) 1 min after commencing perfusion. Note the slowing of the inactivation process. In both cases, the current level returned to zero after the end of the depolarization pulse. The main plot shows the dose–response curve for CgNa (n=41) on τh. Data were fitted (continuous line) by a dose–response function with an IC50 of 1.34±0.4 μM (n=41) and a Hill slope coefficient of 0.6±0.2 (n=41). (C) Effect of CgNa on the current-density–voltage curve. Top: INa produced at different potentials in the presence and absence of CgNa (10 μM, for clarity only the first 5 ms of each record are shown). Currents were produced by voltage pulses to the potentials indicated from a holding potential of −100 mV. Note that CgNa affects the currents at all the voltages tested. Bottom: current-density–voltage relationships (n=6). Perfusion with CgNa (10 μM, closed circles) did not produce significant changes either on the voltage at which the maximum current density was reached or on the reversal potential. (D) Effect of CgNa on steady-state inactivation. Top: representative experiment from which the curves were obtained. Bottom: steady-state inactivation of INa (n=6). The steady-state inactivation parameter (h∞) was determined using the two-pulse protocol shown in the inset. Data obtained at the test pulse were plotted as a function of the pre-pulse potential and fitted to a Boltzmann function (continuous lines). CgNa (10 μM, closed circles) caused a significant 8 mV hyperpolarizing shift in the V½,inact. The slope factor was also significantly changed at 3.5 mV.
In voltage-clamp experiments, the inactivation of TTX-S sodium currents was well fitted by a single exponential function (correlation coefficient ≥ 95%). Accordingly, CgNa slows the inactivation of the TTX-S sodium current. The mean time constant of inactivation (τh) was studied using a 40 ms test pulse at −20 mV from a holding potential (Vh) of −100 mV. Under control conditions, τh was 0.5±0.7 ms, which increased to 0.85±0.15 ms (n=9) in the presence of 10 μM CgNa (Figure 4B). Moreover, the maximum effect was reached within 1 min of exposure to the toxin. The relationship between the current measured at the end of the pulse and the current measured at the peak showed an increase of approx. 100% after application of the toxin. The effect of CgNa on inactivation kinetics were partially reversible by washing the preparation and, at the highest concentration used (10 μM), 71% of recovery occurred following a 5 min washout of CgNa.
The effect of CgNa on the inactivation time course of the TTX-S sodium current was concentration-dependent and thus the dose–response curve was constructed using the fixed-pulse protocol described above. The data were fitted by a dose–response function as follows: y=A1+(A2−A1)/1+10(log IC50−x)P, where A1 is the y value of the bottom plateau, A2 is the y value of the top plateau, log IC50 is the concentration at which the response is halfway between A1 and A2, x is the actual value on the abscissa and P is the Hill slope. The dose–response relationship for the effect of CgNa on the inactivation of the TTX-S sodium current was fitted with a curve that had an IC50 of 1.34±0.4 μM (n=41) and a Hill slope coefficient of 0.6±0.2 (n=41) (Figure 4B). The effect of CgNa (10 μM) on the peak sodium current amplitude was not significant at any of the concentrations tested (paired Student's t test, P>0.05).
For the current–voltage relationships, the current-density–voltage curves were obtained by normalizing the ionic current amplitudes as a function of membrane capacity (Figure 4C). Under control conditions, the maximum current density was reached at −20 mV, and perfusion of CgNa (10 μM) did not produce any significant change in the sodium current density or its reversal potential, at any of the voltages tested (n=6; paired Student's t test, P>0.05).
To study the effects of CgNa on the conductance- and voltage-dependence of steady-state TTX-S sodium current inactivation, a two-pulse protocol from a Vh of −100 mV was used. Using this protocol, a 100 ms conditioning pre-pulse from −120 to +70 mV in 10 mV increments was followed by a 100 ms test pulse at −20 mV (intersweep interval=8 s). The peak currents recorded during the test pulse were normalized as a function of the maximum amplitude value and plotted against the conditioning pre-pulse potential. Normalized currents were well fitted by a Boltzmann function. Application of CgNa (10 μM) produced a significant leftward shift in the steady-state inactivation curve of the TTX-S sodium current (paired Student's t test, P<0.05; Figure 4D). The half-maximal inactivation in control neurons of −65.8±0.6 mV (n=6) was shifted to −73.7±1.4 mV (n=6) in the presence of CgNa. The slope values increased significantly from 7.5±0.3 mV (n=6) under control conditions to 11±0.8 mV (n=6) in the presence of CgNa (paired Student's t test, P<0.05).
The TTX-S sodium-channel conductance was calculated from the equation: gNa=INa/(E−ENa) where I is the current, E is the test pulse voltage and ENa is the reversal potential for sodium. The sodium conductance was plotted as a function of the membrane potential and the ratio was fitted by a Boltzmann equation, for both the control samples and the experimental samples treated with CgNa (10 μM). No difference was observed in the voltage potential at which activation is 0.5 (V½act) under control conditions (−29.5±0.6 mV) or after the application of CgNa (−31.0±0.6 mV) (Student's t test, P>0.05). The slope factors for these curves were 6.3±0.5 mV (n=6) and 6.9±0.5 mV (n=6) respectively.
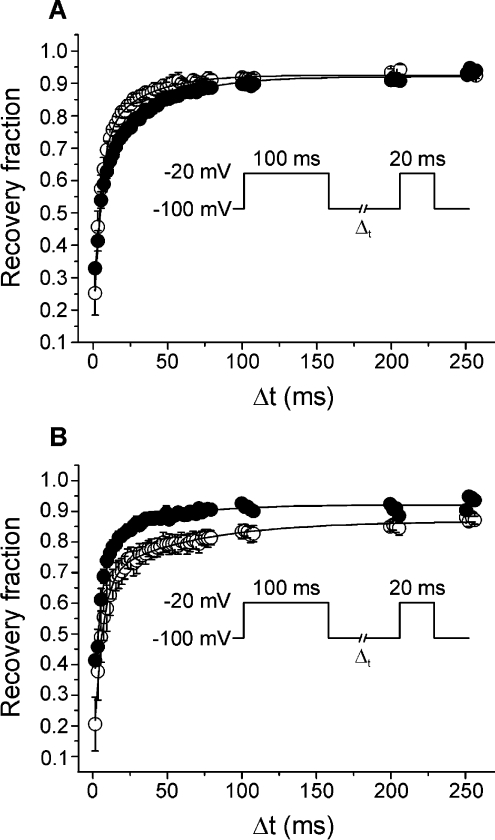
To study the action of CgNa on the rate of recovery from inactivation, a two-pulse protocol was used with an increment in the duration of the interpulse interval. In this protocol, the Vh was either −100 or −80 mV, the test pulses (40 ms) were up to −20 mV, and the interpulse interval (Δt) varied from 2.5 to 1000 ms. Using a Vh of −100 mV, the time course of recovery from inactivation was fitted by a second-order exponential function with a fast time constant (τrec,fast) in control conditions of 5.5±0.3 ms and a slow time constant (τrec,slow) of 26.4±1.2 ms. These figures were not significantly different to those measured after application of CgNa: τrec,fast=5.1±0.3 ms and τrec,slow=25.4±1.1 ms (Figure 5A). In contrast, at a Vh of −80 mV, the time course of recovery from the inactivation of TTX-S sodium current was enhanced by CgNa. Although the τrec,fast was not significantly different between control conditions (6.3±1.4 ms) and CgNa treated conditions (5.8±0.6 ms), the τrec,slow diminished upon exposure to CgNa from 49.6±8.2 ms under control conditions to 20.8±3.5 ms in the presence of CgNa (Figure 5B).
Figure 5. CgNa enhanced the time course of recovery from inactivation.
(A) Recovery curves obtained from the protocol in the inset (Vh=−100 mV). Curves were fitted (continuous lines) to a second order exponential function with a fast (τrec,fast) and a slow (τrec,slow) time constant. CgNa (10 μM, closed circles) did not produce significant changes under these parameters. (B) Using Vh=−80 mV, the τrec,slow decreased significantly from 49.6±8.2 ms in control to 20.8±3.5 ms in the presence of CgNa.
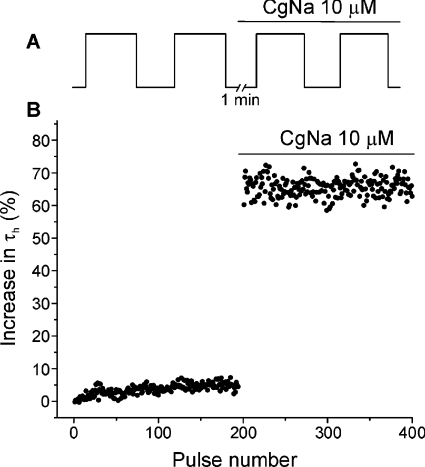
We explored the state of the sodium channel on which CgNa exerts its action with a pair of depolarizing trains of 200 pulses, increasing the voltage to −20 mV from a holding potential of −100 mV (pulse duration of 40 ms, pulse interval of 200 ms). The first (control) and second (test) trains were separated by a rest period of 1 min, during which the cell was held at a holding potential of −100 mV and perfused with 10 μM CgNa. The inactivation time constant (τh) of the sodium current evoked by each pulse was plotted against the pulse number. In the absence of the toxin, τh remained constant during the control train. In contrast, the time constant of the ionic current produced by the first test pulse increased by 59.1±14.6% (n=4) following CgNa application, indicating that the toxin binds preferentially to the channel in the resting state. The effectiveness of CgNa did not increase when the channels were activated by trains of repetitive stimuli, indicating that its action was not dependent on channel usage (Figure 6).
Figure 6. CgNa acts on TTX-S voltage-gated sodium channels in the closed state.
(A) Post-resting protocol employed to obtain the records. The toxin was applied at the beginning of the rest period (1 min) represented by the break. (B) Temporal course of the inactivation time constant τh. These data were obtained from a single cell, and it can be observed that the maximum effect of CgNa was already evident from the first pulse after the rest period. The bar indicates perfusion with 10 μM CgNa.
The effect of CgNa on the TTX-R sodium currents was studied using the same solutions used for TTX-S sodium currents, but with the addition of TTX (300 nM) to the external solution (n=5). CgNa showed no significant effect on TTX-R sodium currents (Figure 7A). Finally, the effect of CgNa (10 μM) on total potassium (IK, n=4; Figure 7B) and total calcium currents (ICa; n=4; Figure 7C) was studied. Similarly, CgNa did not modify either the peak amplitude or the sustained component at the end of the pulse of these currents.
Figure 7. CgNa did not alter TTX-R sodium, potassium or calcium currents.
In all panels, two superimposed traces and two current-density–voltage curves are presented, one under control conditions (open circles) and the other approx. 2 min after toxin perfusion (closed circles). (A) TTX-R sodium currents were isolated in the presence of 300 nM TTX. (B) Current-density–voltage curves for the potassium channels were obtained measuring the maximum current (circles) or the current at the end of the trace (squares) and normalizing these data to membrane capacity. (C) Current-density–voltage curves as obtained for calcium channels.
DISCUSSION
Electrophysiology
CgNa increases the duration of the action potential, owing to the slowing down of TTX-S sodium current inactivation. This action seems to be very selective for the TTX-S sodium channel, since the highest toxin concentration used (10 μM) did not produce any significant effect on other ion currents. The effect of CgNa on the inactivation process takes place within 1 min of application, and it was partially reversible following its removal and wash out. The reversibility of the effects of sea-anemone toxins upon vertebrate or insect sodium channels has been reported by many authors [43–45], in contrast with the irreversible toxicity on crustacean sodium channels [43]. Interestingly, whereas the effects of ATX I and ATX II in crayfish giant axons were irreversible [46], the activity of ATX III in the same preparation was reversible. The different properties of these toxins may be caused by both the characteristics of the compounds themselves and also the structural and functional differences in sodium channels between various tissues and animal phyla.
Addition of the toxin significantly modified the steady-state inactivation of TTX-S sodium channels. The slowing of inactivation kinetics and the reduced voltage-dependence of the steady-state availability curve indicate that CgNa modifies the transition rate of the sodium channel, both from the open to the inactivated state and vice versa. CgNa did not increase the peak currents of sodium channels at the concentrations used here, despite the marked effect of the toxin on sodium-current inactivation which would increase current density. Thus we cannot exclude the possibility that CgNa may also produce some degree of channel occlusion.
In the presence of the toxin, the τh on the first pulse after a rest period increased, and this value remained within a narrow range during successive pulses in the test train. These features are consistent with the action of the toxin not being dependent on channel use. Indeed, these observations indicate that CgNa has a high affinity for the closed state of the channel, and, accordingly, the toxin shows no preference for the sodium channel in the open state. The time course of recovery from sodium current inactivation was not significantly affected by CgNa when studied using a Vh of −100 mV. However, while the fast time constant was not affected by CgNa at a Vh of −80 mV, the slow time constant was enhanced in the presence of CgNa. Previous reports indicate that ApB also significantly enhances the recovery from inactivation of both cardiac and neural sodium-channel isoforms [47]. On the basis of kinetic modifications of the sodium channel, this has been explained as an increase in the direct rate of transition back to the open state or as enhancement of normal transition through the different inactivation states. Since the effect of CgNa on the recovery rate from inactivation was voltage-dependent, the binding of CgNa to sodium channels displayed some type of voltage-dependence, which was suggested previously for α-scorpion toxins [48].
Structural studies
Structural studies analysing the effects of modifying sea-anemone toxins have been carried out by several groups, focusing mainly on mutated ApB. Through such studies, Gly10 and Gly15 of ApB appear to be important for toxin affinity, and Gly20 is an essential folding determinant [49]. Indeed, these three glycine residues are conserved in CgNa. In addition, Arg12, Leu18, Trp33, Lys37, Ile43 and Trp45 contribute significantly to the tight binding affinity of ApB [50–53]. Only two of these residues are strictly conserved in CgNa, Leu18 and Trp33 (Trp31 in CgNa) (Figure 2).
According to previous analyses, the overall alignment of type I and type II toxins shows that there are 11 identical residues and five conservative substituted residues along the peptide [4]. Of those residues, CgNa lacks the invariable Arg14 (replaced by histidine) and the conserved hydrophobic residue at position 41 (replaced by alanine). Overall, CgNa has a greater percentage of exposed negatively charged residues and a lower percentage of exposed hydrophobic residues than is typical for type I and II toxins. These differences are derived mainly from the lack of cationic residues in the flexible loop of residues 8–16 of CgNa and the unique amino acids Asp36, Glu37 and Glu43 in CgNa (instead of Trp45 in ApB), which create an important negatively charged patch in conjunction with the conserved residues Asp7 and Asp9. This patch occupies part of a strongly cationic surface that builds up around Lys37 in ApB [54,55]. As it has been demonstrated previously that Lys37 in ApB can directly interact with Asp1612 of the rat cardiac sodium channel [53], the partial substitution of a positive patch could be one factor that helps to explain the lower potency of CgNa (10 μM tested on ganglion neurons expressing TTX-R sodium channels compared with 9 nM ApB tested on N1E-115 cells). Substitution of Arg12 (serine in CgNa) may further weaken this interaction, as mutation of Arg12 has been shown to reduce the affinity of ApB for both neuronal and cardiac channels [50]. Moreover, there is a substitution of alanine for the conserved hydrophobic residue at position 43 in ApB in CgNa (residue 41 of its primary structure), and an aspartic acic residue is found at position 43 CgNa in comparison with an aromatic residue in the other type I toxins possessing a three-dimensional structure known at present. These changes also disrupt a surface-exposed hydrophobic patch that involves residues previously shown to be relevant for the binding of the toxin to the sodium channel [51,52]. Despite these differences, the CgNa protein is still active and thus the cationic residues in the 8–16 loop, Lys37 and the small surface-exposed hydrophobic patch that includes Ile43 of ApB are not absolutely essential for binding to the neuronal isoform of mammalian voltage-gated sodium channels. Future mutational analysis of CgNa, based on the entire set of structural comparisons discussed above, should contribute to further elucidation of the structural characteristics underlying the biological properties of CgNa and structurally related toxins.
Acknowledgments
This work was supported by a CONACyT (Consejo Nacional de Ciencia y Technología) grant (J200.812-2004), a VIEP-BUAP grant (6/I/SAL/05) and funding from the Dirección General de Investigación Científica y Técnica of Spain (grants BIO2002-0374 and BQU2003-03550-C03-01). Supercomputing time from CESGA (Centro de Supercomputación de Galicia) is also acknowledged.
References
- 1.Aneiros A., Garateix A. Bioactive peptides from marine sources: pharmacological properties and isolation procedures. J. Chromatogr. B. 2004;803:41–53. doi: 10.1016/j.jchromb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Norton T. R. Cardiotonic polypeptides from Anthopleura xanthogrammica (Brandt) and A. elegantissima (Brandt) Fed. Proc. 1981;40:21–25. [PubMed] [Google Scholar]
- 3.Loret E. P., del Valle R. M., Mansuelle P., Sampieri F., Rochat H. Positively charged amino acid residues located similarly in sea anemone and scorpion toxins. J. Biol. Chem. 1994;269:16785–16788. [PubMed] [Google Scholar]
- 4.Norton R. S. Structure and structure–function relationships of sea anemone proteins that interact with the Na+ channel. Toxicon. 1991;29:1051–1084. doi: 10.1016/0041-0101(91)90205-6. [DOI] [PubMed] [Google Scholar]
- 5.Rogers J. C., Qu Y., Tanada T. N., Scheuer T., Catterall W. A. Molecular determinants of high affinity binding of α-scorpion toxin and sea anemone toxin in the S3–S4 extracellular loop in domain IV of the Na+ channel α subunit. J. Biol. Chem. 1996;271:15950–15962. doi: 10.1074/jbc.271.27.15950. [DOI] [PubMed] [Google Scholar]
- 6.Meves H., Simard J. M., Watt D. D. Interactions of scorpion toxins with the sodium channel. Ann. N.Y. Acad. Sci. 1986;479:113–132. doi: 10.1111/j.1749-6632.1986.tb15565.x. [DOI] [PubMed] [Google Scholar]
- 7.Possani L. D., Becerril B., Delepierre M., Tytgat J. Scorpion toxins specific for Na+-channels. Eur. J. Biochem. 1999;264:287–300. doi: 10.1046/j.1432-1327.1999.00625.x. [DOI] [PubMed] [Google Scholar]
- 8.Narahashi T., Moore J. W., Shapiro B. I. Condylactis toxin: interaction with nerve membrane ionic conductances. Science. 1969;163:680–681. doi: 10.1126/science.163.3868.680. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher J. I., Chapman B. E., Mackay J. P., Howden M. E., King G. F. The structure of versutoxin (δ-atracotoxin-Hv1) provides insights into the binding of site 3 neurotoxins to the voltage-gated sodium channel. Structure. 1997;5:1525–1535. doi: 10.1016/s0969-2126(97)00301-8. [DOI] [PubMed] [Google Scholar]
- 10.Little M. J., Zappia C., Gilles N., Connor M., Tyler M. I., Martin-Eauclaire M.-F., Gordon D., Nicholson G. M. δ-Atracotoxins from Australian funnel-web spiders compete with scorpion α-toxin binding but differentially modulate alkaloid toxin activation of voltage-gated sodium channels. J. Biol. Chem. 1998;273:27076–27083. doi: 10.1074/jbc.273.42.27076. [DOI] [PubMed] [Google Scholar]
- 11.Sahara Y., Gotoh M., Konno K., Miwa A., Tsubokawa H., Robinson H. P., Kawai N. A new class of neurotoxin from wasp venom slows inactivation of sodium current. Eur. J. Neurosci. 2000;12:1961–1970. doi: 10.1046/j.1460-9568.2000.00084.x. [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita E., Maejima H., Yamaoka K., Konno K., Kawai N., Shimizu E., Yokote S., Nakayama H., Seyama I. Novel wasp toxin discriminates between neuronal and cardiac sodium channels. Mol. Pharmacol. 2001;59:1457–1463. [PubMed] [Google Scholar]
- 13.Cannon S. C., Corey D. P. Loss of Na+ channel inactivation by anemone toxin (ATX II) mimics the myotonic state in hyperkalaemic periodic paralysis. J. Physiol. 1993;466:501–520. [PMC free article] [PubMed] [Google Scholar]
- 14.Lehmann-Horn F., Jurkat-Rott K. Voltage-gated ion channels and hereditary disease. Physiol. Rev. 1999;79:1317–1372. doi: 10.1152/physrev.1999.79.4.1317. [DOI] [PubMed] [Google Scholar]
- 15.Jurkat-Rott K., Mitrovic N., Hang C., Kouzmekine A., Iaizzo P., Herzog J., Lerche H., Nicole S., Vale-Santos J., Chauveau D., et al. Voltage-sensor sodium channel mutations cause hypokalemic periodic paralysis type 2 by enhanced inactivation and reduced current. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9549–9554. doi: 10.1073/pnas.97.17.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blumenthal K. M., Seibert A. L. Voltage-gated sodium channel toxins: poisons, probes, and future promise. Cell Biochem. Biophys. 2003;38:215–238. doi: 10.1385/CBB:38:2:215. [DOI] [PubMed] [Google Scholar]
- 17.Ständker L., Béress L., Garateix A., Christ T., Ravens U., Salceda E., Soto E., John H., Forssmann W. G., Aneiros A. A new toxin from the sea anemone Condylactis gigantea with effect on sodium channel inactivation. Toxicon. 2006;48:211–220. doi: 10.1016/j.toxicon.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Wunderer G., Eulitz M. Amino-acid sequence of toxin I from Anemonia sulcata. Eur. J. Biochem. 1978;89:11–17. doi: 10.1111/j.1432-1033.1978.tb20890.x. [DOI] [PubMed] [Google Scholar]
- 19.Wunderer G., Fritz H., Wachter E., Machleidt W. Amino-acid sequence of a coelenterate toxin: toxin II from Anemonia sulcata. Eur. J. Biochem. 1976;68:193–198. doi: 10.1111/j.1432-1033.1976.tb10778.x. [DOI] [PubMed] [Google Scholar]
- 20.Scheffler J. J., Tsugita A., Linden G., Schweitz H., Lazdunski M. The amino acid sequence of toxin V from Anemonia sulcata. Biochem. Biophys. Res. Commun. 1982;107:272–278. doi: 10.1016/0006-291x(82)91700-4. [DOI] [PubMed] [Google Scholar]
- 21.Sunahara S., Muramoto K., Tenma K., Kamiya H. Amino-acid sequence of two sea-anemone toxins from Anthopleura fuscoviridis. Toxicon. 1987;25:211–219. doi: 10.1016/0041-0101(87)90243-1. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka M., Hainu M., Yasunobu K. T., Norton T. R. Amino acid sequence of the Anthopleura xanthogrammica heart stimulant, anthopleurin A. Biochemistry. 1977;16:204–208. doi: 10.1021/bi00621a007. [DOI] [PubMed] [Google Scholar]
- 23.Reimer N. S., Yasunobu C. L., Yasunobu K. T., Norton T. R. Amino acid sequence of the Anthopleura xanthogrammica heart stimulant, anthopleurin-B. J. Biol. Chem. 1985;260:8690–8693. [PubMed] [Google Scholar]
- 24.Kelso G. J., Blumenthal K. M. Identification and characterization of novel sodium channel toxins from the sea anemone Anthopleura xanthogrammica. Toxicon. 1998;36:41–51. doi: 10.1016/s0041-0101(97)00064-0. [DOI] [PubMed] [Google Scholar]
- 25.Salceda E., Garateix A., Aneiros A., Salazar H., Lopez O., Soto E. Effects of ApC, a sea anemone toxin, on sodium currents of mammalian neurons. Brain Res. 2006;1110:136–143. doi: 10.1016/j.brainres.2006.06.113. [DOI] [PubMed] [Google Scholar]
- 26.Goudet C., Ferrer T., Galan L., Artiles A., Batista C. F., Possani L. D., Alvarez J., Aneiros A., Tytgat J. Characterization of two Bunodosoma granulifera toxins active on cardiac sodium channels. Br. J. Pharmacol. 2001;134:1195–1206. doi: 10.1038/sj.bjp.0704361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salceda E., Garateix A., Soto E. The sea anemone toxins BgII and GgIII prolong the inactivation time course of the tetrodotoxin-sensitive sodium current in rat dorsal root ganglion neurons. J. Pharmacol. Exp. Ther. 2002;303:1067–1074. doi: 10.1124/jpet.102.038570. [DOI] [PubMed] [Google Scholar]
- 28.Kostyuk P. G., Veselovsky N. S., Tsyndrenko A. Y. Ionic currents in the somatic membrane of rat dorsal root ganglion neurons-I. Sodium currents. Neuroscience. 1981;6:2423–2430. doi: 10.1016/0306-4522(81)90088-9. [DOI] [PubMed] [Google Scholar]
- 29.Roy M. L., Narahashi T. Differential properties of tetrodotoxin-sensitive and tetrodotoxin-resistant sodium channels in rat dorsal root ganglion neurons. J. Neurosci. 1992;12:2104–2111. doi: 10.1523/JNEUROSCI.12-06-02104.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strachan L. C., Lewis R. J., Nicholson G. M. Differential actions of pacific ciguatoxin-1 on sodium channel subtypes in mammalian sensory neurons. J. Pharmacol. Exp. Ther. 1999;288:379–388. [PubMed] [Google Scholar]
- 30a.Bezanilla F., Armstrong C. M. Inactivation of the sodium channel. I. Sodium current experiments. J. Gen. Physiol. 1977;70:549–566. doi: 10.1085/jgp.70.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartels C., Xia T. H., Billeter M., Güntert P., Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 32.Schäfer N. Diploma Thesis. Zürïch, Switzerland: ETH Zürich; 1992. Automatische signalerkenning in heteronuklearen 3D und 4D NMR spektren. [Google Scholar]
- 33.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 33a.Herrmann T., Güntert P., Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 34.Engh R. A., Huber R. Accurate bond and angle parameters for X-ray protein-structure refinement. Acta Crystallogr. Sect. A Found. Crystallogr. 1991;47:392–400. [Google Scholar]
- 35.Pearlman D. A., Case D. A., Caldwell J. W., Ross W. S., Cheatham T. E., Debolt S., Ferguson D., Seibel G., Kollman P. AMBER, a package of computer-programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 36.Reference deleted
- 37.Cornell W. D., Cieplak P., Bayly C. I., Gould I. R., Merz K. M., Ferguson D. M., Spellmeyer D. C., Fox T., Caldwell J. W., Kollman P. A. A 2nd generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 38.Koradi R., Billeter M., Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 39.Wang J., Wang W., Kollman P. A., Case D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graphics Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Laskowski R. A., Rullmann J. A. C., MacArthur M. W., Kaptein R., Thornton J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 41.Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 42.Blumenthal K. Ionic channels as targets for toxins, drugs and genetic diseases. In: Sperelakis N., editor. Cell Physiology Source Book. San Diego: Academic Press; 1989. pp. 389–403. [Google Scholar]
- 43.Salgado V. L., Kem W. R. Actions of three structurally distinct sea anemone toxins on crustacean and insect sodium channels. Toxicon. 1992;30:1365–1381. doi: 10.1016/0041-0101(92)90512-4. [DOI] [PubMed] [Google Scholar]
- 44.Cestele S., Kopeyan C., Oughideni R., Mansuelle P., Granier C., Rochat H. Biochemical and pharmacological characterization of a depressant insect toxin from the venom of the scorpion Buthacus arenicola. Eur. J. Biochem. 1997;243:93–99. doi: 10.1111/j.1432-1033.1997.93_1a.x. [DOI] [PubMed] [Google Scholar]
- 45.Bosmans F., Aneiros A., Tytgat J. The sea anemone Bunodosoma granulifera contains surprisingly efficacious and potent insect-selective toxins. FEBS Lett. 2002;532:131–134. doi: 10.1016/s0014-5793(02)03653-0. [DOI] [PubMed] [Google Scholar]
- 46.Rathmayer W. Sea anemone toxins: tools in the study of excitable membranes. In: Ceccarelli B., Clementi F., editors. Advances in Cytopharmacology. New York: Raven Press; 1979. pp. 335–344. [PubMed] [Google Scholar]
- 47.Benzinger G. R., Tonkovich G. S., Hanck D. A. Augmentation of recovery from inactivation by site-3 Na channel toxins: a single-channel and whole-cell study of persistent currents. J. Gen. Physiol. 1999;113:333–346. doi: 10.1085/jgp.113.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strichartz G. R., Wang G. K. Rapid voltage-dependent dissociation of scorpion α-toxins coupled to Na channel inactivation in amphibian myelinated nerves. J. Gen. Physiol. 1986;88:413–435. doi: 10.1085/jgp.88.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seibert A. L., Liu J., Hanck D. A., Blumenthal K. M. Arg-14 loop of site 3 anemone toxins: effects of glycine replacement on toxin affinity. Biochemistry. 2003;42:14515–14521. doi: 10.1021/bi035291d. [DOI] [PubMed] [Google Scholar]
- 50.Gallagher M. J., Blumenthal K. M. Importance of the unique cationic residues arginine 12 and lysine 49 in the activity of the cardiotonic polypeptide anthopleurin B. J. Biol. Chem. 1994;269:254–259. [PubMed] [Google Scholar]
- 51.Dias-Kadambi B. L., Combs K. A., Drum C. L., Hanck D. A., Blumenthal K. M. The role of exposed tryptophan residues in the activity of the cardiotonic polypeptide anthopleurin B. J. Biol. Chem. 1996;271:23828–23835. doi: 10.1074/jbc.271.39.23828. [DOI] [PubMed] [Google Scholar]
- 52.Dias-Kadambi B. L., Drum C. L., Hanck D. A., Blumenthal K. M. Leucine 18, a hydrophobic residue essential for high affinity binding of anthopleurin B to the voltage-sensitive sodium channel. J. Biol. Chem. 1996;271:9422–9428. doi: 10.1074/jbc.271.16.9422. [DOI] [PubMed] [Google Scholar]
- 53.Benzinger G. R., Kyle J. W., Blumenthal K. M., Hanck D. A. A specific interaction between the cardiac sodium channel and site-3 toxin anthopleurin B. J. Biol. Chem. 1998;273:80–84. doi: 10.1074/jbc.273.1.80. [DOI] [PubMed] [Google Scholar]
- 54.Khera P. K., Benzinger G. R., Lipkind G., Drum C. L., Hanck D. A., Blumenthal K. M. Multiple cationic residues of anthopleurin B that determine high affinity and channel isoform discrimination. Biochemistry. 1995;34:8533–8541. doi: 10.1021/bi00027a003. [DOI] [PubMed] [Google Scholar]
- 55.Khera P. K., Blumenthal K. M. Importance of highly conserved anionic residues and electrostatic interactions in the activity and structure of the cardiotonic polypeptide anthopleurin B. Biochemistry. 1996;35:3503–3507. doi: 10.1021/bi9528457. [DOI] [PubMed] [Google Scholar]
- 56.Wilcox G. R., Fogh R. H., Norton R. S. Refined structure in solution of the sea anemone neurotoxin ShI. J. Biol. Chem. 1993;268:24707–24719. [PubMed] [Google Scholar]